Abstract
The paintings from Tomba della Scimmia, in Tuscany, are representative of the heavy bacterial colonization experienced in most Etruscan necropolises. The tomb remained open until the late 70′s when it was closed because of severe deterioration of the walls, ceiling and paintings after decades of visits. The deterioration is the result of environmental changes and impacts suffered since its discovery in 1846. We show scanning electron microscopy and molecular studies that reveal the extent and nature of the biodeterioration. Actinobacteria, mainly Nocardia and Pseudonocardia colonize and grow on the tomb walls and this process is linked to the availability of organic matter, phyllosilicates (e.g. clay minerals) and iron oxides. Nocardia is found metabolically active in the paintings. The data confirm the specialization of the genera Nocardia and Pseudonocardia in the colonization of subterranean niches.
The Etruscan civilization was characterized by the construction of tombs decorated with impressive paintings1. The Tomba della Scimmia (Tomb of the Monkey) 480–470 BC, near Chiusi, Italy, discovered in 1846 by Alexander François, was named after a monkey on a tree depicted in the entrance chamber (Fig. 1). The tomb is composed of three rooms around an atrium and excavated 8 m below the ground level on a lithological complex of Pliocene sands weakly cemented with intercalations of clay layers and pebble beds. The walls were decorated with paintings representing funeral games. The paintings were made with hematite, Egyptian blue and charcoal black, deposited on a previously prepared thin clay layer2.
Figure 1. Painting in Tomba della Scimmia, Chiusi, Italy.
The current status of the tomb is the result of the accumulation of multiple microenvironmental changes and impacts suffered from the time of its discovery. As many other Etruscan and Roman necropolises, this tomb is included in a tourism net that brings thousands of visitors per year. Tombs receiving visits are exposed to microclimatic modifications due to the presence of visitors, which originated disturbances of the usual steady climate (i.e., switch on/off of the lights, openings of the door, ventilation, etc.).
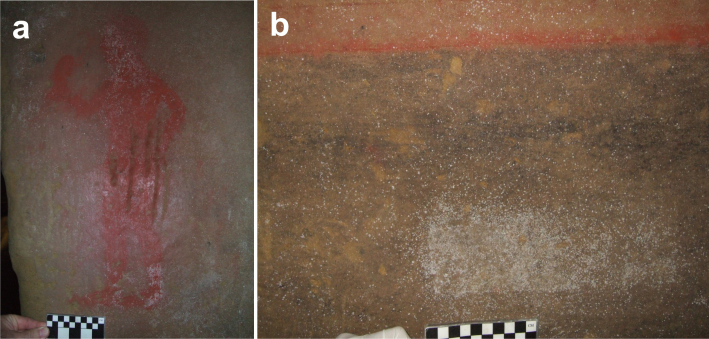
At the beginning of this century, the tomb ceiling presented a detached sand stratum, while material losses occurred between the sands and the underlying beds of pebbles, which was the point where the rock had a lesser cohesion. In addition, between 1971 and 1989 the wall surfaces were partly covered with whitish spots attributed to salt efflorescences, but they were later identified as bacterial colonizations. The walls were treated with Neodesogen (benzalkonium chloride) in a restoration carried out in 1993, and with 2% Preventol R80 (dodecyl dimethyl dichlorobenzyl ammonium chloride) in a further restoration in 2000. Currently the tomb is open to visitors for three days a week and two visits every day and the walls and paintings are heavily colonized by bacteria as shown in Figure 2a,b.
Figure 2. Painting heavily colonized by actinobacteria.
(a) Sample SC3 was taken from the right leg. (b) Sample SC6 was taken from the whitish area.
The aim of this work is to know the reason for such a heavy bacterial colonization, exhibited by most Tuscanian tombs, and the composition of the bacterial communities involved in the deterioration of the paintings of Tomba della Scimmia.
Results
Microscopic study
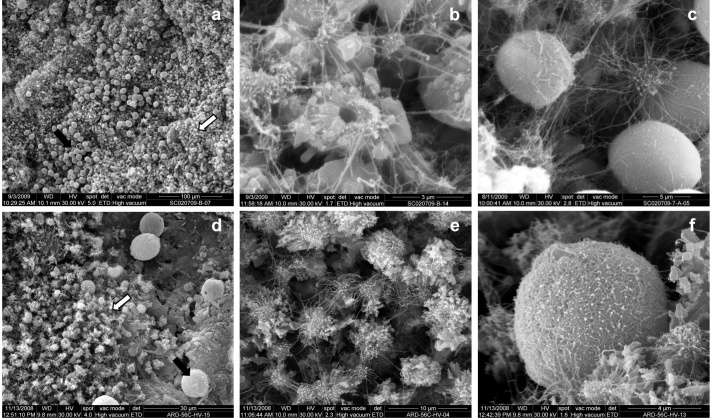
White colonizations were sampled from different rooms. In general, the colonizations are small, from less than 1 mm up to 2–3 mm diameter. They present an outline typically defined, circular to irregular, and have little relief. Unlike other subterranean environments3, the bacterial mats do not act as water condensation points. Environmental scanning electron microscopy (ESEM) showed that the colonizations were formed by a dense network of fine and large bacterial filaments with some extracellular polymeric substances (EPS) and a bed of mineral elements with morphology in rosette or "nest", and spherical elements (Fig. 3a–c). The morphology of the filaments is compatible with those of actinobacteria3 and similar mineral structures were previously found in Altamira Cave, Spain4,5.
Figure 3. Environmental scanning electron micrographs of white colonizations from Tomba della Scimmia (a–c) and from Ardales Cave (d–f).
(a) Bed formed of CaCO3 nest-like aggregates (white arrow) with dispersed spheroidal elements (black arrows) from Tomba della Scimmia. (b) Small patches of CaCO3 nest-like aggregates and filaments from Tomba della Scimmia. (c) Detail of spheroidal elements of CaCO3 coated with filamentous bacteria from Tomba della Scimmia. Similar mineral and biological morphologies were observed in Ardales Cave walls and sediments colonized by Pseudonocardia8 as shown in d–f. (d) A bed of nest-like aggregates and spheroidal elements in Ardales Cave. (e) Small patches of CaCO3 nest-like aggregates and filaments in Ardales Cave. (f) Spheroidal CaCO3 coated with filaments in Ardales Cave.
Two main types of CaCO3 deposits were observed in the white colonizations:
Fabrics with morphology in rosette or "nest" formed by aggregations of small crystals, directly placed on the substratum (Fig. 3a,b). They are composed of calcium carbonate as revealed Energy-Dispersive Spectroscopy (EDS) microanalysis. Average sizes vary between 2 to 4 μm. In detail, they are composed of aggregates of subeuhedral to euhedral calcite crystals, with radial arrangement, sometimes with a pseudohexagonal contour. The nests usually show a central hole (0.5–0.7 μm in diameter). Sometimes they appear coated by a dense network of fine filaments and EPS.
Spheroidal elements averaging between 8 and 10 μm in diameter, composed of calcium carbonate (EDS microanalysis) which appear coated or not by a filamentous biofilm (Fig. 3c). The mineralogical phase is probably vaterite, which typically presents this spheroidal habit and similar dimensions. In many cases, a hole of between 0.5 to 1 μm in diameter was observed on the sphere surface. The biofilm that covers some of the spheroids consist of fine bacterial filaments and EPS. Surfaces not covered by the biofilms are irregular and sometimes with idiomorphic crystal morphologies that resemble rhombohedral Calcite and presented corroded surfaces.
In this tomb, the morphologies of the bacterial and mineral structures were quite similar to those of Ardales Cave, Spain, colonized by Pseudonocardia. In fact, the micrographs showed in Figure 3d–f revealed a striking similarity between the white colonizations and mineral deposits from both sites. ESEM data suggests the active participation of actinobacteria in the bioinduction of CaCO3 deposits, as discussed previously by other authors3,4,5.
DGGE study
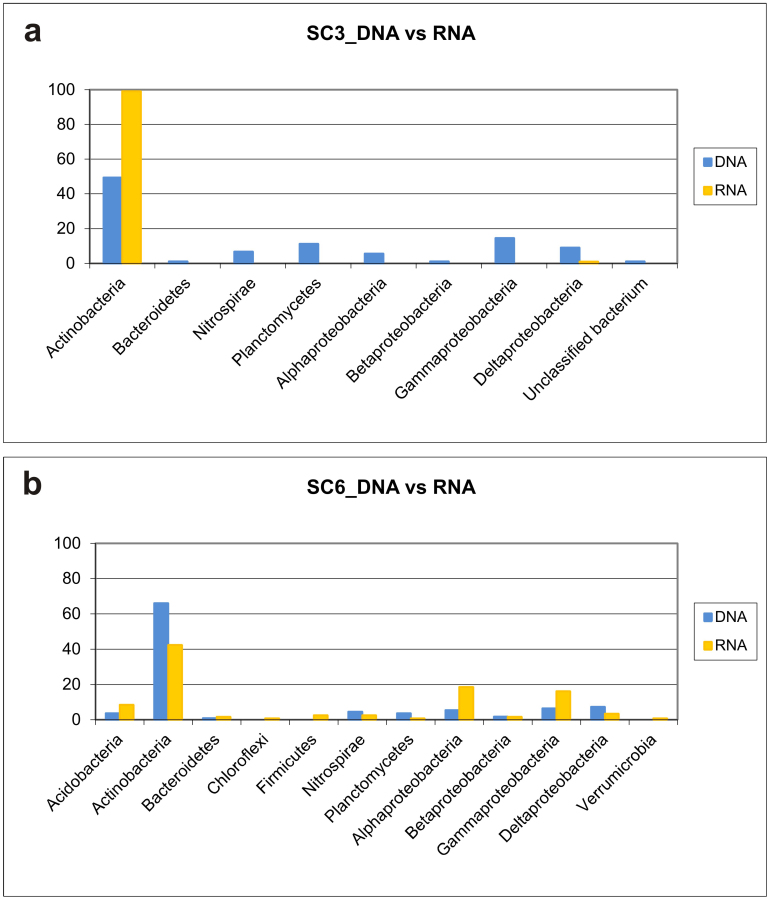
Comparative DGGE analyses of the total (DNA) and metabolically active (RNA) bacterial communities6,7 from two white colonizations are shown in Supplementary Figure 1. Sample SC3 was taken from a figure painted with hematite (Fig. 2a) and sample SC6 from the atrium wall in the dark area below the paintings (Fig. 2b). DGGE showed some common bands, particularly in sample SC6-DNA and -RNA, but the abundance and distribution of representative bands between SC3 and SC6 samples were different. In sample SC3, it was clearly shown the predominance of a single band from Nocardia in the RNA pattern, but Pseudonocardia and other actinobacteria were identified in the DNA pattern.
Distribution of 16 rRNA gene clone sequences among different phyla
A total of 414 non-chimeric sequences were obtained from the two samples. The sequences found in each sample are reported in Supplementary Tables S1–S4. Rarefaction curves (Supplementary Fig. S2) indicated that the full extent of diversity was not reached. A deep coverage of bacterial communities in samples from historical sites is almost impossible due to the natural limitations imposed to samplings by the protection of cultural heritage sites.
The distribution of different phyla among the samples is summarized in Figure 4. Only one very major phylogenetic group belonging to Actinobacteria was encountered in the clone libraries, which represented 49.4 and 66% of DNA sequences, and 99 and 42.4% of RNA sequences for samples SC3 and SC6, respectively. Besides Actinobacteria, only Planctomycetes and Proteobacteria dominated in both samples, either as total or metabolically active members. Planctomycetes (11.2%) and Gammaproteobacteria (14.6%) attained some importance in SC3-DNA, and Alphaproteobacteria (18.6%) and Gammaproteobacteria (16.1%) in SC6-RNA.
Figure 4. Distribution of major phylogenetic groups in the 16S rRNA gene clone library constructed from Tomba della Scimmia SC3 and SC6 samples.
A survey on the sequences obtained from the two samples and the nearest published relatives (Supplementary Tables S1–S4) revealed that 38.6% corresponded to uncultured bacteria from caves and shelters, 26.3% to sequences retrieved from polluted soils and 16.2% to sequences from different soil types. The data points to the presence of a community specialized in soil/subterranean niches able to cope with recalcitrant organic matter and pollutants as nutrient sources.
Discussion
The phylum Actinobacteria constitutes a significant part of the bacterial population in subterranean environments8,9,10. Previous studies on different Spanish and Italian caves revealed that the special microclimatic conditions together with nutrient availability and the nature of the organic matter are important factors controlling the activity of Actinobacteria in caves9, which have a major role in biogeochemical processes, namely biodeterioration and bioprecipitation of minerals, as recently discussed3,4,5,8,9. It is remarkable that 99% of RNA sequences retrieved from sample SC3 were from Nocardia.
The phylum Planctomycetes and the classes Alphaproteobacteria and Gammaproteobacteria, also with some abundance in the samples, are commonly reported in polluted subterranean environments11.
The genus Nocardia is frequently isolated from soils12 and clinical specimens10. In the last few years, members of Nocardia were isolated from European and Asian caves10. Although the primary reservoir of Nocardia asteroides was thought to be the soil13, N. asteroides was common in the air of all the halls and galleries in Castañar de Ibor Cave, Spain, but absent in the air outside the cave (unpublished data). Literature data show a worldwide distribution of members of the genus Nocardia in subterranean environments.
The genus Pseudonocardia is also common in subterranean environments, such as gold mine caves10. Pseudonocardia halophobica was found in Thai caves14. Pseudonocardia spp. were reported in Malta catacombs15, Carlsbad Cavern, New Mexico, USA16 and in white colonizations in the Spanish Altamira, Ardales and Santimamiñe caves3,8,17. A previous study on these Spanish caves showed that the DGGE patterns of the metabolically active bacterial communities were composed of a major, almost exclusive, band corresponding to Pseudonocardia, in amounts as high as 85.6% and 65.7% for Ardales and Santimamiñe caves, respectively8.
The presence and abundance of Nocardia and Pseudonocardia in subterranean environments may be driven by:
Organic matter. Cave colonization by Nocardia and Pseudonocardia were related with agricultural and/or livestock activities on the top soil8,9,18. The organic carbon content in most Italian top soils (86.4% of total land area) is ≤ 2%19. Soil organic matter is composed of biodegraded lignin and humic substances, among other macromolecules20,21. Subterranean dripping waters contain these macromolecules as dissolved organic matter (DOM), which can reach the paintings. Nocardia (e.g. Nocardia erythropolis, Nocardia corallina and Nocardia opaca) and Pseudonocardia spp. use humic substances as sole carbon and nitrogen sources22,23,24. In addition, Nocardia has been shown to degrade lignin in soil25. Barton et al.16 found that 80% of the total community of a limestone formation in Carlsbad Cavern was represented by Pseudonocardia. These authors suggested that organic matter in this cave is of a phenolic and aromatic nature, as previously observed in other caves18.
Phyllosilicates. Phyllosilicates and particularly clay minerals have a high adsorbance potential for macromolecules26. DOM from dripping waters may be adsorbed to clay, hindering the loss of nutrient in an oligotrophic environment, and in this way may be accessed by bacteria. Colonization and growth of Pseudonocardia is favoured by the presence of clayey substrata in the Spanish caves of Ardales and Santimamiñe8. In Carlsbad Cavern, to the limestone that supported the Pseudonocardia were associated clay particles16. Etruscan paintings were applied on a thin layer of clay2. The stimulation of clays on the colonization and metabolic activity of Nocardia was well-known time ago27.
Iron oxides. Hematite was used as red pigment for the paintings2. Iron is required for an optimal growth of Nocardia28,29. Hematite and clays can adsorbe humic substances, thus facilitating the access of microorganisms to carbon sources30.
In conclusion, the abundant actinobacterial colonization of Tomba della Scimmia is promoted by the presence of organic matter and clays on the walls. Iron oxide represents an additional factor favouring growth. This type of colonization was also observed in other Tuscanian tombs with similar constructive features (e.g. Tomba del Colle in the Poggio Renzo Necropolis, Tomba della Pellegrina, near Tomba della Scimmia, etc.). The actinobacteria are involved in the bioprecipitation of minerals4,5,6 (Fig. 3), among other biogeochemical processes, as well as in the biodeterioration of the paintings. In fact, actinobacteria have a broad ability to produce acids from most carbohydrates and cause mineral leaching31.
The identification of active microorganisms7,17,32 can provide clues for an effective control and should be the target in cleaning and restoration processes, when possible. Biocides treatments in caves where used in the past and currently are critically discussed because they seem not to be effective or at least have not long-term action. If we consider the intimate contact between bacteria and mineral layer (Fig. 3), a removal of the dead bacterial biomass, after treatment of the paintings is impossible without damaging the paint layer. This dead biomass will support the growth of saprophyte fungi as secondary invaders as soon as the biocide is degraded or even the biocide used as a nutrient source by some other bacteria32.
Due to the particular Etruscan pictorial technique (application of pigments on a fine clay layer), the only possible action is controlling the microclimate, visits, and the input of DOM to the interior of the tomb by acting on top soil grasses and plants (non agricultural use, seasonal harvest of spontaneous grasses, etc.) in order to reduce organic matter decomposition. Similar actions were carried out in Altamira Cave with remarkable results18. Biocide treatments are not recommended in subterranean environments32,33 because they are not effective at short and mid-term, promote further secondary colonizations and increase microbial biodiversity.
Methods
Sample collection
Sample SC3 and SC6 were collected from Tomba della Scimmia on 2nd July, 2009. Both samples represented macroscopic white colonizations located on the paintings (SC3) and on a place without paintings (SC6) (Fig. 2a,b). The samples were taken by scraping off the colonizations with a sterile scalpel. One mL of RNA-later was added to a set of tubes. Upon collection, the samples were stored on ice and processed or frozen when arrived to the laboratory.
Nucleic acid extraction and PCR amplification
Total nucleic acids from the samples were extracted using the method described by Griffiths et al.34. Extracted nucleic acids were resuspended in 50 μl of sterile ultrapure water (Sigma-Aldrich). For RNA extraction, a 25 μl aliquot was subjected to a DNase digestion step using an RNase-Free DNase (Qiagen), and the RNA was purified using the RNeasy MinElute Cleanup kit (Qiagen), according to the manufacturer’s instructions. Complementary DNA (cDNA) from this RNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen) with a single specific primers: 907R (5′-CCCCGTCAATTCATTTGAGTTT-3′) for bacterial 16S rRNA gene35, Arch 1000R (5′-GGCCATGCACYWCYTCTC-3′) for archaeal 16S rRNA gene36 and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) for fungal internal transcribed spacer (ITS) regions37, at a temperature of 42°C for 50 min. Amplification of cDNA was performed with the bacteria-specific primers, 616F (5′-AGAGTTTGATYMTGGCTCAG-3′)38 and 907R35, the archaea-specific primers, Arch 340F (5′-CCCTAYGGGGYGCASCAG-3′) and Arch 1000R36, and fungi-specific primers, ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS437. One or two μl of cDNA, diluted 1/10 were used as a template. PCR conditions were the same for DNA. Amplifications of archaea and fungi from samples SC3 and SC6 were unsuccessful.
PCR reactions were performed in a Bio-Rad iCycler thermal cycler (Bio-Rad). The PCR reaction mixture (1 ml) consisted of 100 μl of PCR buffer (Biotools), 30 μl of MgCl2 (50 mM, Biotools), 10 μl of each primer (50 mM), 100 μl of dNTPs (100 mM, Bioline), 5 μl of Taq polymerase (Biotools) and 750 μl of sterile ultrapure water. PCR reactions were performed in 0.2 mL PCR tubes containing 25 or 50 μL of reaction mixture and from 0.5 to 2 μl of DNA template (pure or diluted to 2 ng/μl). PCR amplifications were performed using the following thermal conditions: 94°C for 2 min; 35 cycles of 94°C for 15 s, 55°C for 15 s (50°C for ITS regions), 72°C for 2 min; and a final step of 72°C for 10 min. Positive and negative controls were included in all amplification experiments. All amplification products were purified with a JETquick PCR Purification Spin kit (Genomed) and stored at −20°C for further analysis.
Cloning and sequencing
DNA libraries of PCR amplified products were constructed using the pGEM-T Easy Vector (Promega) and then transformed into One Shot Max Efficiency DH5a-T1 chemically competent Escherichia coli (Invitrogen) according to manufactureś instructions. Transformants were randomly picked and transferred to multiwell plates containing Luria–Bertani medium supplemented with 100 μg·mL−1 ampicillin and 15% w/v glycerol and stored at −80°C. We constructed four libraries. On average 100 clones from each library was sequenced at Macrogen Inc., Seoul, Korea, using the universal bacterial primer 616F.
Sequence analyses
Sequences were checked for chimera by chimera.slayer as implemented in the software package mothur39. Putative chimeric sequences were excluded from further analysis and 414 sequences were included in phylogenetic analyses. Sequences were aligned using mothur. After this analysis, all sequences were compared to the non-redundant database of sequences deposited at the National Center for Biotechnology (NCBI) and EzTaxon40 using BLASTN algorithm41. Aligned sequences were clustered into operational taxonomic units (OTUs) using mothur, with a 97% sequence identity cutoff. Rarefaction curves were also obtained using mothur. All sequences were submitted to GenBank with consecutive accession numbers HF58493-HF58701.
Bacterial community fingerprints
Fingerprints of the total and metabolically active microbial communities were obtained by denaturing gradient gel electrophoresis (DGGE) according to Muyzer et al.42. A nested-PCR reaction was performed with the primers 341F (5′-CCTACGGGAGGCAGCAG-3′; with a GC-rich tail added at its 5′-end) and 518R (5′-ATTACCGCGGCTGCTGG-3′). PCR amplifications were performed using the following thermal conditions: 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 55°C for 15 s and 72°C for 30 s with a final extension at 72°C for 30 min. DGGE was carried out using a DCODE™ System (Bio-Rad).
Environmental scanning electron microscopy
Textural and microestructural characterization of different white colonizations were performed using an Inspect-S50 low-vacuum environmental scanning electron microscope (FEI Company, Japan) that includes an energy-dispersive spectroscopy probe. First the samples were observed and described under controlled low-vacuum conditions. Further, the samples were gold sputter covered (gold coater EMITECH K550Y) and observed under high vacuum conditions to improve the photographic quality and EDS microanalysis.
Author Contributions
C.S.-J. conceived, planned the projects and wrote the manuscript. V.J. collected samples, supervised phylogenetic analysis and performed statistical analyses. M.D.-H. extracted DNA and RNA, performed DGGE and sequence analyses. S.C. and S.S.-M. conducted ESEM analyses and interpreted the micrographs. L.L. advised M.D.-H. in laboratory analyses. P.P. and P.T. organized the sampling trips, provided access to the tomb, supplied historical data on the paintings and discussed on the conservation methods.
Supplementary Material
Supplementary Information
Acknowledgments
M.D.-H. was supported by a JAE Research Fellowship from the Spanish Council of Scientific Research and S.C. by a Juan de la Cierva contract. This research was supported with funding provided to the projects CGL2010-17183, 201030E011 and Consolider CSD2007-00058.
Accession codes: Sequences were deposited at GenBank under the accession numbers HF58493-HF58701.
References
- Steingraber S. Abundance of Life: Etruscan Wall Painting (Arsenale, Verona, 2006). [Google Scholar]
- Colombini M. P., Giachi G., Pallechi P. & Ribechini E. T. Tecniche pittoriche utilizzate nelle tombe etrusche di Chiusi e Sarteano. In: Pittura Etrusca. Problemi e Prospettive. (ed Minetti A.) 162–167 (Protagon Ed. Toscani Siena, 2003). [Google Scholar]
- Cuezva S., Sanchez-Moral S., Saiz-Jimenez C. & Cañaveras J. C. Microbial communities and associated mineral fabrics in Altamira Cave, Spain. Int. J. Speleol. 38, 83–92 (2009). [Google Scholar]
- Cañaveras J. C. et al. On the origin of fiber calcite crystals in moonmilk deposits. Naturwissenschaften 93, 27–32 (2006). [DOI] [PubMed] [Google Scholar]
- Cuezva S. et al. The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 81, 281–290 (2012). [DOI] [PubMed] [Google Scholar]
- Piñar G., Saiz-Jimenez C., Schabereiter-Gurtner C., Blanco-Varela M. T., Lubitz W. & Rölleke S. Archaeal communities in two disparate deteriorated ancient wall paintings, detection, identification and temporal monitoring by DGGE. FEMS Microbiol. Ecol. 37, 45–54 (2001). [Google Scholar]
- Gonzalez J. M., Portillo M. C. & Saiz-Jimenez C. Metabolically active Crenarchaeota in Altamira Cave. Naturwissenschaften 93, 42–45 (2006). [DOI] [PubMed] [Google Scholar]
- Stomeo F., Portillo M. C., Gonzalez J. M., Laiz L. & Saiz-Jimenez C. Pseudonocardia in white colonizations in two caves with Paleolithic paintings. Int. Biodeterior. Biodegrad. 62, 483–486 (2008). [Google Scholar]
- Groth I., Vetermann R., Schuetze B., Schumann P. & Saiz-Jimenez C. Actinomycetes in karstic caves of Northern Spain (Altamira and Tito Bustillo). J. Microbiol. Methods 36, 115–122 (1999). [DOI] [PubMed] [Google Scholar]
- Jurado V. et al. Pathogenic and opportunistic microorganisms in caves. Int. J. Speleol. 39, 15–24 (2010). [Google Scholar]
- Miller A. Z. et al. Enigmatic reticulated filaments in subsurface granite. Environ. Microbiol. Rep. 4, 596–603 (2012). [DOI] [PubMed] [Google Scholar]
- Goodfellow M. & Williams S. T. Ecology of actinomycetes. Ann. Rev. Microbiol. 37,189–216 (1983). [DOI] [PubMed] [Google Scholar]
- Yamamura H., Hayakawa M., Nakagawa Y. & Iimura Y. Characterization of Nocardia asteroides isolates from different ecological habitats on the basis of repetitive extragenic palindromic-PCR fingerprinting. Appl. Environ. Microbiol. 70, 3149–3151 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaew N., Pathom-aree W. & Lumyong S. Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetologica 23, 21–26 (2009). [Google Scholar]
- De Leo F., Iero A., Zammit G. & Urzì C. Chemoorganotrophic bacteria isolated from biodeteriorated surfaces in cave and catacombs. Int. J. Speleol. 41, 125–136 (2012). [Google Scholar]
- Barton H. A., Taylor N. M., Kreate M. P., Springer A. C., Oehrle S. A. & Bertog J. L. The impact of host rock geochemistry on bacterial community structure in oligotrophic cave environments. Int. J. Speleol. 36, 93–104 (2007). [Google Scholar]
- Portillo M. C., Saiz-Jimenez C. & Gonzalez J. M. Molecular characterization of total and metabolically active bacterial communities of ‘‘white colonizations’’ in the Altamira Cave, Spain. Res. Microbiol. 160, 41–47 (2009). [DOI] [PubMed] [Google Scholar]
- Saiz-Jimenez C. et al. Paleolithic art in peril: Policy and science collide at Altamira Cave. Science 334, 42–43 (2011). [DOI] [PubMed] [Google Scholar]
- Rusco E., Jones R. & Bidoglio G. Organic matter in the soils of Europe: Present status and future trends. Report EUR 20556.Accessed at http://eusoils.jrc.ec.europa.eu/esdb_archive/eusoils_docs/other/ESF_OM7.pdf on 19 February 2013.
- Martin F., Saiz-Jimenez C. & Cert A. Pyrolysis-gas chromatography-mass spectrometry of soil humic fractions. II. The high boiling point compounds. Soil Sci. Soc. Am. J. 43, 309–312 (1979). [Google Scholar]
- Saiz-Jimenez C., Hawkins B. L. & Maciel G. E. Cross polarization, magic-angle spinning 13C nuclear magnetic resonance spectroscopy of soil humic fractions. Org. Geochem. 9, 277–284 (1986). [Google Scholar]
- Steinbrenner K. & Mundstock I. Untersuchungen zum Huminstoffabbau durch Nokardien. Arch. Acker. Pflanzenbau Bodenk. 19, 243–255 (1975). [Google Scholar]
- Sidorenko O. D., Aristarkhova V. I. & Chernikov V. A. Changes in the composition and properties of humic acids brought about by the action of microorganisms of the genus Nocardia. Biol. Bull. Acad. Sci. USSR 5, 150–155 (1978). [PubMed] [Google Scholar]
- Solntseva I. E. Certain energy aspects of microorganism-induced transformation of humic substances. In Humic Substances in the Global Environment and Implications on Human Health (eds Senesi N., & Miano T. M.) 373–379 (Elsevier, Amsterdam, 1994). [Google Scholar]
- Tuomela M., Vikman M., Hatakka A. & Itävaara M. Biodegradation of lignin in a compost environment, a review. Bioresour. Technol. 72, 169–183 (2000). [Google Scholar]
- Evans L. T. & Russell E. W. The adsorption of humic and fulvic acids by clays. J. Soil Sci. 10, 119–132 (1959). [Google Scholar]
- Martin J. P., Filip Z. & Haider K. Effect of montmorillonite and humate on growth and metabolic activity of some actinomycetes. Soil Biol. Biochem. 8, 409–413 (1976). [Google Scholar]
- Heim A. H. & Lechevalier H. Effect of iron, zinc, manganese and calcium on the growth of various strains of Streptomyces. Mycologia 48, 628–636 (1956). [Google Scholar]
- Webley D. M. The effect of deficiency of iron, zinc and manganese on the growth and morphology of Nocardia opaca. J. Gen. Microbiol. 23, 87–92 (1960). [DOI] [PubMed] [Google Scholar]
- Murphy E. M., Zachara J. M. & Smith S. C. Influence of mineral-bound humic substances on the sorption of hydrophobic organic compounds. Environ. Sci. Technol. 24, 1507–1516 (1990). [DOI] [PubMed] [Google Scholar]
- Abdulla H. Bioweathering and biotransformation of granitic rock minerals by actinomycetes. Microb. Ecol. 58, 753–761 (2009). [DOI] [PubMed] [Google Scholar]
- Martin-Sanchez P. M., Nováková A., Bastian F., Alabouvette C. & Saiz-Jimenez C. Use of biocides for the control of fungal outbreaks in subterranean environments: The case of the Lascaux Cave in France. Environ. Sci. Technol. 46, 762–3770 (2012). [DOI] [PubMed] [Google Scholar]
- Saiz-Jimenez C. Painted Materials. In Cultural Heritage Microbiology (eds Mitchell R., & McNamara C. J.) 3–13 (ASM Press, Washington, DC, 2010). [Google Scholar]
- Griffiths R. I., Whiteley A. S., O’Donnell A. G. & Bailey M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A., Wawer C., Muyzer G. & Ramsing N. B. Distribution of sulphate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and DGGE of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62,1405–1415 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner S., Andersson A. F., Alonso-Sáez L. & Bertilsson S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84, 12–18 (2011). [DOI] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S. & Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, A Guide to Methods and Applications (eds Innis M. A., Gelfand D. H., Sninsky J. J., & White T. J.) 315–322 (Academic Press, New York, 1990). [Google Scholar]
- Zimmermann J., Gonzalez J. M., Ludwig W. & Saiz-Jimenez C. Detection and phylogenetic relationships of a highly diverse uncultured acidobacterial community on paleolithic paintings in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiol. J. 22, 379–388 (2005). [Google Scholar]
- Schloss P. C. et al. Introducing mothur: Open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Muyzer G., de Waal E. C. & Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information