Abstract
Introduction
There are now over 30 confirmed loci predisposing to rheumatoid arthritis (RA). Studies have been largely undertaken in patients with anticyclic citrullinated peptide (anti-CCP) positive RA, and some genetic associations appear stronger in this subgroup than in anti-CCP negative disease, although few studies have had adequate power to address the question. The authors therefore investigated confirmed RA susceptibility loci in a large cohort of anti-CCP negative RA subjects.
Methods
RA patients and controls, with serological and genetic data, were available from UK Caucasian patients (n=4068 anti-CCP positive, 2040 anti-CCP negative RA) and 13,009 healthy controls. HLA-DRB1 genotypes and 36 single nucleotide polymorphisms were tested for association between controls and anti-CCP positive or negative RA.
Results
The shared epitope (SE) showed a strong association with anti-CCP positive and negative RA, although the effect size was significantly lower in the latter (effect size ratio=3.18, p<1.0E-96). A non-intronic marker at TNFAIP3, GIN1/C5orf30, STAT4, ANKRD55/IL6ST, BLK and PTPN22 showed association with RA susceptibility, irrespective of the serological status, the latter three markers remaining significantly associated with anti-CCP negative RA, after correction for multiple testing. No significant association with anti-CCP negative RA was detected for other markers (eg, AFF3, CD28, intronic marker at TNFAIP3), though the study power for those markers was over 80%.
Discussion
In the largest sample size studied to date, the authors have shown that the strength of association, the effect size and the number of known RA susceptibility loci associated with disease is different in the two disease serotypes, confirming the hypothesis that they might be two genetically different subsets.
Introduction
Based on the presence or absence of anticyclic citrullinated peptide (anti-CCP) antibodies, rheumatoid arthritis (RA) can be classified into anti-CCP positive and anti-CCP negative RA. Anti-CCP antibodies have been widely shown to be strong predictors of disease severity and radiological damage.1 2 It is currently a matter of some debate as to whether anti-CCP positive and anti-CCP negative RA are two distinct entities or represent two different subsets of one and the same disease.3–5 Linkage and association analysis revealed the shared epitope (SE) to be associated only with anti-CCP positive RA and not with anti-CCP negative RA.6 A study in twin pairs has shown that the estimated heritability of anti-CCP negative RA is 66% (95% CI 21% to 82%), similar to the heritability of anti-CCP positive RA, estimated at 68% (95% CI 55% to 79%).7 In the same study, the SE was found to explain 18% of the genetic component of RA susceptibility in anti-CCP positive RA but only 2.4% in anti-CCP negative RA.
Several studies have investigated putative associations between different human leucocyte antigen (HLA) alleles or single nucleotide polymorphisms (SNPs) within the HLA region and predisposition to anti-CCP negative RA,5 6 8–11 with contradictory results. A large meta-analysis, across four European populations, found only a weakly significant association between several HLA alleles and anti-CCP negative RA, but observed marked geographical differences.12 Lack of consistency between studies might therefore be explained by a different sample size and power, by geographical differences in allele frequencies and association patterns or by different study designs or definitions of HLA genotypes (two vs four-digit typing, different classifications for the SE). A large study, performed on Caucasians of Northern European descent, investigated several HLA-DRB1 susceptibility and protective models and SE subgroups for association with RA after stratification by autoantibody status. Significant associations between several HLA-DRB1 alleles and anti-CCP negative RA were found.13 Together, these findings strengthen the hypothesis that genetic factors predisposing to anti-CCP positive RA are different to those predisposing to anti-CCP negative RA.
A recent large meta-analysis brought the number of confirmed non-HLA RA susceptibility loci to 31.14 However, most studies on the identification of RA susceptibility loci published to date have been performed in largely anti-CCP positive RA cohorts, thereby biasing the search of RA susceptibility loci towards genetic variants predisposing to anti-CCP positive RA. Importantly, most studies have been underpowered to identify anti-CCP negative RA predictors.
Very few studies have systematically compared the genetic basis of anti-citrullinated protein autoantibody (ACPA)-positive to ACPA-negative RA outside the HLA region. In a genome-wide association study (GWAS) recently performed in 774 anti-CCP negative RA patients, 1147 anti-CCP positive RA patients and 1079 common controls,15 no SNP achieved genome-wide significance in the comparison between anti-CCP negative RA and controls, while the PTPN22 gene was associated with anti-CCP positive RA, together with hundreds of SNPs located within the HLA locus on chromosome 6.
Candidate gene association studies and a subsequent meta-analysis have confirmed an association of STAT4 polymorphisms with anti-CCP positive and negative RA.16 17 IRF5 polymorphisms have been shown in independent studies to be more strongly associated with anti-CCP negative RA than with anti-CCP positive RA in Caucasians,18 19 while this differential association is controversial in Asians.20 21 No other SNPs have been convincingly associated with anti-CCP negative RA. Interestingly, the association of PTPN22 polymorphisms with anti-CCP negative RA is controversial, with some investigators reporting association in anti-CCP negative patients.22 23 For example, a recent study investigating the usefulness of data derived from electronic health records tested multiple non-HLA RA susceptibility markers in 871 anti-CCP positive RA patients, 378 anti-CCP negative RA patients and 1212 common healthy controls.24 Only PTPN22 showed an association with anti-CCP negative RA with a p value<0.05. Due to the small sample size of the anti-CCP negative subgroup, conclusions could be made only for SNPs in aggregate rather than for individual SNPs. The authors conclude that there is a partial overlap between the genetic basis of anti-CCP positive and anti-CCP negative RA.
We hypothesised that currently known RA susceptibility SNPs would show a differential association pattern in anti-CCP negative RA compared with anti-CCP positive RA. Therefore, we tested the 31 RA confirmed susceptibility loci for association with RA in a dataset comprising between 1935 and 3827 anti-CCP positive RA patients, between 808 and 1918 anti-CCP negative RA patients and between 11468 and 12392 healthy controls per genetic marker, the largest number of anti-CCP negative patients studied so far in this setting.
Methods
Data collection, SNP selection and genotyping
Confirmed RA susceptibility SNPs were selected from the large meta-analysis by Stahl et al.14 Most of the SNPs had been already genotyped in RA cases and controls as part of several other projects conducted at our laboratory. Cohorts and patient characteristics are presented in the online supplementary table S1; genotyping and quality control procedures have been described elsewhere.22 25–27 All patients and controls were Caucasians originating from the UK satisfying the 1987 American College of Rheumatology classification criteria for RA. Only RA cases with an available anti-CCP status, as determined with the second generation CCP (CCP2) assay, were included in the analysis. Several studies/cohorts did not contain any controls. Therefore, anti-CCP positive RA cases or anti-CCP negative RA cases from different studies were pooled together and compared with controls. If a SNP reported in the study of Stahl was not available in a cohort, a SNP proxy was selected to fulfil the following requirements: linkage disequilibrium (r2) ≥0.90 with the original SNP and a maximum of three different SNPs for one specific locus across all datasets. If these requirements could not be fulfilled for one dataset, it was excluded from the analysis for that particular locus. Therefore, different numbers of cases and controls were available for analysis for different loci. The following HLA-DRB1 genotypes were considered as SE alleles: ‘0101’ ‘0102’ ‘0104’ ‘0105’ ‘0107’ ‘0108’ ‘0110’ ‘0111’ ‘0401’ ‘0404’ ‘0405’ ‘0408’ ‘0409’ ‘0410’ ‘0413’ ‘0416’ ‘0419’ ‘0421’ ‘0423’ ‘0426’ ‘0428’ ‘0429’ ‘0430’ ‘0433’ ‘0434’ ‘0435’ ‘0438’ ‘0440’ ‘0442’ ‘0443’ ‘0445’ ‘1001’ ‘1113’ ‘1126’ ‘1134’ ‘1402’ ‘1409’ ‘1413’ ‘1417’ ‘1419’ ‘1420’ ‘1421’ ‘1429’ ‘1430’ ‘1431’ ‘1432’ ‘1434’ ‘1441’ ‘1446’ ‘1447’ ‘1448’.
Statistical analysis
Multinomial logistic regression was applied to compute OR, 95% CI and p values for association between the minor allele at every locus and either anti-CCP-positive (ORccp-positive) or anti-CCP-negative RA (ORccp-negative), assuming additivity on the log-odds scale (ie, every locus was coded as 0,1 or 2 corresponding to the copy number of the minor allele). The minor allele was defined according to the allele frequency in the total population, including cases and controls. To test for differences between ORccp-positive and ORccp-negative, the linear combination β+ – β-, where β+ is log(ORccp-positive) and β- is log(ORccp-negative) was calculated, along with its standard error. This enables a p value for the difference in association to be calculated.
Due to a high pretest probability for association in anti-CCP positive RA, p values were not corrected for multiple testing in this subset. Based on previously published data, 32 of the 35 non-HLA SNPs tested have been confirmed to represent independent effects. Therefore, without assuming any prior probability for association in the anti-CCP negative RA subset, which is a stringent assumption, the Bonferroni-corrected significance threshold would be 1.6E-03. Statistical analysis was performed with Stata V.10.1 (Stata Statistical Software: Release 10; Stata Corp., College Station, Texas, USA).
Results
Thirty-one confirmed RA susceptibility loci,14 some of which contain independent effects, were considered for analysis. Together, this represents a total of 36 markers, plus the SE. Actual SNPs and proxys are presented for each locus in table 1. The combination of datasets available led to a total number of 4068 anti-CCP-positive, 2040 anti-CCP-negative RA and 13009 healthy UK controls. The actual number of cases and controls varies for every locus from 1935 to 3827 for anti-CCP positive RA, from 808 to 1918 for anti-CCP negative RA and from 11468 to 12392 for healthy controls. Basic cohort characteristics are presented in the online supplementary table S1. The results of the association analysis are presented in table 2. As expected, every locus showed an association with anti-CCP positive RA with a p value<0.05. The SE and the corresponding tag SNP were highly associated with anti-CCP positive RA with an OR of 4.08 and 2.68, respectively. Three other markers at AFF3, CD28, PTPN22 and two at the TNFAIP3 locus reached genome-wide significance (<5E-08) in anti-CCP positive RA with OR of 1.17, 1.18, 1.91, 1.29 and 1.45, respectively. By contrast, only six non-HLA loci in total reach a p value below 0.05 in anti-CCP negative RA: TNFAIP3, GIN1/C5orf30, STAT4, ANKRD55/IL6ST, BLK and PTPN22. The three last loci remained significant at the Bonferroni corrected threshold of 1.6E-03. The SE, and its tag SNP, show an association with anti-CCP negative RA with an OR of 1.28 and 1.15, respectively. The online supplementary table S2 shows association results stratified by rheumatoid factor and anti-CCP positivity. The probability of obtaining, by chance, at least 7/36 associated loci at a significance level of 0.05 is 1.8E-03. Therefore, under a prior hypothesis of no association, there is still a significant accumulation of RA susceptibility loci associated with anti-CCP negative RA.
Table 1.
Single nucleotide polymorphism (SNP) markers and their proxys for the independent rheumatoid arthritis susceptibility loci considered for analysis. The shared epitope is not defined by SNP markers, but by a list of 4-digit HLA-DRB1 alleles described in the methods section
| Chromosome | Locus name | Single nucleotide polymorphism markers | ||
|---|---|---|---|---|
| 1 | CD2/CD58 | rs11586238 | ||
| 1 | FCGR2A | rs12746613 | rs10494360 | |
| 1 | MMEL1/TNFRSF14 | rs10910099 | rs3890745 | |
| 1 | PTPN22 | rs2476601 | rs6679677 | |
| 1 | PTPRC | rs10919563 | rs1932435 | |
| 2 | AFF3 locus 1 | rs1160542 | rs11676922 | rs9653442 |
| 2 | AFF3 locus 2 | rs10865035 | ||
| 2 | CD28 | rs1980422 | ||
| 2 | CTLA4 | rs3087243 | rs231804 | |
| 2 | REL | rs13031237 | ||
| 2 | SPRED2 | rs934734 | rs17534670 | |
| 2 | STAT4 | rs7574865 | ||
| 3 | DNASE1L3/PXK | rs13315591 | rs9813011 | |
| 4 | IL2/IL21 | rs6822844 | rs13151961 | |
| 4 | RBPJ | rs874040 | rs10517086 | |
| 5 | ANKRD55/IL6ST | rs6859219 | ||
| 5 | GIN1/C5orf30 | rs26232 | rs35797 | |
| 6 | CCR6 | rs3093023 | rs6907666 | rs3093024 |
| 6 | HLA-DRB1 0401 tag | rs6910071 | ||
| 6 | PRDM1 | rs548234 | ||
| 6 | TAGAP | rs394581 | rs169858 | |
| 6 | TNFAIP3 locus 1 | rs6920220 | rs2327832 | |
| 6 | TNFAIP3 locus 2 | rs13207033 | rs10499194 | |
| 6 | TNFAIP3 locus 3 | rs5029937 | rs5029939 | |
| 7 | IRF5 | rs10488631 | rs12531711 | |
| 8 | BLK | rs2736340 | ||
| 9 | CCL21 locus 1 | rs951005 | ||
| 9 | CCL21 locus 2 | rs2812378 | rs10814138 | |
| 9 | TRAF1/C5 | rs3761847 | ||
| 10 | IL2RA locus 1 | rs706778 | rs10795791 | rs7072793 |
| 10 | IL2RA locus 2 | rs2104286 | ||
| 10 | PRKCQ | rs4750316 | rs10796045 | |
| 11 | RAG1/TRAF6 | rs540386 | rs5030437 | rs1046864 |
| 12 | KIF5A/PIP4K2C | rs1678542 | rs11172254 | |
| 20 | CD40 | rs4810485 | rs1569723 | |
| 22 | IL2RB | rs3218253 | rs3218258 | |
Table 2.
OR with 95% CI and p value for association of the minor allele at every locus with anticyclic citrullinated peptide (anti-CCP) positive or anti-CCP negative rheumatoid arthritis (RA). The minor allele is defined according to the frequency in the total population, including cases and controls.
| UK anti-CCP positive RA | UK anti-CCP negative RA | Comparison | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | Locus Name | OR | 95% CI | p Value | N cases | N cont | OR | 95% CI | p Value | N cases | N cont | ORpos/ORneg | p Value |
| 1 | CD2/CD58 | 1.11 | 1.05 to 1.18 | 3.68E-04 | 3827 | 11468 | 1.05 | 0.97 to 1.13 | 0.242 | 1918 | 11468 | 1.06 | 0.183 |
| 1 | FCGR2A | 1.16 | 1.07 to 1.25 | 2.28E-04 | 3467 | 11468 | 1.08 | 0.96 to 1.20 | 0.195 | 1563 | 11468 | 1.08 | 0.241 |
| 1 | MMEL1/TNFRSF14 | 0.88 | 0.83 to 0.93 | 2.12E-05 | 3485 | 12394 | 0.97 | 0.89 to 1.05 | 0.426 | 1574 | 12394 | 0.91 | 0.045 |
| 1 | PTPN22 | 1.91 | 1.77 to 2.05 | 3.91E-65 | 3813 | 12398 | 1.20 | 1.07 to 1.33 | 1.34E-03 | 1894 | 12398 | 1.60 | 7.11E-15 |
| 1 | PTPRC | 0.91 | 0.84 to 0.99 | 0.022 | 3600 | 11427 | 0.96 | 0.86 to 1.08 | 0.504 | 1580 | 11427 | 0.94 | 0.384 |
| 2 | AFF3 locus 1 | 1.17 | 1.11 to 1.23 | 5.36E-09 | 3777 | 11832 | 1.05 | 0.98 to 1.13 | 0.165 | 1881 | 11832 | 1.11 | 0.008 |
| 2 | AFF3 locus 2 | 1.17 | 1.09 to 1.25 | 2.64E-06 | 2378 | 8425 | 1.06 | 0.97 to 1.17 | 0.187 | 991 | 8425 | 1.10 | 0.086 |
| 2 | CD28 | 1.18 | 1.12 to 1.26 | 3.15E-08 | 3826 | 11469 | 1.08 | 1.00 to 1.17 | 0.055 | 1912 | 11469 | 1.09 | 0.049 |
| 2 | CTLA4 | 0.91 | 0.87 to 0.96 | 9.32E-04 | 3626 | 11975 | 0.94 | 0.88 to 1.02 | 0.121 | 1592 | 11975 | 0.97 | 0.475 |
| 2 | REL | 1.13 | 1.07 to 1.20 | 1.58E-05 | 3196 | 10878 | 1.02 | 0.95 to 1.11 | 0.554 | 1437 | 10878 | 1.11 | 0.028 |
| 2 | SPRED2 | 0.91 | 0.86 to 0.97 | 3.69E-03 | 2706 | 9032 | 0.97 | 0.89 to 1.06 | 0.565 | 1132 | 9032 | 0.94 | 0.198 |
| 2 | STAT4 | 1.13 | 1.06 to 1.21 | 1.34E-04 | 3401 | 11822 | 1.11 | 1.02 to 1.20 | 0.015 | 1824 | 11822 | 1.02 | 0.656 |
| 3 | DNASE1L3/PXK | 1.13 | 1.02 to 1.24 | 0.014 | 3445 | 11486 | 0.93 | 0.80 to 1.08 | 0.338 | 1432 | 11486 | 1.21 | 0.020 |
| 4 | IL2/IL21 | 0.88 | 0.82 to 0.95 | 1.03E-03 | 3402 | 11767 | 0.95 | 0.86 to 1.04 | 0.264 | 1814 | 11767 | 0.93 | 0.211 |
| 4 | RBPJ | 1.14 | 1.07 to 1.21 | 1.98E-05 | 3195 | 10879 | 1.02 | 0.94 to 1.11 | 0.681 | 1434 | 10879 | 1.12 | 0.021 |
| 5 | ANKRD55/IL6ST | 0.85 | 0.78 to 0.92 | 9.62E-05 | 2377 | 8428 | 0.80 | 0.70 to 0.90 | 2.42E-04 | 991 | 8428 | 1.07 | 0.357 |
| 5 | GIN1/C5orf30 | 0.88 | 0.83 to 0.94 | 4.86E-05 | 3372 | 11260 | 0.88 | 0.80 to 0.95 | 2.32E-03 | 1410 | 11260 | 1.01 | 0.835 |
| 6 | CCR6 | 1.13 | 1.07 to 1.19 | 1.48E-05 | 3423 | 11313 | 0.99 | 0.92 to 1.07 | 0.851 | 1508 | 11313 | 1.14 | 0.004 |
| 6 | HLA-DRB1 0401 tag | 2.68 | 2.50 to 2.88 | 9.97E-169 | 2378 | 8428 | 1.15 | 1.03 to 1.28 | 9.55E-03 | 991 | 8428 | 2.33 | 6.30E-47 |
| 6 | HLA-DRB1 SE | 4.08 | 3.64 to 4.56 | 1.18E-131 | 2366 | 1352 | 1.28 | 1.14 to 1.45 | 5.18E-05 | 1315 | 1352 | 3.18 | 2.56E-97 |
| 6 | PRDM1 | 1.06 | 1.00 to 1.12 | 0.041 | 3827 | 11469 | 1.04 | 0.97 to 1.12 | 0.275 | 1916 | 11469 | 1.02 | 0.692 |
| 6 | TAGAP | 0.90 | 0.85 to 0.95 | 1.57E-04 | 3824 | 11466 | 0.97 | 0.90 to 1.04 | 0.416 | 1914 | 11466 | 0.92 | 0.068 |
| 6 | TNFAIP3 locus 1 | 1.29 | 1.21 to 1.37 | 3.70E-16 | 3659 | 12392 | 1.09 | 1.00 to 1.18 | 0.048 | 1862 | 12392 | 1.18 | 3.52E-04 |
| 6 | TNFAIP3 locus 2 | 0.89 | 0.84 to 0.95 | 2.72E-04 | 3425 | 11923 | 0.93 | 0.85 to 1.02 | 0.109 | 1497 | 11923 | 0.96 | 0.383 |
| 6 | TNFAIP3 locus 3 | 1.45 | 1.28 to 1.65 | 1.33E-08 | 3414 | 12021 | 0.91 | 0.73 to 1.13 | 0.374 | 1460 | 12021 | 1.60 | 7.36E-05 |
| 7 | IRF5 | 1.12 | 1.02 to 1.23 | 0.016 | 2739 | 9043 | 1.09 | 0.95 to 1.24 | 0.213 | 1175 | 9043 | 1.03 | 0.696 |
| 8 | BLK | 1.09 | 1.03 to 1.16 | 3.77E-03 | 3490 | 11481 | 1.15 | 1.05 to 1.25 | 1.53E-03 | 1525 | 11481 | 0.95 | 0.332 |
| 9 | CCL21 locus 1 | 0.92 | 0.85 to 1.00 | 0.048 | 3199 | 10878 | 1.01 | 0.91 to 1.13 | 0.833 | 1439 | 10878 | 0.91 | 0.145 |
| 9 | CCL21 locus 2 | 1.13 | 1.07 to 1.20 | 7.68E-06 | 3642 | 11936 | 1.04 | 0.97 to 1.12 | 0.274 | 1855 | 11936 | 1.09 | 0.046 |
| 9 | TRAF1/C5 | 1.10 | 1.04 to 1.16 | 7.54E-04 | 3135 | 11544 | 1.02 | 0.94 to 1.10 | 0.698 | 1447 | 11544 | 1.08 | 0.073 |
| 10 | IL2RA locus 1 | 1.14 | 1.06 to 1.23 | 3.21E-04 | 1935 | 5477 | 1.07 | 0.96 to 1.19 | 0.224 | 808 | 5477 | 1.07 | 0.240 |
| 10 | IL2RA locus 2 | 0.88 | 0.83 to 0.93 | 2.71E-05 | 3806 | 12396 | 0.97 | 0.90 to 1.05 | 0.450 | 1896 | 12396 | 0.91 | 0.033 |
| 10 | PRKCQ | 0.87 | 0.82 to 0.94 | 1.18E-04 | 3657 | 12427 | 0.95 | 0.86 to 1.04 | 0.281 | 1624 | 12427 | 0.92 | 0.132 |
| 11 | RAG1/TRAF6 | 0.92 | 0.85 to 0.99 | 0.032 | 3440 | 11379 | 0.98 | 0.88 to 1.09 | 0.723 | 1556 | 11379 | 0.94 | 0.285 |
| 12 | KIF5A/PIP4K2C | 0.90 | 0.85 to 0.95 | 1.81E-04 | 3688 | 12452 | 0.95 | 0.88 to 1.02 | 0.135 | 1881 | 12452 | 0.95 | 0.237 |
| 20 | CD40 | 0.89 | 0.84 to 0.95 | 4.36E-04 | 3640 | 11971 | 1.02 | 0.94 to 1.11 | 0.594 | 1858 | 11971 | 0.87 | 0.005 |
| 22 | IL2RB | 1.15 | 1.08 to 1.21 | 3.15E-06 | 3811 | 12392 | 1.08 | 1.00 to 1.16 | 0.057 | 1900 | 12392 | 1.06 | 0.164 |
N cases, cont: number of cases or controls with available genotype. Comparison ORpos/ORneg: effect size ratio between the OR in CCP-positive and CCP-negative RA, with the p value for its significance. The CI of the effect size OR is not shown to avoid overloading the table
Due to the different number of cases in anti-CCP positive and negative RA, simply comparing p values would be misleading. Therefore, the significance of the observed difference in association between anti-CCP positive and negative RA was addressed by computing a comparison OR (or effect size OR) and its corresponding p value (column ‘comparison’ in table 2). This allows the classification of the loci into three distinct categories (table 3 and figure 1): the HLA-DRB1 SE, PTPN22 and one marker at TNFAIP3 are associated with anti-CCP positive and negative RA, but show a clear differential association with an effect size significantly higher in anti-CCP positive RA (category 1). Other loci, like C5orf30 or STAT4 are associated with RA irrespective of the serological status, with the effect size not differing significantly between subsets (category 2: anti-CCP independent associations). A third category comprises anti-CCP positive specific loci with no significant association detected in anti-CCP negative RA; however, the p value for the effect size ratio is below 0.05. The remaining loci could not be classified into one of these three categories, because although they are associated with anti-CCP positive RA with a p value<0.05, the effect in anti-CCP-negative RA is not significantly different from that in anti-CCP-positive RA, nor significantly different from the null. This last situation can only be explained by a lack of power. Indeed, for the vast majority of markers in category 3, the study has, in the anti-CCP negative subgroup, a power between 60% and 90% at the 0.05 significance level to detect an association of the same effect size as observed in the anti-CCP positive subgroup, while power drops to between 15% and 50% for most of the unclassifiable markers. As an example in category 3, an association of an effect size of 1.17 would be detected for AFF3 locus 1 with a power of 89.3% at the 0.05 significance level for a minor allele frequency (MAF) of 45.5% in controls. An association of a larger effect size of 1.45 would be detected for the intronic TNFAIP3 marker with a power of 81.6% at the 0.05 significance level for a MAF of 3.5% in controls. The detection power drops for smaller effects around 1.13, but is still 64.4% CCL21 locus 2 (MAF in controls 34.2%). However, although CD2/CD58 has been genotyped in 1918 anti-CCP negative cases, it is not possible to classify it as being associated with anti-CCP positive, negative or both because the power to detect an effect of 1.11 in anti-CCP negative with a MAF of 24% in controls is only 46.1%.
Table 3.
Schematic classification of RA susceptibility loci into three categories depending on their association pattern in anti-CCP positive and negative RA
| Category | Associations | Locus Name |
|---|---|---|
| 1 | Both CCP positive and negative RA, stronger in CCP positive RA | HLA-DRB1 SE, PTPN22, TNFAIP3 locus 1 |
| 2 | Both CCP positive and negative RA, equally strong in both | ANKRD55, BLK, C5orf30, STAT4 |
| 3 | CCP positive RA only, significant difference between CCP positive and negative RA | AFF3 locus 1, CCR6, CCL21 locus 2, IL2RA locus 2, CD28, CD40, PXK, REL, RBPJ, TNFRSF14, TNFAIP3 locus 3 |
| Not classifiable | CCP positive RA only, but no significant difference between CCP positive and negative RA | All others |
CCP, cyclic citrullinated peptide; RA, rheumatoid arthritis.
Figure 1.
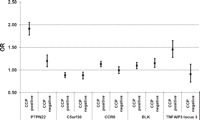
OR and 95% CI for different single nucleotide polymorphism association patterns. PTPN22: significant association in both serotypes, significantly different. C5orf30: significant association in both serotypes with the same effect size. CCR6: only associated in anticyclic citrullinated peptide (anti-CCP) positive rheumatoid arthritis (RA). BLK: associated in both serotypes, effect size slightly, but not significantly, larger in anti-CCP negative RA. TNFAIP3 locus 3: only associated in anti-CCP positive RA, highly significant difference. PTPN22, C5orf30, CCR6 are prototypic examples illustrating the three categories presented in table 3.
Of note, is the fact, that the effect size of all but two loci, ANKRD55 and BLK (figure 1), is larger in anti-CCP positive RA than in anti-CCP negative RA. However, due to relatively wide CIs, the effect size ratio is not significant, so these two loci are both classified in category 2.
Interestingly, the three known independent effects at TNFAIP3 are classified in at least two different categories (table 3). At this locus, the most profound discordance between anti-CCP positive and negative RA is seen for the intronic marker (figure 1), which displays a genome-wide significant association of a ‘large’ effect size in anti-CCP positive RA with an OR above one (OR 1.45, 95% CI 1.28 to 1.65, p=1.33×10−8), representing a risk factor for this disease. However, no significant association is detected in anti-CCP negative RA (OR 0.91, 95% CI 0.73 to 1.13, p=0.37). The effect size ratio is highly significant (OR 1.60, 95% CI 1.27 to 2.02, p=7.36×10−5).
Discussion
With a number of anti-CCP-negative RA patients ranging from 808 to 1918 per locus, our study represents the largest genetic study on anti-CCP negative RA to date. The 31 confirmed RA susceptibility loci identified so far have been primarily established in anti-CCP positive RA, and the meta-analysis by Stahl et al14 included only seropositive RA. The effect size and strength of association of the loci identified in the meta-analysis by Stahl et al and in the anti-CCP positive RA subset of this study are very consistent. Slight differences might be related to the difference of power for certain loci and to the use, in some instances, of several different proxies for one locus. It should be noted that we defined the minor allele frequency according to the frequency in the total population, including cases and controls. Since the minor allele frequency is close to 50% for SPRED2 in the meta-analysis by Stahl, where the minor allele frequency is based on controls only, the G allele is the minor allele, while it is the major allele in this study. This results in an inversion of the OR for association with anti-CCP positive RA. All loci tested here are associated with anti-CCP positive RA with p<0.05, and five loci reach genome-wide significance (<5E-08).
Interestingly, we show a strong and highly significant association of the SE with anti-CCP negative RA; the association remains present after stratification for rheumatoid factor (online supplementary table S2), which is consistent with the findings of a large study performed in patients from the same genetic background.13 However, several smaller studies, performed in populations of different origins, did not detect significant association, likely due to a lack of power or ethnic differences. A large European meta-analysis, taking only 2-digit typing into account across several different European populations, concluded that there were weak, but no robust associations between any HLA-DRB1 alleles and anti-CCP negative RA.12 The number of anti-CCP negative RA patients tested in our study is much higher than that in the meta-analysis or any other study, and confirms that the SE is associated with anti-CCP negative RA in UK Caucasians.
Compared with the results in anti-CCP positive RA, where all loci show evidence for association, only seven loci achieve a p value below 0.05 in the anti-CCP negative subgroup. However, this number is higher than reported to date, and represents a statistically significant overlap (p=0.0018). Among the loci tested here, 32 non-HLA loci have been convincingly shown to be independently associated in anti-CCP positive RA. Therefore, without assuming any prior probability for association, the Bonferroni-corrected significance threshold would be 1.6E-03. Three non-HLA loci would remain significantly associated with anti-CCP negative RA after the Bonferroni correction: ANKRD55/IL6ST, BLK and PTPN22.
Interestingly, both BLK and STAT4 are confirmed systemic lupus erythematosus (SLE) susceptibility loci and it would be interesting to investigate the overlap of other SLE loci with anti-CCP negative disease. Our findings confirm those of the previous reports showing association in both anti-CCP positive and negative RA subgroups at the STAT4 locus. IRF5, which has been found to be associated with anti-CCP negative RA by previous investigators, is not associated with anti-CCP negative RA in this study. However, it should be noted that the current study only had 19.6% power to detect an association of an effect size of 1.12 at the 0.05 significance level. In addition, a differential association of this marker with RA and its different serotypes in different ethnic groups, or gene-environment interactions, might further account for the lack of association seen here in anti-CCP negative patients.
The effect size of BLK and ANKRD55/IL6ST is larger in anti-CCP negative RA than in anti-CCP positive RA, although this difference does not reach statistical significance. All other loci show a higher effect size in anti-CCP positive RA. This can be explained by the fact that loci tested here have been primarily identified in cohorts containing mainly or exclusively anti-CCP positive patients.
The genetic difference between the two RA serotypes can be particularly illustrated by the association pattern of the intronic locus at TNFAIP3: it is strongly associated with anti-CCP positive RA, but not with anti-CCP negative RA, despite a detection power of over 80%; the difference in OR is statistically significant. The intronic locus at TNFAIP3 therefore represents a risk factor for anti-CCP positive RA, but not for anti-CCP negative RA in this analysis.
The present study shows a genetic contrast between anti-CCP positive and anti-CCP negative RA and allows the classification of known RA susceptibility SNPs in different categories. The first category comprises markers associated with both subsets, but their effect size is significantly larger in anti-CCP positive RA. Although anti-CCP negative RA could not be clearly divided into distinct clinical subphenotypes in a recent study in the Netherlands,28 it might still comprise several genetically and serologically different subsets, based for example on the presence of ACPA, other than anti-CCP antibodies.29 The second category contains SNPs, similarly associated in both anti-CCP positive and negative RA, with the same effect size. The third category comprises SNPs associated with anti-CCP positive RA, but not with anti-CCP negative RA and the effect size ratio is statistically significant. A lack of association of some markers, while others are associated with disease irrespective of the serological status, suggests that RA susceptibility markers might cluster to different molecular pathways, some associated with autoantibody production, others not.
In summary, among 33 independent genetic loci tested in this study, 18 could be classified into three different categories, according to their association pattern, while 15 could not, mainly due to lack of power. Seven markers show an association with anti-CCP negative RA, while 11 others are unlikely to be associated. The use of a multinomial logistic regression analysis leading to three p values, as described here (anti-CCP positive RA, anti-CCP negative RA, effect size ratio), represents a straightforward method to classify markers into three meaningful categories or to exclude them from classification, if only one p value out of three is significant. This latter situation occurs mainly when the power is low. The accuracy of classification depends on the definition of the significance threshold. If a Bonferroni corrected p value is used for classification, markers with p values between 0.05 and 1.6E-03 would change category. Markers with highly significant associations and effect size ratio like PTPN22 can be considered as accurately categorised. Future studies might identify more categories; for example, for makers associated exclusively with anti-CCP negative RA, or displaying a larger effect size in anti-CCP negative RA than in anti-CCP positive RA.
Despite this being the largest sample of anti-CCP negative cases studied to date, the main limitation remains lack of power for many markers. This is particularly pertinent to the loci, which could not be classified into one of the three categories presented here. Larger sample sizes will be required to explore these loci more fully. Six thousand five hundred anti-CCP negative patients and 11 000 controls would be required to detect an effect size of 1.10 with a power of 80% for a marker present at a MAF of 28% in controls (average MAF of RA susceptibility loci in controls reported in the study by Stahl et al14).
The low number of anti-CCP positive RA susceptibility loci associated with anti-CCP negative RA highlights the need for a well-powered GWAS for the discovery of yet unknown anti-CCP negative RA specific loci. The current study presents genetic differences and similarities between anti-CCP positive and anti-CCP negative RA. Although the two disease serotypes show significant differences in disease course and severity, anti-CCP antibodies are currently not used to guide treatment decisions in clinical practice. However, the results presented here highlight the need for genetic analyses of susceptibility, severity and treatment response to consider the two serotypes both separately and together for future investigations.
Supplementary Material
Acknowledgments
SV was initially supported by a grant from the Swiss National Science Foundation, grant no. PBGEP3-129009; he is currently supported by a research grant from the Swiss Foundation for Medical-Biological Scholarships (SSMBS), managed by the Swiss National Science Foundation (grant reference number PASMP3_134380). This grant is financed by a donation of Novartis to the SSMBS. The authors thank Arthritis Research UK (grant ref 17552) and the NIHR Manchester Biomedical Research Centre for support.
Footnotes
Contributors Data analysis: SV, DP and JB. Statistical analysis: SV and ML. Manuscript preparation: SV, SE, AB and JW.
Funding: Swiss National Science Foundation/Swiss Foundation for Medical-Biological Scholarships/Arthritis Research UK
Competing interests: None.
Patient Consent: Obtained.
Ethics approval: The University of Manchester Ethics Committee approved this study.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Bukhari M, Thomson W, Naseem H, et al. The performance of anti-cyclic citrullinated peptide antibodies in predicting the severity of radiologic damage in inflammatory polyarthritis: results from the Norfolk Arthritis Register. Arthritis Rheum 2007;56:2929–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglin E, Johansson T, Sundin U, et al. Radiological outcome in rheumatoid arthritis is predicted by presence of antibodies against cyclic citrullinated peptide before and at disease onset, and by IgA-RF at disease onset. Ann Rheum Dis 2006;65:453–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Helm-van Mil AH, Huizinga TW. Advances in the genetics of rheumatoid arthritis point to subclassification into distinct disease subsets. Arthritis Res Ther 2008;10:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daha NA, Toes RE. Rheumatoid arthritis: Are ACPA-positive and ACPA-negative RA the same disease? Nat Rev Rheumatol 2011;7:202–3 [DOI] [PubMed] [Google Scholar]
- 5.Ohmura K, Terao C, Maruya E, et al. Anti-citrullinated peptide antibody-negative RA is a genetically distinct subset: a definitive study using only bone-erosive ACPA-negative rheumatoid arthritis. Rheumatology (Oxford) 2010;49:2298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huizinga TW, Amos CI, van der Helm-van Mil AH, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum 2005;52:3433–8 [DOI] [PubMed] [Google Scholar]
- 7.van der Woude D, Houwing-Duistermaat JJ, Toes RE, et al. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum 2009;60:916–23 [DOI] [PubMed] [Google Scholar]
- 8.Irigoyen P, Lee AT, Wener MH, et al. Regulation of anti-cyclic citrullinated peptide antibodies in rheumatoid arthritis: contrasting effects of HLA-DR3 and the shared epitope alleles. Arthritis Rheum 2005;52:3813–18 [DOI] [PubMed] [Google Scholar]
- 9.Lundström E, Källberg H, Smolnikova M, et al. Opposing effects of HLA-DRB1*13 alleles on the risk of developing anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum 2009;60:924–30 [DOI] [PubMed] [Google Scholar]
- 10.Verpoort KN, van Gaalen FA, van der Helm-van Mil AH, et al. Association of HLA-DR3 with anti-cyclic citrullinated peptide antibody-negative rheumatoid arthritis. Arthritis Rheum 2005;52:3058–62 [DOI] [PubMed] [Google Scholar]
- 11.Ding B, Padyukov L, Lundström E, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum 2009;60:30–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Woude D, Lie BA, Lundström E, et al. Protection against anti-citrullinated protein antibody-positive rheumatoid arthritis is predominantly associated with HLA-DRB1*1301: a meta-analysis of HLA-DRB1 associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in four European populations. Arthritis Rheum 2010;62:1236–45 [DOI] [PubMed] [Google Scholar]
- 13.Mackie SL, Taylor JC, Martin SG, et al. A spectrum of susceptibility to rheumatoid arthritis within HLA-DRB1: stratification by autoantibody status in a large UK population. Genes Immun 2012;13:120–8 [DOI] [PubMed] [Google Scholar]
- 14.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 2010;42:508–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padyukov L, Seielstad M, Ong RT, et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann Rheum Dis 2011;70:259–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YH, Woo JH, Choi SJ, et al. Association between the rs7574865 polymorphism of STAT4 and rheumatoid arthritis: a meta-analysis. Rheumatol Int 2010;30:661–6 [DOI] [PubMed] [Google Scholar]
- 17.Orozco G, Alizadeh BZ, Delgado-Vega AM, et al. Association of STAT4 with rheumatoid arthritis: a replication study in three European populations. Arthritis Rheum 2008;58:1974–80 [DOI] [PubMed] [Google Scholar]
- 18.Dieguez-Gonzalez R, Calaza M, Perez-Pampin E, et al. Association of interferon regulatory factor 5 haplotypes, similar to that found in systemic lupus erythematosus, in a large subgroup of patients with rheumatoid arthritis. Arthritis Rheum 2008;58:1264–74 [DOI] [PubMed] [Google Scholar]
- 19.Sigurdsson S, Padyukov L, Kurreeman FA, et al. Association of a haplotype in the promoter region of the interferon regulatory factor 5 gene with rheumatoid arthritis. Arthritis Rheum 2007;56:2202–10 [DOI] [PubMed] [Google Scholar]
- 20.Kim YJ, Park JH, Kim I, et al. Putative role of functional interferon regulatory factor 5 (IRF5) polymorphism in rheumatoid arthritis in a Korean population. J Rheumatol 2008;35:2106–18 [DOI] [PubMed] [Google Scholar]
- 21.Shimane K, Kochi Y, Yamada R, et al. A single nucleotide polymorphism in the IRF5 promoter region is associated with susceptibility to rheumatoid arthritis in the Japanese population. Ann Rheum Dis 2009;68:377–83 [DOI] [PubMed] [Google Scholar]
- 22.Morgan AW, Thomson W, Martin SG, et al. Reevaluation of the interaction between HLA-DRB1 shared epitope alleles, PTPN22, and smoking in determining susceptibility to autoantibody-positive and autoantibody-negative rheumatoid arthritis in a large UK Caucasian population. Arthritis Rheum 2009;60:2565–76 [DOI] [PubMed] [Google Scholar]
- 23.Pierer M, Kaltenhäuser S, Arnold S, et al. Association of PTPN22 1858 single-nucleotide polymorphism with rheumatoid arthritis in a German cohort: higher frequency of the risk allele in male compared to female patients. Arthritis Res Ther 2006;8:R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011;88:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plant D, Thomson W, Lunt M, et al. The role of rheumatoid arthritis genetic susceptibility markers in the prediction of erosive disease in patients with early inflammatory polyarthritis: results from the Norfolk Arthritis Register. Rheumatology (Oxford) 2011;50:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plant D, Prajapati R, Hyrich KL, et al. Replication of association of the PTPRC gene with response to anti-tumor necrosis factor therapy in a large UK cohort. Arthritis Rheum 2012;64:665–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Rooy DP, Willemze A, Mertens B, et al. Can anti-cyclic citrullinated peptide antibody-negative RA be subdivided into clinical subphenotypes? Arthritis Res Ther 2011;13:R180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klareskog L, Rönnelid J, Lundberg K, et al. Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol 2008;26:651–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


