Abstract
Background. The persistent presence of antiphospholipid antibodies (APA) may lead to the development of primary or secondary antiphospholipid syndrome. Although the genetic basis of APA has been suggested, the identity of the underlying genes is largely unknown. In this study, we have performed a genome-wide association study (GWAS) in an effort to identify susceptibility loci/genes for three main APA: anticardiolipin antibodies (ACL), lupus anticoagulant (LAC), and anti-β 2 glycoprotein I antibodies (anti-β 2GPI). Methods. DNA samples were genotyped using the Affymetrix 6.0 array containing 906,600 single-nucleotide polymorphisms (SNPs). Association of SNPs with the antibody status (positive/negative) was tested using logistic regression under the additive model. Results. We have identified a number of suggestive novel loci with P < E − 05. Although they do not meet the conservative threshold of genome-wide significance, many of the suggestive loci are potential candidates for the production of APA. We have replicated the previously reported associations of HLA genes and APOH with APA but these were not the top loci. Conclusions. We have identified a number of suggestive novel loci for APA that will stimulate follow-up studies in independent and larger samples to replicate our findings.
1. Introduction
Antiphospholipid antibodies (APA) are a heterogeneous group of antibodies that are detected in a variety of conditions, including primary antiphospholipid syndrome (APS) and systemic lupus erythematosus (SLE) [1]. The term antiphospholipid antibodies is a misnomer as APA present in autoimmune disease, like SLE, do not bind to phospholipids but recognize phospholipid-binding proteins [2]. Patients with persistent APA who develop pregnancy complications or thrombosis are considered to have primary APS and those who develop these complications in the presence of autoimmune disease are classified having secondary APS. Since the definition of APS is not limited to a single APA assay, it is required to measure more than one APA. Indeed, currently recognized laboratory criteria for APS include having one or more of three APA, including anticardiolipin antibodies (ACL), lupus anticoagulant (LAC), or anti-β 2 glycoprotein I antibodies (anti-β 2GPI) in conjunction with the presence of thrombosis or pregnancy loss [3].
Although the genetic basis of APA [4] and APS [5] has been suggested, the underlying genetic factors have not been clearly established. Understanding the genetic bases of various APA may help to delineate the mechanisms for APS. The objective of this study was to perform a genome-wide association study (GWAS) in an effort to identify loci/genes for the three main APA, namely, ACL, LAC, and anti-β 2GPI.
2. Subjects and Methods
2.1. Subjects
A subset of individuals from our larger GWAS of SLE (unpublished data) that had the ACL (n = 670), LAC (n = 708), and anti-β 2GPI (n = 496) measurements available were used in this study. All individuals were women of European ancestry. The study participants included both SLE cases and controls and their characteristics are given in Table 1. Our controls were apparently healthy individuals that were recruited from blood bank. We measured APA in our controls but they were not characterized for primary APS due to our study design that is focused on identifying genes for SLE and APA. Furthermore, there were only 28 individuals with APS, and this small number was not considered to be appropriate for a GWAS analysis. All subjects provided written informed consent and the study was approved by the Institutional Review Board.
Table 1.
Characteristics of study participants with three antiphospholipid antibodies in the GWAS dataset*.
| ACL | LAC | Anti-β 2GPI | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| (n = 183) | (n = 487) | (n = 127) | (n = 581) | (n = 136) | (n = 360) | |
| Mean age ± SD | 46.92 ± 11.41 | 46.19 ± 10.85 | 45.64 ± 11.35 | 46.40 ± 11.36 | 45.91 ± 10.68 | 46.79 ± 11.18 |
| SLE cases (%) | 58.5 | 58.1 | 70.8 | 56.3 | 71.3 | 51.9 |
| Controls (%) | 41.5 | 41.9 | 29.2 | 43.7 | 28.7 | 48.1 |
*ACL: anticardiolipin antibodies; LAC: lupus anticoagulant; Anti-β 2GPI: anti-β 2 glycoprotein I antibodies.
2.2. Antiphospholipid Antibodies
The presence of ACL (IgG > 15 GPL units, IgM > 10 MPL units, IncStar, Stillwater, MN, USA), LAC (partial thromboplastin time or Russell's viper venon time with mix) and anti-β 2GPI (QUANTA Lite β 2GPI screen, INOVA Diagnostics, Inc. San Diego, CA, USA) was tested in sera or plasma obtained from the study subjects. The three APA (ACL, LAC, and anti-β 2GPI) were classified into antibody-positive and antibody-negative groups based on manufacturer's protocols.
2.3. Genotyping and Quality Control (QC)
DNA samples were genotyped using the Affymetrix Genome-Wide Human SNP Array 6.0 containing 906,600 SNPs at Expression Analysis, Durham, NC, USA. All samples used in this study passed strict quality control measurements in our larger GWAS. Exclusion criteria included samples with poor performance (<95% average call rate across the array), poorly performing markers (44,592 with <95% call rate across all samples genotyped), and markers with significant deviation from Hardy-Weinberg equilibrium (P ≤ 1E − 06) and with low minor allele frequency (MAF <0.01). Population stratification analysis was conducted using a multidimensional scaling method implemented in PLINK. SNPs falling within the genomic regions with abnormal linkage disequilibrium patterns and structural variations (hg18; chr2: 130–140 Mb, chr6: 24–36 Mb, chr8: 8–12 Mb, chr11: 42–58 Mb, and chr17: 40–43 Mb) were excluded from the principal component (PC) analysis but were included in subsequent association analysis. First 4 components were determined to be relevant for the determination of population origin based on visual examination of PC plots and were used as covariates in the association statistics.
2.4. Association Analysis
The three APA (ACL, LAC, and anti-β 2GPI) were classified into antibody-positive and antibody-negative groups based on manufacturer's protocols. Association of SNPs with the antibody status was tested using logistic regression under the additive model. Considering the effect of SNPs on the antibody status may be confounded by the disease status (SLE) and other demographic variables (age, BMI, smoking), we used the stepwise regression method to select the most parsimonious set of covariates for each dependent variable. The analysis for each antibody was adjusted for the disease status (SLE) and the first four principal components. In addition, the ACL and LAC analyses were adjusted for smoking and BMI, respectively. R and/or PLINK statistical software programs were used for all analyses performed for this study.
3. Results
3.1. Quantile-Quantile Plots of the GWAS Data
The genome-wide association analysis was performed on 670 individuals with ACL, 708 individuals with LAC and 496 individuals with anti-β 2GPI (Table 1) who were genotyped using the Affymetrix Genome-Wide Human SNP Array 6.0. Figure 1 shows the quantile-quantile plots for comparisons of observed and expected P values distribution for ACL, LAC, and anti-β 2GPI. For all three APA, the distribution of observed P values conformed to the null distribution until the tail of the distribution where it deviated, indicating no evidence of significant population stratification but evidence of genetic association.
Figure 1.
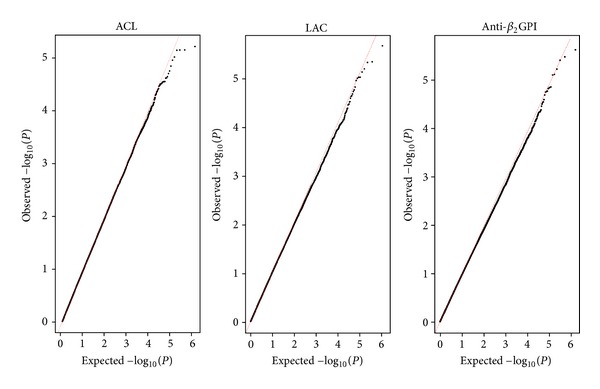
Quantile-quantile plots of the observed versus the expected P values for ACL, LAC, and Anti-β 2GPI.
3.2. Association with Anticardiolipin Antibodies (ACL)
Figure 2 shows the genome-wide P values for ACL in a Manhattan plot and the top loci with P < 1E − 04 are presented in Table 2. Three top SNPs with P < 1E − 05 were observed. The most significant SNP, rs6889746 (P = 6.02E − 06), was located upstream of PELO (Pelota homolog) on chromosome 5q11.2. The next top SNP, rs6681460 (P = 6.98E − 06), was present in SGIP1 (SH3-domain GRB2-like-intercation protein1) on chromosome 1p31.3. There was a total of 28 SNPs in this region with P < 1E − 03. The next top SNP, rs12204683 (P = 7.02E − 06), resided downstream of LCA5 on chromosome 6q14.1.
Figure 2.
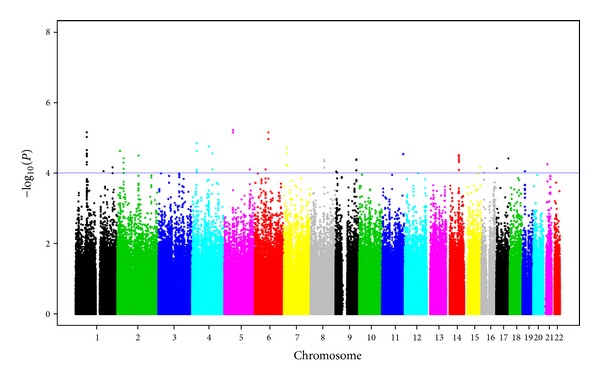
Manhattan plot showing the genome-wide association P values with anticardiolipin antibodies (ACL). Blue line indicates P = 1E − 04.
Table 2.
Genetic loci associated with the occurrence of ACL with P < 1E − 04*.
| CHR | Gene | Lead SNP | BP | Total SNPs | MAF | OR | P | |
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | |||||||
| 5 | PELO | rs6889746 | 51742663 | 3 | 0.3534 | 0.5 | 1.776 | 6.02E − 06 |
| 1 | SGIP1 | rs6681460 | 66895645 | 28 | 0.3755 | 0.5168 | 1.827 | 6.98E − 06 |
| 6 | LCA5 | rs12204683 | 80212978 | 4 | 0.2378 | 0.3701 | 1.877 | 7.02E − 06 |
| 4 | MIR4275 | rs17642174 | 28160435 | 6 | 0.1355 | 0.2346 | 2.021 | 1.42E − 05 |
| 4 | C4orf37 | rs13134014 | 99323902 | 21 | 0.1517 | 0.2542 | 1.928 | 1.76E − 05 |
| 7 | BZW2 | rs6961256 | 16697717 | 3 | 0.01261 | 0.05587 | 5.214 | 1.96E − 05 |
| 2 | FAM49A | rs6753768 | 16483918 | 11 | 0.2836 | 0.1648 | 0.5016 | 2.37E − 05 |
| 4 | MAD2L1 | rs10518344 | 1.21E + 08 | 3 | 0.05136 | 0.1178 | 2.696 | 2.77E − 05 |
| 11 | KIRREL3 | rs1793667 | 1.26E + 08 | 1 | 0.2789 | 0.3966 | 1.781 | 2.89E − 05 |
| 14 | YLPM1 | rs2241275 | 74321193 | 7 | 0.435 | 0.5587 | 1.754 | 3.06E − 05 |
| 14 | PROX2 | rs4899536 | 74386367 | 2 | 0.4331 | 0.5559 | 1.752 | 3.28E − 05 |
| 2 | ATL2 | rs6749177 | 38531205 | 11 | 0.4958 | 0.3659 | 0.576 | 3.77E − 05 |
| 17 | PRPSAP1 | rs11077813 | 71848140 | 1 | 0.2704 | 0.1564 | 0.5072 | 3.82E − 05 |
| 9 | TTLL11 | rs10985483 | 1.24E + 08 | 4 | 0.3703 | 0.25 | 0.5571 | 4.09E − 05 |
| 8 | ZBTB10 | rs406629 | 81620942 | 6 | 0.3795 | 0.2599 | 0.5501 | 4.26E − 05 |
| 21 | LINC00317 | rs2827107 | 22175926 | 3 | 0.2128 | 0.3156 | 1.847 | 5.63E − 05 |
| 7 | NUPL2 | rs10232205 | 23197079 | 1 | 0.09244 | 0.02793 | 0.2465 | 6.22E − 05 |
| 1 | DUSP10 | rs11118750 | 2.2E + 08 | 4 | 0.2174 | 0.3287 | 1.753 | 6.88E − 05 |
| 15 | MCTP2 | rs1863095 | 92917093 | 1 | 0.3224 | 0.4511 | 1.665 | 6.90E − 05 |
| 17 | SHPK | rs222790 | 3478239 | 1 | 0.2051 | 0.3097 | 1.793 | 7.30E − 05 |
| 6 | KHDRBS2 | rs2752976 | 63486388 | 11 | 0.431 | 0.5447 | 1.688 | 7.88E − 05 |
| 5 | GLRA1 | rs154111 | 1.51E + 08 | 14 | 0.4569 | 0.3399 | 0.5875 | 7.91E − 05 |
| 9 | MIR548AA1 | rs4836873 | 1.24E + 08 | 4 | 0.3651 | 0.2514 | 0.5746 | 8.29E − 05 |
| 1 | LOC100505918 | rs16860501 | 1.67E + 08 | 1 | 0.09119 | 0.1648 | 2.116 | 8.89E − 05 |
| 19 | OR7A10 | rs4808564 | 14825095 | 1 | 0.05126 | 0.1117 | 2.449 | 8.91E − 05 |
| 9 | C9orf46 | rs4742085 | 5340548 | 2 | 0.3501 | 0.4678 | 1.696 | 9.02E − 05 |
| 9 | PTPRD | rs2484741 | 10477877 | 7 | 0.2977 | 0.1983 | 0.5439 | 9.79E − 05 |
| 16 | PDXDC1 | rs3198697 | 15037441 | 1 | 0.3658 | 0.4859 | 1.646 | 9.83E − 05 |
*CHR: chromosome; Gene: a plausible biological candidate gene in the locus or the nearest annotated gene to the lead SNP; Lead SNP: most significant SNP in the gene region; BP: base-pair position of the lead SNP; Total SNPs: total number of SNPs with P < 1E − 03 in the gene region; MAF: minor allele frequencies in antibody-negative and antibody-positive groups; OR: odds ratio; P: P-values for the test.
3.3. Association with Lupus Anticoagulant (LAC)
The Manhattan plot for LAC is shown in Figure 3 and the top hits with P < 1E − 04 are given in Table 3. The most significant SNP, rs1978968, was observed in MICAL3 on chromosome 22q11.21 (P = 2.21E − 06) and there were additional 7 significant SNPs in this region with P < 1E − 03. The next significant SNP was observed on chromosome 2p12 in FAM176A (rs17011455, P = 4.70E − 06). However, no other SNP with P < 1E − 03 was observed in this region. The third significant SNP, rs17791782, was observed in DSTN on chromosome 20p12.1 (P = 6.54E − 06).
Figure 3.
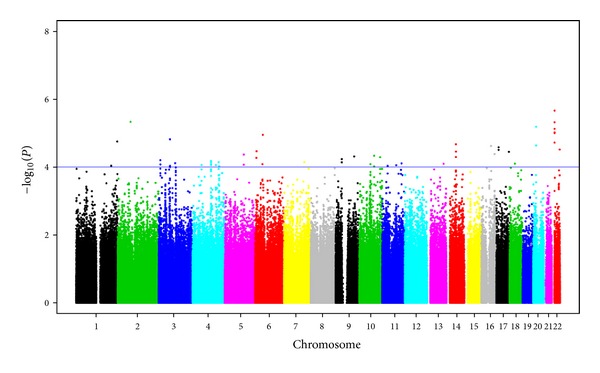
Manhattan plot showing the genome-wide association P values with lupus anticoagulant (LAC). Blue line indicates P = 1E − 04.
Table 3.
Genetic loci associated with the occurrence of lupus anticoagulant (LAC) with P < 1E − 04*.
| CHR | Gene | Lead SNP | BP | Total SNPs | MAF | OR | P | |
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | |||||||
| 22 | MICAL3 | rs1978968 | 16828113 | 8 | 0.2144 | 0.3553 | 2.235 | 2.21E − 06 |
| 2 | FAM176A | rs17011455 | 75643997 | 1 | 0.01661 | 0.06967 | 5.211 | 4.70E − 06 |
| 20 | DSTN | rs17791782 | 17514069 | 2 | 0.07867 | 0.1721 | 2.628 | 6.54E − 06 |
| 6 | SUPT3H | rs9472374 | 44904278 | 1 | 0.01957 | 0.07083 | 4.77 | 1.14E − 05 |
| 3 | LRIG1 | rs4549225 | 66850149 | 10 | 0.3735 | 0.5246 | 1.867 | 1.54E − 05 |
| 1 | SMYD3 | rs7527610 | 2.45E + 08 | 2 | 0.005236 | 0.04098 | 10.19 | 1.77E − 05 |
| 14 | PELI2 | rs754314 | 55822350 | 8 | 0.04833 | 0.1167 | 3.111 | 2.13E − 05 |
| 20 | BFSP1 | rs16999416 | 17489830 | 1 | 0.08494 | 0.1777 | 2.412 | 2.32E − 05 |
| 16 | NDRG4 | rs11862356 | 57067820 | 1 | 0.05846 | 0.1393 | 2.745 | 2.40E − 05 |
| 17 | CDRT15P1 | rs7208809 | 13678048 | 2 | 0.06806 | 0.1516 | 2.573 | 2.62E − 05 |
| 22 | FAM19A5 | rs9615320 | 47219591 | 2 | 0.1065 | 0.2008 | 2.333 | 3.07E − 05 |
| 6 | SNRNP48 | rs17398435 | 7549105 | 1 | 0.06392 | 0.1446 | 2.589 | 3.45E − 05 |
| 14 | OTX2 | rs12897597 | 56314078 | 1 | 0.1531 | 0.2667 | 2.097 | 3.53E − 05 |
| 17 | RBFOX3 | rs16972153 | 74719776 | 1 | 0.04974 | 0.1148 | 2.925 | 3.59E − 05 |
| 16 | MAF | rs9935211 | 78440577 | 5 | 0.05467 | 0.1311 | 2.692 | 4.18E − 05 |
| 5 | YTHDC2 | rs6865651 | 1.13E + 08 | 2 | 0.1848 | 0.3058 | 2.005 | 4.32E − 05 |
| 10 | LDB3 | rs4934256 | 88490345 | 2 | 0.05507 | 0.123 | 2.783 | 4.68E − 05 |
| 9 | KLF4 | rs1888617 | 1.1E + 08 | 1 | 0.2657 | 0.4009 | 1.935 | 4.92E − 05 |
| 10 | TACC2 | rs12773310 | 1.24E + 08 | 6 | 0.3536 | 0.219 | 0.4945 | 5.19E − 05 |
| 6 | LY86 | rs9328374 | 6536628 | 1 | 0.1875 | 0.307 | 1.989 | 5.40E − 05 |
| 9 | ZCCHC7 | rs7031314 | 37366122 | 3 | 0.2204 | 0.3375 | 1.924 | 5.90E − 05 |
| 3 | SETD5 | rs17050346 | 9456593 | 5 | 0.01926 | 0.06967 | 4.047 | 6.34E − 05 |
| 4 | COL25A1 | rs13104799 | 1.1E + 08 | 6 | 0.2245 | 0.1107 | 0.4106 | 6.68E − 05 |
| 7 | C7orf58 | rs12537243 | 1.2E + 08 | 1 | 0.09178 | 0.1736 | 2.289 | 7.22E − 05 |
| 4 | RBM46 | rs7687314 | 1.56E + 08 | 6 | 0.4474 | 0.5902 | 1.808 | 7.25E − 05 |
| 9 | LOC100506710 | rs10973184 | 37056617 | 1 | 0.09895 | 0.1885 | 2.252 | 7.33E − 05 |
| 3 | EPHA6 | rs4318565 | 97564200 | 1 | 0.03369 | 0.08607 | 3.32 | 7.77E − 05 |
| 11 | LOC283143 | rs1393275 | 1.15E + 08 | 3 | 0.05026 | 0.1261 | 2.595 | 7.90E − 05 |
| 18 | MAPRE2 | rs573269 | 30863716 | 1 | 0.3129 | 0.4385 | 1.816 | 8.05E − 05 |
| 13 | FARP1 | rs285031 | 97584032 | 1 | 0.1489 | 0.2438 | 2.092 | 8.11E − 05 |
| 10 | ANXA2P3 | rs10822492 | 66751936 | 15 | 0.3439 | 0.219 | 0.5075 | 8.32E − 05 |
| 6 | TBCC | rs11759402 | 42831787 | 4 | 0.1658 | 0.2686 | 1.945 | 8.32E − 05 |
| 4 | LNX1 | rs6831173 | 54085908 | 4 | 0.1337 | 0.2377 | 1.962 | 8.80E − 05 |
| 4 | SLC7A11 | rs10440463 | 1.39E + 08 | 3 | 0.2073 | 0.3238 | 1.88 | 8.85E − 05 |
| 1 | HHAT | rs1028383 | 2.09E + 08 | 1 | 0.2248 | 0.3475 | 1.882 | 9.29E − 05 |
| 11 | WT1 | rs2207549 | 32325033 | 2 | 0.4202 | 0.2833 | 0.5274 | 9.37E − 05 |
| 4 | FAM198B | rs17036867 | 1.59E + 08 | 12 | 0.02747 | 0.08621 | 3.54 | 9.54E − 05 |
*CHR: chromosome; Gene: a plausible biological candidate gene in the locus or the nearest annotated gene to the lead SNP; Lead SNP: most significant SNP in the gene region; BP: base-pair position of the lead SNP; Total SNPs: total number of SNPs with P < 1E − 03 in the gene region; MAF: minor allele frequencies in antibody-negative and antibody-positive groups; OR: odds ratio; P: P-values for the test.
3.4. Association with Anti-β 2 Glycoprotein I Antibodies (Anti-β 2GPI)
Five loci on four chromosomes were observed at P < 1E − 05 for association with anti-β 2GPI (Figure 4, Table 4). The top SNP (rs10492418) at P = 2.05E − 06 was observed on chromosome 13q33.3 in MYO16. This chromosome also harbors another locus for anti-β 2GPI at 13q14.11 (rs9315762, P = 6.68E − 06), near a region expressing long intergenic nonprotein coding RNAs. The second most significant SNP, rs11975235, was observed in PDE1C on chromosome 7p14.3 (P = 2.88E − 06). The third most significant SNP was observed upstream of TANK on chromosome 2q24.2 (rs2357982, P = 3.38E − 06) that also harbored 12 additional significant SNPs with P < 1E − 03.
Figure 4.
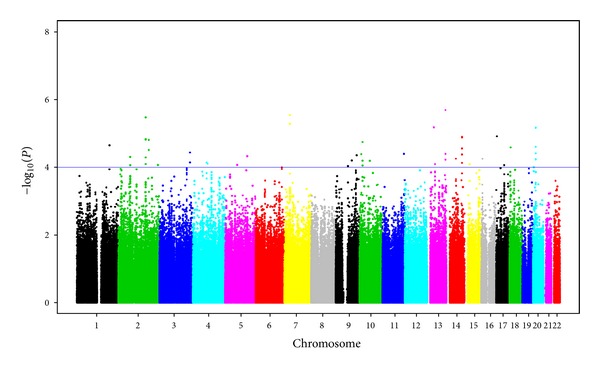
Manhattan plot showing the genome-wide association P values with anti-β 2 glycoprotein I antibodies (Anti-β 2GPI). Blue line indicates P = 1E − 04.
Table 4.
Genetic loci associated with the occurrence of anti-β 2GPI antibodies with P < 1E − 04*.
| CHR | Gene | Lead SNP | BP | Total SNPs | MAF | OR | P | |
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | |||||||
| 13 | MYO16 | rs10492418 | 108178727 | 5 | 0.4292 | 0.6016 | 2.172 | 2.05E − 06 |
| 7 | PDE1C | rs11975235 | 32156065 | 4 | 0.4843 | 0.311 | 0.4675 | 2.88E − 06 |
| 2 | TANK | rs2357982 | 161594349 | 13 | 0.2359 | 0.3885 | 2.189 | 3.38E − 06 |
| 13 | FLJ42392 | rs9315762 | 39639907 | 2 | 0.1535 | 0.2901 | 2.255 | 6.68E − 06 |
| 20 | MACROD2 | rs6080100 | 15951406 | 7 | 0.2655 | 0.4275 | 2.086 | 6.86E − 06 |
| 17 | CAMKK1 | rs758642 | 3733656 | 1 | 0.3314 | 0.4837 | 2.038 | 1.23E − 05 |
| 14 | ITPK1 | rs8021497 | 92637371 | 6 | 0.2059 | 0.3508 | 2.101 | 1.26E − 05 |
| 2 | SESTD1 | rs10186547 | 179921914 | 2 | 0.3853 | 0.5391 | 2.009 | 1.56E − 05 |
| 10 | CACNB2 | rs12356676 | 18687510 | 6 | 0.2587 | 0.3943 | 2.134 | 1.80E − 05 |
| 18 | LRRC30 | rs9965173 | 7201755 | 4 | 0.07887 | 0.1718 | 2.647 | 2.59E − 05 |
| 3 | PEX5L | rs9856007 | 181225431 | 3 | 0.2514 | 0.3817 | 1.976 | 3.68E − 05 |
| 11 | TMEM45B | rs10894119 | 129081436 | 1 | 0.1835 | 0.293 | 2.168 | 4.05E − 05 |
| 10 | SFTA1P | rs1000039 | 10597879 | 1 | 0.02254 | 0.08779 | 4.19 | 4.13E − 05 |
| 9 | OR1J1 | rs2778636 | 124270913 | 2 | 0.2641 | 0.3931 | 1.955 | 4.43E − 05 |
| 5 | RAPGEF6 | rs17671387 | 130911895 | 1 | 0.04507 | 0.1183 | 3.186 | 4.71E − 05 |
| 2 | GMCL1 | rs4241261 | 69917164 | 3 | 0.4761 | 0.3308 | 0.5278 | 4.93E − 05 |
| 14 | C14orf101 | rs7153196 | 56256660 | 2 | 0.362 | 0.229 | 0.4996 | 5.58E − 05 |
| 9 | WNK2 | rs10821084 | 94991443 | 1 | 0.07627 | 0.1718 | 2.425 | 6.33E − 05 |
| 10 | SLC16A9 | rs7082987 | 61007793 | 1 | 0.3663 | 0.2317 | 0.4886 | 6.46E − 05 |
| 4 | GDEP | rs11730315 | 81020089 | 2 | 0.1648 | 0.2829 | 2.075 | 7.24E − 05 |
| 4 | ARHGAP24 | rs17010960 | 86938938 | 2 | 0.02254 | 0.07634 | 4.326 | 7.85E − 05 |
| 13 | HTR2A | rs582385 | 46343995 | 4 | 0.1624 | 0.2824 | 2.02 | 8.09E − 05 |
| 15 | FMN1 | rs2444955 | 31199305 | 2 | 0.2211 | 0.355 | 1.974 | 8.10E − 05 |
| 5 | CARTPT | rs16869487 | 70976850 | 1 | 0.02841 | 0.09542 | 3.602 | 8.59E − 05 |
| 2 | NEU2 | rs11695991 | 233608333 | 1 | 0.0169 | 0.0687 | 4.802 | 8.63E − 05 |
| 17 | CA10 | rs203076 | 47354484 | 7 | 0.2944 | 0.1603 | 0.4729 | 8.70E − 05 |
| 9 | C9orf135 | rs1389124 | 71669176 | 3 | 0.09943 | 0.1985 | 2.286 | 9.23E − 05 |
| 20 | SMOX | rs1764996 | 4070805 | 2 | 0.02817 | 0.07634 | 4.014 | 9.93E − 05 |
*CHR: chromosome; Gene: a plausible biological candidate gene in the locus or the nearest annotated gene to the lead SNP; Lead SNP: most significant SNP in the gene region; BP: base-pair position of the lead SNP; Total SNPs: total number of SNPs with P < 1E − 03 in the gene region; MAF: minor allele frequencies in antibody-negative and antibody-positive groups; OR: odds ratio; P: P-values for the test.
3.5. Association with Presence of Two or More Antibodies
In addition to the single-antibody analyses described above, we also performed an association analysis between individuals who were positive for two or more antibodies (n = 100) versus individuals who were negative for all three antibodies (n = 227). Table 5 shows the results of top loci with P < 1E − 04. Interestingly, five of these loci (SESTD1, CACNB2, TANK, TMEM45B, and FMN1) overlapped with those observed in the anti-β 2GPI analysis (see Table 4) and two (DSTN and BFSP1) overlapped with those observed in the LAC analysis (see Table 3). Although the most significant locus, DYNLRB2 (P = 1.44E − 06), was not among the top loci detected in any of the single-antibody analyses, the second most significant locus, SESTD1 (P = 6.08E − 06), also showed association with anti-β 2GPI.
Table 5.
Genetic loci associated with the occurrence of two or more antiphospholipid antibodies (ACL, LAC, or Anti-β 2GPI) with P < 1E − 04*.
| CHR | Gene | Lead SNP | BP | Total SNPs | MAF | OR | P | |
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | |||||||
| 16 | DYNLRB2 | rs8060581 | 78750106 | 6 | 0.02466 | 0.1406 | 6.714 | 1.44E − 06 |
| 2 | SESTD1 | rs13403289 | 179924976 | 2 | 0.3857 | 0.5833 | 2.423 | 6.08E − 06 |
| 1 | DNAH14 | rs3913653 | 223603694 | 11 | 0.543 | 0.3421 | 0.43 | 1.09E − 05 |
| 10 | CACNB2 | rs10828616 | 18710023 | 6 | 0.2175 | 0.3698 | 2.538 | 1.11E − 05 |
| 18 | EPB41L3 | rs7238186 | 5469093 | 2 | 0.213 | 0.3854 | 2.327 | 2.61E − 05 |
| 1 | MAGI3 | rs11102625 | 113750976 | 5 | 0.4355 | 0.6146 | 2.298 | 2.75E − 05 |
| 2 | TANK | rs13010671 | 161593338 | 5 | 0.07442 | 0.1882 | 3.437 | 3.17E − 05 |
| 7 | CNTNAP2 | rs12113442 | 145329124 | 1 | 0.1054 | 0.2188 | 2.938 | 3.43E − 05 |
| 18 | ZNF519 | rs8093228 | 13989380 | 4 | 0.2152 | 0.3646 | 2.326 | 4.85E − 05 |
| 3 | FAM198A | rs7624799 | 43020807 | 3 | 0.1592 | 0.2969 | 2.466 | 5.00E − 05 |
| 20 | DSTN | rs17791782 | 17514069 | 2 | 0.07883 | 0.1927 | 3.086 | 5.06E − 05 |
| 20 | BFSP1 | rs16999416 | 17489830 | 1 | 0.08371 | 0.2031 | 2.93 | 5.63E − 05 |
| 13 | ANKRD20A9P | rs7319595 | 18392986 | 1 | 0.3484 | 0.5213 | 2.337 | 5.75E − 05 |
| 15 | TLE3 | rs10518889 | 68337482 | 4 | 0.2838 | 0.4427 | 2.231 | 6.17E − 05 |
| 22 | CLTCL1 | rs8135222 | 17672446 | 1 | 0.1749 | 0.3281 | 2.293 | 6.19E − 05 |
| 11 | TMEM45B | rs10894119 | 129081436 | 1 | 0.1789 | 0.3158 | 2.432 | 6.25E − 05 |
| 1 | GADD45A | rs787480 | 67868460 | 1 | 0.2207 | 0.08854 | 0.3088 | 6.57E − 05 |
| 20 | SPTLC3 | rs6105044 | 13065747 | 2 | 0.352 | 0.1979 | 0.4138 | 7.08E − 05 |
| 6 | HIVEP1 | rs6908010 | 12325985 | 2 | 0.4753 | 0.3073 | 0.4688 | 7.11E − 05 |
| 7 | CUX1 | rs427534 | 101673424 | 3 | 0.4439 | 0.2656 | 0.4569 | 7.28E − 05 |
| 10 | MSMB | rs7094791 | 51229942 | 1 | 0.3857 | 0.2396 | 0.4081 | 7.35E − 05 |
| 8 | TPD52 | rs10090469 | 81396522 | 1 | 0.1099 | 0.2292 | 2.668 | 7.43E − 05 |
| 14 | LOC100506433 | rs698322 | 47597316 | 1 | 0.3914 | 0.2344 | 0.4277 | 7.58E − 05 |
| 6 | MMS22L | rs1206164 | 97703068 | 1 | 0.2511 | 0.4219 | 2.212 | 8.26E − 05 |
| 12 | CDK17 | rs11108526 | 95379901 | 2 | 0.02691 | 0.08333 | 5.95 | 8.40E − 05 |
| 11 | PDGFD | rs4754095 | 103278377 | 3 | 0.2851 | 0.1436 | 0.3771 | 8.83E − 05 |
| 15 | FMN1 | rs2444955 | 31199305 | 2 | 0.1951 | 0.3474 | 2.408 | 8.89E − 05 |
| 10 | SORCS1 | rs4918273 | 108715485 | 11 | 0.3597 | 0.5319 | 2.064 | 8.92E − 05 |
*CHR: chromosome; Gene: a plausible biological candidate gene in the locus or the nearest annotated gene to the lead SNP; Lead SNP: most significant SNP in the gene region; BP: base-pair position of the lead SNP; Total SNPs: total number of SNPs with P < 1E − 03 in the gene region; MAF: minor allele frequencies in antibody-negative (negative for ALC, ACL and anti-β 2GPI) and antibody-positive (positive for at least two of ALC, ACL or anti-β 2GPI) groups; OR: odds ratio; P: P-values for the test.
3.6. Association of Extended Major Histocompatibility Complex (xMHC) Region and Apolipoprotein H (APOH) with APA
Previously, several studies have reported genetic association of the human leukocyte antigen (HLA) genes located at the MHC locus on chromosome 6p21 with the presence of APA [6]. Likewise, since β 2GPI is the main target antigen for APA, genetic variation in its gene, APOH, is expected to be associated with the occurrence of APA. Although no SNPs from either the HLA genes or APOH were among the top GWAS SNPs with P < 1E − 04 (Tables 2–5), the xMHC region revealed 104, 191, and 108 significant SNPs (P < 0.05) to be associated with ACL, LAC, and anti-β 2GPI, respectively. Table 6 lists significant SNPs with P < 0.01 in the MHC region for the three APA examined. Most significant SNPs were observed in or near HLA-DPB1, HLA-DPB2, HLA-DPA1, HLA-DQA1, HLA-DQA2, and HLA-DMA. Noteworthy, some SNPs were associated with more than one APA. For example, among the SNPs located upstream of HLA-DQA2, rs9275765 and rs9275772 were associated with LAC (P = 7.86E − 04) and anti-β 2GPI (P = 3.15E − 03), rs9275793 with LAC (P = 8.84E − 04) and anti-β 2GPI (P = 3.10E − 03), and rs9276298 with LAC (P = 1.33E − 03) and anti-β 2GPI (P = 5.23E − 03). Likewise, rs2395357 near HLA-DPB2 showed association with ACL (P = 4.34E − 04) and LAC (P = 1.09E − 02) and rs11539216 in HLA-DMA with ACL (P = 9.96E − 04) and LAC (P = 9.29E − 03). Of the 21 QC-passed SNPs present in or near APOH, six revealed nominal associations with anti-β 2GPI, and the Trp316Ser variant (rs1801690) was the most significant SNP (P = 3.12E − 03) (Table 7). Two additional SNPs also showed nominal associations with LAC (P = 0.026, 0.027).
Table 6.
Significant SNPs with P < 0.01 in the MHC region on chromosome 6 for ACL, LAC, and Anti-β 2GP1*.
| Gene | SNP | P |
|---|---|---|
| ACL | ||
|
| ||
| HLA-DPB1 | rs3128918 | 0.00028 |
| HLA-DPB2 | rs2395357 | 0.00043 |
| HLA-DMA | rs11539216 | 0.00099 |
| HLA-DQB2 | rs10484564 | 0.00536 |
| GNL1 | rs9295888 | 0.00758 |
| GNL1 | rs9295873 | 0.00794 |
| HLA-DOA | rs4713603 | 0.0081 |
| RPP21 | rs1548515 | 0.00842 |
| GNL1 | rs9461607 | 0.00863 |
| GNL1 | rs17411480 | 0.00863 |
| RPP21 | rs9261821 | 0.00863 |
| RPP21 | rs9261850 | 0.00863 |
| RPP21 | rs9261854 | 0.00863 |
| RPP21 | rs9261855 | 0.00863 |
| RPP21 | rs1548513 | 0.00863 |
| RPP21 | rs9261925 | 0.00863 |
| RPP21 | rs9261926 | 0.00863 |
| BRD2 | rs17840186 | 0.00939 |
| RPP21 | rs9261799 | 0.00955 |
|
| ||
| LAC | ||
|
| ||
| TAP2 | rs1044043 | 0.00029 |
| HLA-DQA1 | rs642093 | 0.00032 |
| AIF1 | rs2736177 | 0.00041 |
| HLA-DQA2 | rs9275765 | 0.00078 |
| HLA-DQA2 | rs9275772 | 0.00078 |
| HLA-DQA2 | rs9275793 | 0.00088 |
| HLA-DQA1 | rs9272346 | 0.00130 |
| HLA-DQA2 | rs9276298 | 0.00133 |
| HLA-DQA1 | rs9272219 | 0.00167 |
| C6orf10 | rs3129934 | 0.00168 |
| HLA-DQA1 | rs9272535 | 0.00179 |
| HLA-DRB1 | rs674313 | 0.00215 |
| HLA-DRB1 | rs502771 | 0.00227 |
| HLA-DRB1 | rs9270986 | 0.00250 |
| AIF1 | rs2857597 | 0.00257 |
| HCG26 | rs2516516 | 0.00282 |
| HLA-DRB1 | rs615672 | 0.00295 |
| HLA-DRB1 | rs502055 | 0.00300 |
| HLA-DQA1 | rs9272723 | 0.00329 |
| LOC100294145 | rs9276915 | 0.00352 |
| C6orf10 | rs2894254 | 0.00372 |
| UBD | rs9368606 | 0.00403 |
| C6orf10 | rs3129900 | 0.00458 |
| MCCD1 | rs2734573 | 0.00553 |
| PRRC2A | rs1046080 | 0.00618 |
| C6orf10 | rs7767325 | 0.00623 |
| C6orf15 | rs2517448 | 0.00627 |
| C6orf10 | rs3132928 | 0.00640 |
| HLA-H | rs3132722 | 0.00675 |
| HLA-DQB2 | rs2857210 | 0.00685 |
| HLA-DRA | rs3129868 | 0.00731 |
| ATP6V1G2-DDX39B/DDX39B | rs933208 | 0.00740 |
| NFKBIL1 | rs2857605 | 0.00755 |
| TRIM26 | rs3132671 | 0.00757 |
| HLA-DQB1 | rs3129716 | 0.00770 |
| MSH5/MSH5-C6orf26 | rs3131379 | 0.00849 |
| MSH5/MSH5-C6orf26 | rs3130484 | 0.00901 |
| PSMB9 | rs9276832 | 0.00907 |
| BTNL2 | rs2213581 | 0.00922 |
| HLA-DMA | rs11539216 | 0.00929 |
| ATP6V1G2-DDX39B/DDX39B | rs3093978 | 0.00957 |
| C6orf10 | rs2143461 | 0.00986 |
| TUBB | rs3095330 | 0.00990 |
| LOC100294145 | rs4959119 | 0.00992 |
| TRIM26 | rs2517611 | 0.00993 |
|
| ||
| Anti-β 2GPI | ||
|
| ||
| HLA-DPB2 | rs9277916 | 0.00147 |
| HLA-DQA2 | rs9275793 | 0.00310 |
| HLA-DQA2 | rs9275765 | 0.00315 |
| HLA-DQA2 | rs9275772 | 0.00315 |
| HLA-DPA1 | rs3130182 | 0.00331 |
| HLA-DQB1 | rs9469220 | 0.00372 |
| HLA-DPB2 | rs4711314 | 0.00378 |
| HLA-DQA2 | rs2647089 | 0.00463 |
| HLA-DQA2 | rs9276298 | 0.00523 |
| HLA-DQB1 | rs9275356 | 0.00526 |
| HLA-DQA2 | rs17615250 | 0.00710 |
| HLA-DQA2 | rs9275618 | 0.00807 |
*ACL: anticardiolipin antibodies; LAC: lupus anticoagulant; Anti-β 2GPI: anti-β 2 glycoprotein I antibodies; Gene: a plausible biological candidate gene in the locus or the nearest annotated gene to the SNP; SNP: single-nucleotide polymorphism; P: P-values for the test.
Table 7.
Odds ratios and P-values for the association analysis of APOH SNPs on chromosome 17 with ACL, LAC, and anti-β 2GPI*.
| SNP | BP | ACL | LAC | Anti-β 2GPI | |||
|---|---|---|---|---|---|---|---|
| OR | P | OR | P | OR | P | ||
| rs1801690 | 61638747 | 0.7655 | 0.4048 | 0.6444 | 0.2614 | 2.461 | 0.003122 |
| rs17769836 | 61663751 | 0.8968 | 0.4457 | 1.004 | 0.9821 | 0.5978 | 0.004849 |
| rs2873966 | 61642435 | 0.9519 | 0.7168 | 0.9858 | 0.9262 | 0.6224 | 0.005407 |
| rs7215391 | 61662484 | 0.8932 | 0.4391 | 0.984 | 0.9227 | 0.6146 | 0.008069 |
| rs8073418 | 61678134 | 0.9639 | 0.7759 | 0.8949 | 0.455 | 0.6661 | 0.01061 |
| rs8064837 | 61673165 | 0.9833 | 0.8915 | 1.011 | 0.9388 | 1.404 | 0.02235 |
| rs10491174 | 61685021 | 1.017 | 0.9335 | 0.765 | 0.2665 | 1.487 | 0.07256 |
| rs2215413 | 61679959 | 0.9418 | 0.6397 | 0.842 | 0.2435 | 0.829 | 0.221 |
| rs16958979 | 61654321 | 0.8586 | 0.5569 | 0.4332 | 0.02739 | 1.375 | 0.2253 |
| rs8178822 | 61655991 | 0.8748 | 0.6127 | 0.407 | 0.0258 | 1.379 | 0.2345 |
| rs12452959 | 61635526 | 0.8991 | 0.5748 | 1.196 | 0.3965 | 1.275 | 0.2541 |
| rs4791079 | 61640002 | 1.128 | 0.3573 | 1.206 | 0.2028 | 1.181 | 0.2802 |
| rs3176975 | 61641219 | 0.9723 | 0.8552 | 0.9923 | 0.9649 | 1.209 | 0.2858 |
| rs8066294 | 61673500 | 1.094 | 0.5597 | 1.014 | 0.9365 | 1.162 | 0.3981 |
| rs17763430 | 61635203 | 0.998 | 0.9904 | 0.8987 | 0.5811 | 0.8448 | 0.4051 |
| rs17690171 | 61633319 | 0.9893 | 0.9435 | 1.172 | 0.3479 | 1.141 | 0.4528 |
| rs16959003 | 61671199 | 1.105 | 0.6095 | 0.8378 | 0.4548 | 1.14 | 0.5639 |
| rs735866 | 61670208 | 1.206 | 0.3459 | 0.8419 | 0.4765 | 1.117 | 0.642 |
| rs7208089 | 61689374 | 1.091 | 0.5465 | 0.7956 | 0.1906 | 1.08 | 0.6476 |
| rs7222710 | 61630703 | 1.089 | 0.5011 | 1.016 | 0.9143 | 0.942 | 0.6896 |
| rs6933 | 61638692 | 1.126 | 0.3408 | 1.068 | 0.6431 | 1.02 | 0.8948 |
*APOH: apolipoprotein H; SNP: single-nucleotide polymorphism; BP: base-pair position; OR: odds ratio; P: P-values. ACL: anticardiolipin antibodies; LAC: lupus anticoagulant; Anti-β 2GPI: anti-β 2 glycoprotein I antibodies.
4. Discussion
The persistent presence of APA, such as ACL, LAC, or anti-β 2GPI, may lead to the development of antiphospholipid syndrome (APS), which may occur alone (primary APS) or in the presence of an autoimmune disease (secondary APS). Although the genetic basis of APA and APS has been suggested [4, 5], the precise identity of the causative genes is largely unknown. Here we report the first GWAS focused on identifying the susceptibility loci/genes for the occurrence of three main APA, namely, ACL, LAC, and anti-β 2GPI.
Initially, we performed separate genome-wide analyses for the three APA because the antigen specificity of APA is highly heterogeneous and each APA may have different genetic determinants. This seems to be confirmed in our GWAS results where none of the top loci for the three APA overlapped (see Tables 2–4). However, a single-antibody analysis may include individuals who are positive for more than one antibody in the antibody-positive group or may include individuals in the antibody-negative group who are positive for another antibody, which might have an effect on the genetic association outcome. In order to address this potential problem, we performed an additional genome-wide analysis on individuals who were positive for two or more APA as they presumably would have a higher genetic load of APA susceptibility genes and compared them with those who were negative for all three APA tested. Noteworthy, seven of the top loci observed in the latter analysis overlapped with the top loci observed in the individual analyses of anti-β 2GPI and LAC (see Table 5). Although none of the observed top loci in any analysis met the strict criteria for genome-wide level of significance (P < 5E − 08), we have identified a number of suggestive genomic regions with P < E − 05 that are worthy of follow-up studies in independent samples. They include loci harboring DYNLRB2 (P = 1.44E − 06) and SESTD1 (P = 6.08E − 06) for individuals positive for at least two APA; PELO (P = 6.02E − 06), SGIP1 (P = 6.98E − 06), and LCA5 (P = 7.02E − 06) for ACL; MICAL3 (P = 2.21E − 06), FAM176A (P = 4.70E − 06), and DSTN (P = 6.54E − 06) for LAC; and MYO16 (P = 2.05E − 06), PDE1C (P = 2.88E − 06), TANK (P = 3.38E − 06), FLJ42392 (P = 6.68E − 06), and MACROD2 (P = 6.86E − 06) for anti-β 2GPI.
While many of these loci are of unknown function in antibody production, some of them harbor candidate genes known to be involved in immune response and thus may be relevant to the production of APA. For example, DYNLRB2 is involved in immune signaling and genetic variation in this gene is associated with tuberculosis susceptibility [7]. SESTD1 binds several phospholipid species [8] and may thus serve as an autoantigen for APA. TANK (TRAF family member-associated NFKB activator) is believed to be important in type 1 interferon production [9] and has been suggested to play a role in hepatitis B and C infections [10, 11]. The MYO16 (myosin XVI) locus has recently been implicated in diabetic nephropathy [12–14]. Interestingly, the presence of APA or APS is a strong risk factor for nephropathy [15–17] and one study has suggested that anti-β 2GPI may be protective against lupus nephritis and renal damage [18]. FAM176A (a.k.a TMEM166) has been implicated in autophagy and apoptosis [19], two mechanisms with suggested roles in autoimmunity [20, 21].
Before the GWAS era, the focus of genetic studies on APA was mainly on candidate genes, with a major emphasis on HLA genes located at the MHC locus and to some extent on APOH. Since none of our top hits included SNPs from either the HLA genes or APOH, we examined the extent of association signals in these genomic regions. Indeed, we found a number of promising significant SNPs near or in various HLA genes to be associated with ACL, LAC, and anti-β 2GPI (see Table 6). Our findings are consistent with previous reports that also found multiple associations of HLA genes with these autoantibodies [6]. Previous findings regarding the association of APOH coding SNPs with APA have been inconsistent because of the conflicting reports [6, 22]. In our sample, we found six APOH SNPs to be associated with anti-β 2GPI and the most significant SNP was rs1801690 (Trp316Ser) (see Table 7) that is located in the 5th domain of β 2GPI affecting the phospholipid-binding site [23]. Another coding SNP in APOH, rs3176975 (Val247Leu), that has been reported to be associated with APS [22], showed only a modest trend for association in our sample (odds ratio 1.21; P = 0.286). The replication of previously reported HLA and APOH findings with similar association signals serve as positive controls for our GWAS. On the other hand, it also indicates that HLA and APOH are not among the top loci for APA and thus our focus should be on the identification and characterization of other genes that are more relevant to the production of APA.
In conclusion, to the best of our knowledge, this is the first GWAS that has attempted to delineate the genetic basis of three main APA, namely, ACL, LAC, and anti-β 2GPI. Although we did not identify loci meeting the conservative threshold of genome-wide significance, we have identified a number of suggestive novel loci for APA that will stimulate follow-up studies in independent and larger sample sets to replicate our findings. The main limitations of our study include relatively small sample size and lack of a replication sample; however, our top SNPs provide a select group of suggestive candidate loci/genes that can easily be tested for replication by other research groups, which would also enable a subsequent meta-analysis with increased power.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
This study was supported by the US National Institutes of Health, Grants HL092397, HL088648, AR057028, AR046588, AR057338, HD066139, AR02318, AR30492, AR48098, AR30692, and RR025741, and by a Grant from the Lupus Foundation of America.
References
- 1.Gharavi AE, Wilson WA, Wallace DJ. Antphospholipid antibodies. In: Hahn BH, editor. Dubois'Lupus Erythematosus. 5th. Baltimore, Md, USA: Williams & Wilkins; 1997. pp. 471–491. [Google Scholar]
- 2.Roubey RAS. Update on antiphospholipid antibodies. Current Opinion in Rheumatology. 2000;12(5):374–378. doi: 10.1097/00002281-200009000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Pierangeli SS, De Groot PG, Dlott J, et al. “Criteria” aPL tests: report of a task force and preconference workshop at the 13th International Congress on Antiphospholipid Antibodies, Galveston, Texas, April 2010. Lupus. 2011;20(2):182–190. doi: 10.1177/0961203310395055. [DOI] [PubMed] [Google Scholar]
- 4.Mackworth-Young C, Chan J, Harris N. High incidence of anticardiolipin antibodies in relatives of patients with systemic lupus erythematosus. Journal of Rheumatology. 1987;14(4):723–726. [PubMed] [Google Scholar]
- 5.Matthey F, Walshe K, Mackie IJ, Machin SJ. Familial occurrence of the antiphospholipid syndrome. Journal of Clinical Pathology. 1989;42(5):495–497. doi: 10.1136/jcp.42.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horita T, Merrill JT. Genetics of antiphospholipid syndrome. Current rheumatology reports. 2004;6(6):458–462. doi: 10.1007/s11926-004-0025-0. [DOI] [PubMed] [Google Scholar]
- 7.Png E, Alisjahbana B, Sahiratmadja E, et al. A genome wide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC Medical Genetics. 2012;13:p. 5. doi: 10.1186/1471-2350-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miehe S, Bieberstein A, Arnould I, Ihdene O, Rütten H, Strübing C. The phospholipid-binding protein SESTD1 is a novel regulator of the transient receptor potential channels TRPC4 and TRPC5. Journal of Biological Chemistry. 2010;285(16):12426–12434. doi: 10.1074/jbc.M109.068304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shitao L, Lingyan W, Michael B, et al. Mapping of dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song QL, He XX, Yang H, et al. Association of TANK gene polymorphism with outcomes of hepatitis B virus infection in a Chinese Han population. Viral Immunology. 2012;25:73–78. doi: 10.1089/vim.2011.0053. [DOI] [PubMed] [Google Scholar]
- 11.Mosbruger TL, Duggal P, Goedert JJ, et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. Journal of Infectious Diseases. 2010;201(9):1371–1380. doi: 10.1086/651606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nature Genetics. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 13.Maeda S, Araki SI, Babazono T, et al. Replication study for the association between four loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. Diabetes. 2010;59(8):2075–2079. doi: 10.2337/db10-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezzolesi MG, Poznik GD, Skupien J, et al. An intergenic region on chromosome 13q33.3 is associated with the susceptibility to kidney disease in type 1 and 2 diabetes. Kidney International. 2011;80(1):105–111. doi: 10.1038/ki.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gigante A, Gasperini ML, Cianci R, et al. Antiphospholipid antibodies and renal involvement. American Journal of Nephrology. 2009;30(5):405–412. doi: 10.1159/000235941. [DOI] [PubMed] [Google Scholar]
- 16.Tektonidou MG. Renal involvement in the antiphospholipid syndrome (APS) - APS nephropathy. Clinical Reviews in Allergy and Immunology. 2009;36(2-3):131–140. doi: 10.1007/s12016-008-8112-z. [DOI] [PubMed] [Google Scholar]
- 17.Silvarino R, Sant F, Espinosa G, et al. Nephropathy associated with antiphospholipid antibodies in patients with systemic lupus erythematosus. Lupus. 2011;20:721–729. doi: 10.1177/0961203310397410. [DOI] [PubMed] [Google Scholar]
- 18.Mehrani T, Petri M. IgM anti-β 2 glycoprotein I is protective against lupus nephritis and renal damage in systemic lupus erythematosus. Journal of Rheumatology. 2011;38(3):450–453. doi: 10.3899/jrheum.100650. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Yu C, Lu Y, et al. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis. 2007;12:1489–1502. doi: 10.1007/s10495-007-0073-9. [DOI] [PubMed] [Google Scholar]
- 20.Pierdominici M, Vomero M, Barbati C, et al. Role of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosus. FASEB Journal. 2012;26:1400–1412. doi: 10.1096/fj.11-194175. [DOI] [PubMed] [Google Scholar]
- 21.Mũoz LE, Lauber K, Schiller M, Manfredi AA, Herrmann M. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nature Reviews Rheumatology. 2010;6(5):280–289. doi: 10.1038/nrrheum.2010.46. [DOI] [PubMed] [Google Scholar]
- 22.Chamorro A-J, Marcos M, Miron-Canelo J-A, et al. Val247Leu beta2-glycoprotein-I allelic variant is associated with antiphospholipid syndrome: systemic review and meta-analysis. Autoimmunity Reviews. 2012;11:705–712. doi: 10.1016/j.autrev.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Sanghera DK, Wagenknecht DR, McIntyre JA, Kamboh MI. Identification of structural mutations in the fifth domain of apolipoprotein H (β 2-glycoprotein I) which affect phospholipid binding. Human Molecular Genetics. 1997;6(2):311–316. doi: 10.1093/hmg/6.2.311. [DOI] [PubMed] [Google Scholar]


