Abstract
Cancer therapies are limited by the development of drug resistance, and mutations in drug targets is one of the main reasons for developing acquired resistance. The adequate knowledge of these mutations in drug targets would help to design effective personalized therapies. Keeping this in mind, we have developed a database “CancerDR”, which provides information of 148 anti-cancer drugs, and their pharmacological profiling across 952 cancer cell lines. CancerDR provides comprehensive information about each drug target that includes; (i) sequence of natural variants, (ii) mutations, (iii) tertiary structure, and (iv) alignment profile of mutants/variants. A number of web-based tools have been integrated in CancerDR. This database will be very useful for identification of genetic alterations in genes encoding drug targets, and in turn the residues responsible for drug resistance. CancerDR allows user to identify promiscuous drug molecules that can kill wide range of cancer cells. CancerDR is freely accessible at http://crdd.osdd.net/raghava/cancerdr/
Cancer is a global health problem and a leading cause of deaths worldwide. Both developed and developing countries are affected by this devastating disease. Though we have treatment options for cancer, especially when it is in early stage, but the mortality rate is still high all across the globe. Chemotherapy is one of the principal modes of treatment for cancer patients, which mainly includes cytotoxic drugs, and kills fast proliferating cells, a common feature of all cancer types. One of the limitations of the chemotherapy is that it also kills the normal fast dividing cells causing serious side effects in patients. In order to reduce the side effects, targeted therapies have been developed, which target a specific molecule or pathway differentially expressed in cancer cells. Despite advances in the targeted therapy, still cancer treatment is not effective. There are many reasons behind the failure of cancer treatments that include; (i) acquired drug resistance, and (ii) multiple molecular types of cancer. Recent analysis, based on patterns of DNA mutations and RNA expression in 2000 specimens, revealed 10 molecular types of breast cancer1. In addition, cancer is characterized by extensive genetic and epigenetic alterations2,3 and mutations in drug targets may also be responsible for increased drug resistance4.
Drug resistance is a common cause of treatment failure in cancer. This problem is similar to human immunodeficiency virus (HIV), where frequent mutations in drug targets are responsible for the development of drug resistant HIV5. Recently, it has been hypothesized that cancer, similar to HIV, should be managed by personalized medicine6. In past, attempts have been made to manage cancer treatment based on genomics and proteomics (expression) profiles7,8,9,10. In case of HIV, drug resistance has been tackled based on mutations in drug targets11,12,13. To the best of our knowledge, no attempts have been made to manage drug resistance in cancer based on mutations in drug targets. This study is the first attempt in this direction, where we have collected and compiled valuable information to manage drug resistance in cancer based on mutations in drug targets.
Results
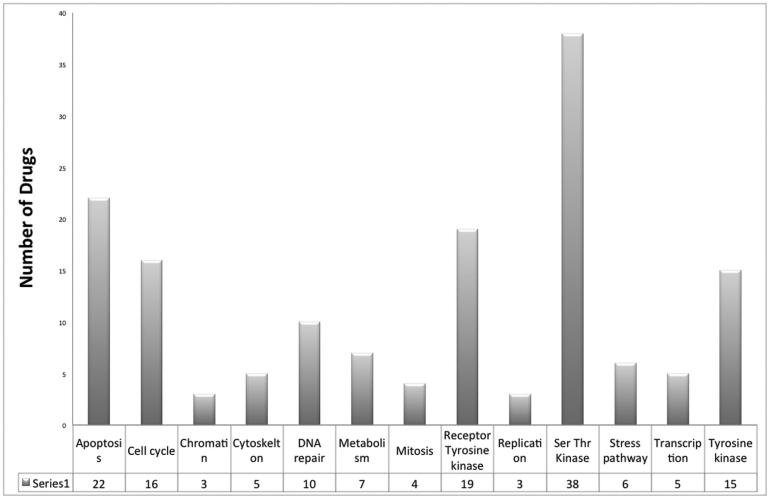
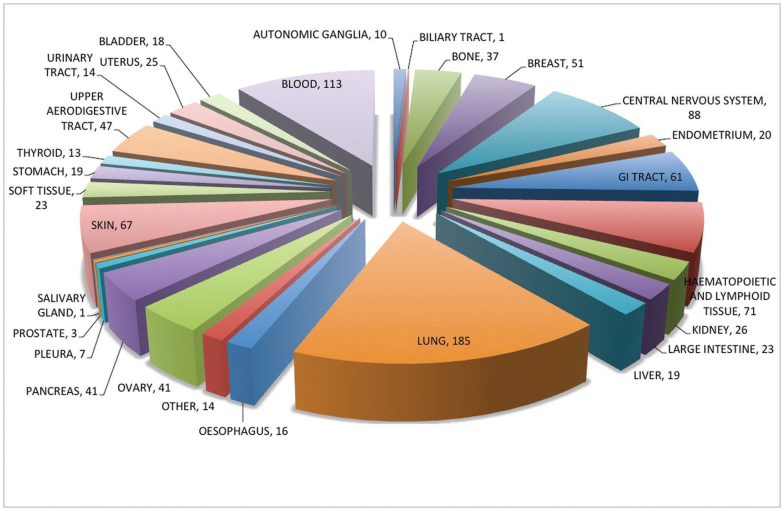
CancerDR is an attempt in the direction of personalized medicine for cancer therapy. We have collected the pharmacological profiling of 148 anti-cancer drugs (36 FDA approved drugs, 48 drugs in clinical trials and 64 experimental drugs). Among these, 130 drugs have been used in targeted therapy, while rest 18 are cytotoxic drugs. These drugs target wide range of biomarkers and pathways like, apoptosis, cell cycle, DNA repair, transcription, protein kinases (tyrosine or Ser/Thr) etc. Most of the drug targets belong to Ser/Thr kinase class of protein kinases (Figure 1). Cancer cell lines used for pharmacological profiling belong to 29 major tissue types like autonomic ganglia, biliary tract, central nervous system etc. Among these, most of the cell lines belong to lung (185) and blood (113) tissue type (Figure 2).
Figure 1. Distribution of anti-cancer drugs in various target classes.
Figure 2. Schematic diagram showing distribution of various cancer cell lines in tissue types.
In cancer therapy, it is very important to understand which drug will be effective against a specific cancer type. CancerDR provides powerful tools to tackle this problem on the basis of pharmacological profiling data of anti-cancer drugs on cancer cell lines. Clustering module of CancerDR clusters the cell lines on the basis of IC50 values. This clustering facility allows user to identify drugs that are more effective against a particular cancer cell line or tissue type. In addition to this, user can cluster the cell lines of a particular tissue type on the basis of IC50 values to identify the drug sensitivity of that tissue type. Similarly, clustering of drugs can also be done. By clustering of cell lines, one can predict the drugs that are effective/sensitive against major type of cancers, for example, cell lines belong to the lung tissue type are most sensitive to paclitaxel (Supplementary Table 1).
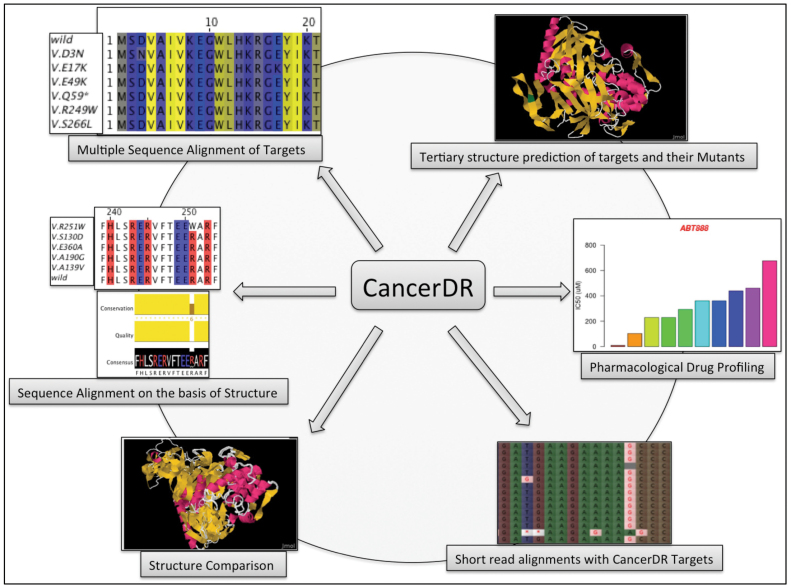
Mutation in drug targets is one of the major causes for acquired drug resistance in case of cancer. Information of drug sensitivity and mutations in drug targets will be helpful for developing prediction models for predicting mutations responsible for drug resistance in cancer. Aim of CancerDR is to maintain pharmacological profiling data of anti-cancer drugs, which will facilitate researchers to understand the effect of mutations in drug targets on acquired drug resistance. In the era of next-generation sequencing (NGS), it is possible to sequence the whole genome of cancer patient, and thus, it is possible to detect mutations in drug targets of defined patient subsets. Based on these mutations, one may identify anti-cancer drugs that will be effective/sensitive for defined patient subsets. The chances of success of patient-specific drug seems much higher than the drugs tested randomly. In order to facilitate users to identify mutations in drug targets, NGS mapping tool has been integrated in CancerDR that allows mapping of short reads, contigs, and sequences on drug targets. In clinical scenario, this tool may assist scientists in identifying the drug(s), which will be most effective and vice versa, by identifying the type of mutations present in drug targets. Possible applications of CancerDR are shown in Figure 3.
Figure 3. Schematic diagram showing various applications of CancerDR.
Mutation in drug targets causes the structural changes, which may be responsible for acquired drug resistance. Thus, understanding of these structural changes in drug targets/mutants may be helpful to manage the drug resistance problem in cancer. To address this issue, we have predicted the tertiary structure of all the drug targets, and their mutants/variants and aligned these structures. Thus, user can identify the structural deviation due to each kind of mutation. Along with this, we have provided facility to predict and compare the structure of user's query protein sequence with the protein structures available in CancerDR.
Discussion
Though considerable progress has been achieved in the field of cancer therapeutics, but acquired resistance to anti-cancer drugs remains a major obstacle in the successful treatment of cancer. Keeping this crucial problem in mind, we have developed CancerDR database, which provides comprehensive information of pharmacological profiling of 148 anti-cancer drugs across different cancer cell lines. Information related to the drug targets, and their gene sequences have also been incorporated. In addition, we have tried to link mutations in the drug targets with acquired drug resistance. All the tools and information provided in CancerDR will facilitate the concept of the personalized medicine. By analyzing genetic alterations in drug targets, and pharmacological profiles of drugs, user can design or select the best therapeutic options for a particular cancer type. Besides improving the therapeutics, personalized medicine approach would reduce the unnecessary blunt treatment of the cancer. One of the short comings of CancerDR is that all the information about pharmacological drug profile is based on cancer cell lines which deviate it, a little bit, from the actual scenario of drug resistance in cancer. However, in future, efforts will be made to collect drug profile data of the cancer patients.
Methods
Data collection and compilation
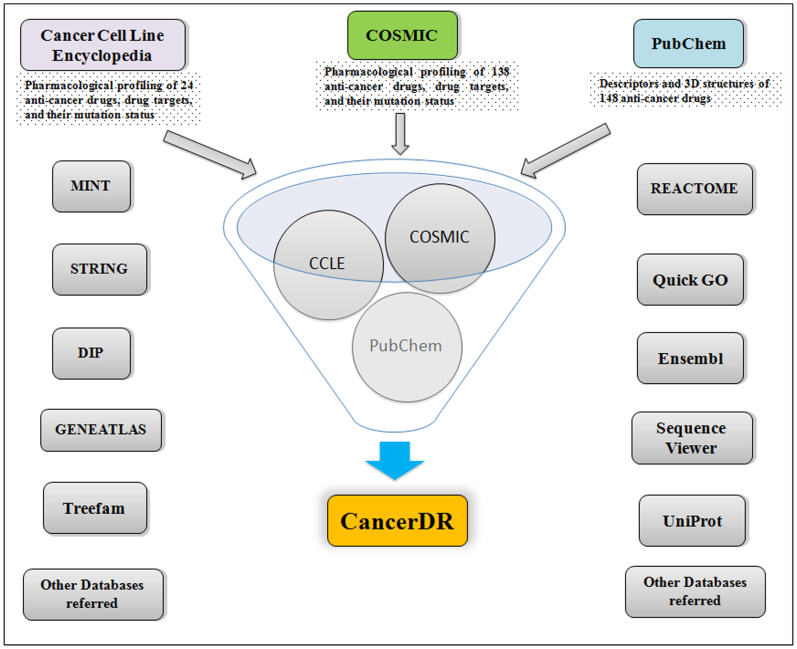
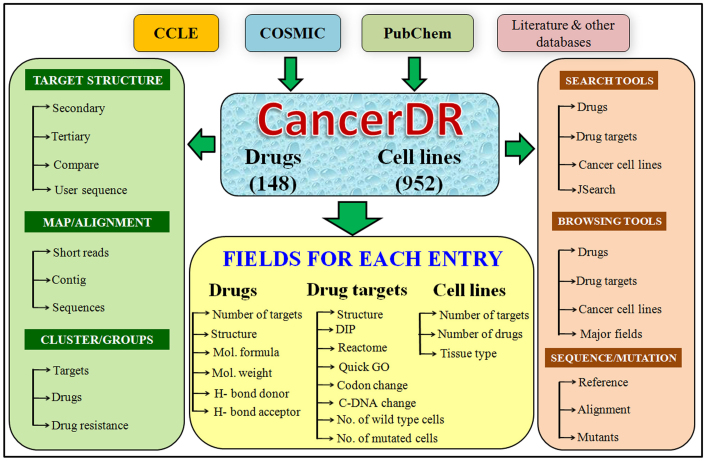
Aim of CancerDR is to collect and compile the pharmacological profiling of anti-cancer drugs on different cancer cell lines in relation to the mutation status of the drug target genes. For this, we have collected the pharmacological profiling data of 148 anti-cancer drugs on 952 cancer cell lines from COSMIC14 and CCLE15 databases. In release 2 of Genomics of drug sensitivity in cancer (one of the projects in COSMIC), 138 anti-cancer drugs targeting a wide range of therapeutic targets, were screened on 714 cancer cell lines, and in CCLE, 24 drugs were screened on 503 cancer cell lines. We focused on 116 drug targets, and their mutation status in each cancer cell line, which was collected from the hybrid capture sequencing data of 947 cancer cell lines available on CCLE website. In CancerDR, 1356 unique mutations in 116 drug targets were reported and all these mutations were mapped on their respective protein sequences. Other information like gene ontology, pathways, phylogeny about the drug targets were collected from various resources and compiled in CancerDR. In addition to this, we have collected the variants of target proteins reported in UniProt. We have also collected the information about the anti-cancer drugs from PubChem16, and Therapeutic Target Database17. For drugs, which were not available in any of the databases, we made their structures in PubChem editor, and calculated their descriptors by ChemAxon software18. Procedure of curation in CancerDR is shown in Figure 4.
Figure 4. Schematic representation of procedure of curation in CancerDR.
Database architecture and web interface
CancerDR is built on Apache HTTP server 2.2 with MySQL 5.1.47 at the back end, and the PHP 5.2.9, HTML and JavaScript at the front end. Apache, MySQL, and PHP are preferred as these are open-source software and platform independent. The architecture of CancerDR database is shown in Figure 5.
Figure 5. Schematic illustartion of architecture of CancerDR.
Organization of data
Primary data
Primary data includes information about the drugs, cell lines, and drug targets, which has been compiled from various resources. It contains 952 cancer cell lines, which were used for pharmacological profiling in CCLE and COSMIC databases, with additional information. Pharmacological profiling of 148 anti-cancer drugs has been compiled along with their chemical properties, and target proteins of these anti-cancer drugs are provided in primary data. Important databases (mentioned elsewhere) are also referred for target proteins.
Secondary data
Secondary data was derived form the primary data, which mainly includes tertiary and assigned secondary structure of target proteins. Structures were predicted by HHsuite 2.0 software19, which performs HMM-HMM-based lightning-fast iterative sequence search and DSSP20 respectively. For the structure prediction of mutants, Modeller, 9.10 21 was used. These predicted structures were used for structure-structure comparison of target proteins by Mustang22 Sequence alignment. These modelled structures were further subjected to PROCHECK23 software to identify the allowed and disallowed regions in Ramachandran plot. Few mutants and variants of drug targets were smaller in size (less than 25 amino acids), so their structures were generated by PEPstr webserver24. In addition, phylogenetic trees were also generated by using clustalw-2.0.10 software25.
Implementation of tools
Data searching
CancerDR is integrated with a user-friendly interface for extracting useful information from the database. Search option enables user to retrieve the information about drugs, drug targets and cancer cell lines. It allows users to select fields they wish to display in their results. In the fields to be displayed, selection check boxes are provided for mutation status (at cDNA, codon and protein level), predicted 3D structure of target, status of cell lines in which the target is mutated or wild type, links for protein-protein interaction databases (e.g. DIP, STRING and MINT), enzyme and pathway databases (e.g. REACTOME) and gene ontology from EMBL-EBI (e.g. QuickGO). In drug search module, user can search different properties of drugs (e.g. molecular weight, polarizability, volume, etc.). Targets of these drugs have been provided along with link to PubChem database for further details. Jmol applet link is also available to view 3D structure of drugs.
Data browsing
We have designed powerful browsing facility that allows users to browse data using various options. A brief description of interfaces designed for browsing is as follows:
Major field. This interface allows the user to browse database on the following three major fields: (i) tissue types; (ii) therapeutic target class; and (iii) type of mutants. In tissue types, user can find out the cell lines belonging to a particular tissue type, and drug sensitivity of each drug against them. Anti-cancer drugs were sub-divided according to the therapeutic target class and user can browse according to each class. For each target, type of mutants, their respective cell lines, and IC50 values are also provided.
Drug targets. For each drug target, we have collected the comprehensive information in the form of external databases links. User can explore the networks, pathways, interactions with other proteins, phylogenetic relations with nearby homologs, mapping on human genome, etc. Cell lines in which particular target is mutated have also been included.
Cell lines and drugs. Information regarding cancer cell lines used for various pharmacological assays and the list of drugs tested against these cell lines along with their IC50 values were collected and compiled. Chemical properties of each drug and their structures have also been compiled.
Alignment/Mutation
This section has been integrated to assist users to analyse variations/mutations in target gene sequences and their structures as well. The description of various modules is as follows:
Total align. This option allows users to visualize multiple sequence alignment of drug target and its natural variants as well as cancer mutants in user-friendly format using Jalview26. This option is very important for identification of mutations in cancer mutants responsible for drug resistance. It also allows users to visualize the tertiary structure along with multiple sequence alignment.
Custom align. This tool helps users to align selected mutants of any target and/or the user's query sequence, which can be seen in Jalview, interactively. By clicking on the target, user can see the list of drugs tested against that target and further selection of the drug enlists the mutants of that target against that drug. User can align more than one mutant and query sequence as well.
Mutants. This tool allows users to find out the reported mutants of a selected target at three levels (i.e. amino acid level, cDNA level, and codon level).
Structural alignment. This tool is helpful to align the tertiary structure of each target with their mutants/variants (using MUSTANG-3.2.1 software) to show the structural deviation occurred by mutations. The interface also displays the sequence alignment along with structure alignment.
Target structure
We have predicted the tertiary structure of all targets, their variants, and their mutants as well. Secondary structural state of each amino acid is also provided. Jmol applet is integrated to find out the effect of mutation on target structure. This tool also provides the facility to compare two or more mutants of a particular target to find out the structural deviation. The experimentally validated structures of each target available in Protein Data Bank (PDB) are also provided. User can also predict the structures of their own target/protein sequences.
Clusters/Groups
This module enables the users to cluster the cell lines or drugs according to the range of drug sensitivity (IC50). Two kinds of ranges are used in CancerDR. First, in which ranges are made in multiples of sensitivity reference. Sensitivity reference is the lowest IC50 value reported for particular drug or cell line. In second type of clustering, absolute range is used i.e. R1: 0–0.001 μM, R2: 0.001–0.005 μM, R3: 0.005–0.025 μM, R4: 0.025–0.125 μM, R5: 0.125–0.625 μM, R6: 0.625–15 μM, R7: 15–390 μM, R8: greater than 390 μM. Clustering can be done either according to the tissue types in which cell lines of particular tissue type will be clustered or according to cell lines having one or more mutations in drug targets.
Map/Alignment
This is an important web interface, which is helpful for users to identify genetic variations/mutations in user defined query sequence(s). User may also submit NGS data (short reads/contigs) directly and interface will map this NGS data to drug targets.
Mapping of short reads. Due to advancement in sequencing technologies, it is feasible to sequence whole transcriptome, exome, genome of cancer patient using NGS techniques. This sequencing data (short reads) can be used to identify sensitive and resistant drugs in a cancer patient based on mutation in drug targets. CancerDR allows users to map/align their short reads on any drug target in this database using software packages BWA27 and SAMtool28. In order to visualize alignment, we have integrated Tablet viewer29 in CancerDR.
Mapping of sequence contigs. The genome assemblers assemble short reads obtained from NGS and produce long sequences called contigs. It is important to find out the genes in the contigs for further analysis. We have developed a module that allows user to submit their contigs to CancerDR. Our server first predicts genes/proteins in contigs using Augustus30 and then aligns these genes against all cancer drug targets using BLAST31.
Sequences. This module allows users to compare any gene or protein sequence with cancer drug targets provided in the database. We have integrated BLAST search tool in this module. It allows users to submit one or more genes or protein sequences in FASTA format for performing BLAST search against the cancer targets.
Download
Download module provides the facility to download sequences, alignments and structures present in CancerDR. User can download sequences or structures of drug targets manually as well as automatically. This database also provides Rsync facility so that user can synchronize or update information.
Update of CancerDR
We have included the most recent data available at CCLE and COSMIC websites in CancerDR. We will try to incorporate the new releases as soon as they will be available in public. Web server allows the user to submit his/her own information by using the submission form available at CancerDR website. However, before including in CancerDR, our team will scrutinize the authentication of the data.
Author Contributions
R.K. collected and organized the data. R.K. and K.C. developed web interface. K.C. integrated the alignment tools. S.G. performed the clustering of cell lines and drugs. H.S. predicted the tertiary structure of proteins. S.K. integrated the NGS tools. P.K. collected the information about the drugs. A.G. contributed in manuscript writing. G.P.S.R. conceived the idea and coordinated the project.
Supplementary Material
Supplementary information
Acknowledgments
Authors are thankful to funding agencies Council of Scientific and Industrial Research (project Open Source Drug Discovery and GENESIS BSC0121) and Department of Biotechnology (project BTISNET), Govt. of India for financial support.
References
- Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronbaek K., Hother C. & Jones P. A. Epigenetic changes in cancer. APMIS 115, 1039–59 (2007). [DOI] [PubMed] [Google Scholar]
- Sadikovic B., Al-Romaih K., Squire J. A. & Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics 9, 394–408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Identification of the MEK1(F129L) activating mutation as a potential mechanism of acquired resistance to MEK inhibition in human cancers carrying the B-RafV600E mutation. Cancer Res 71, 5535–45 (2011). [DOI] [PubMed] [Google Scholar]
- Zdanowicz M. M. The pharmacology of HIV drug resistance. Am J Pharm Educ 70, 100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. & Lengauer T. Managing drug resistance in cancer: lessons from HIV therapy. Nat Rev Cancer 12, 494–501 (2012). [DOI] [PubMed] [Google Scholar]
- Chin L., Andersen J. N. & Futreal P. A. Cancer genomics: from discovery science to personalized medicine. Nat Med 17, 297–303 (2011). [DOI] [PubMed] [Google Scholar]
- Jain K. K. Role of nanobiotechnology in developing personalized medicine for cancer. Technol Cancer Res Treat 4, 645–50 (2005). [DOI] [PubMed] [Google Scholar]
- Mok T. S. Personalized medicine in lung cancer: what we need to know. Nat Rev Clin Oncol 8, 661–8 (2011). [DOI] [PubMed] [Google Scholar]
- Tursz T., Andre F., Lazar V., Lacroix L. & Soria J. C. Implications of personalized medicine--perspective from a cancer center. Nat Rev Clin Oncol 8, 177–83 (2011). [DOI] [PubMed] [Google Scholar]
- Rhee S. Y. et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31, 298–303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R. J. et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 25, 1197–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M. W., Liu T. F. & Shafer R. W. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology 55, 98–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E W. Y., Thiessen P.A. & Bryant S.H. PubChem: Integrated Platform of Small Molecules and Biological Activities. Annual Reports in Computational Chemistry Volume 4 (2008). [Google Scholar]
- Chen X., Ji Z. L. & Chen Y. Z. TTD: Therapeutic Target Database. Nucleic Acids Res 30, 412–5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Chem Base was used for structure searching and chemical database access and management. J Chem(5.10) (2012). [Google Scholar]
- Remmert M., Biegert A., Hauser A. & Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9, 173–5 (2012). [DOI] [PubMed] [Google Scholar]
- Kabsch W. & Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–637 (1983). [DOI] [PubMed] [Google Scholar]
- Eswar N. et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5, Unit 5 6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagurthu A. S., Whisstock J. C., Stuckey P. J. & Lesk A. M. MUSTANG: a multiple structural alignment algorithm. Proteins 64, 559–74 (2006). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: aprogram to check the stereochemical quality of protein structures. J Appl Cryst 26, 283–291 (1993). [Google Scholar]
- Kaur H., Garg A. & Raghava G. P. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept Lett 14, 626–31 (2007). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–80 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troshin P. V., Procter J. B. & Barton G. J. Java bioinformatics analysis web services for multiple sequence alignment--JABAWS:MSA. Bioinformatics 27, 2001–2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAM tools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I. et al. Tablet--next generation sequence assembly visualization. Bioinformatics 26, 401–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Steinkamp R., Waack S. & Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32, W309–12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol 215, 403–10 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information