SUMMARY
Activating mutations in NOTCH1, an essential regulator of T cell development, are frequently found in human T cell acute lymphoblastic leukemia (T-ALL). Despite important advances in our understanding of Notch signal transduction, the regulation of Notch functions in the nucleus remains unclear. Using immunoaffinity purification, we identified NOTCH1 nuclear partners in T-ALL cells and showed that, beyond the well-characterized core activation complex (ICN1-CSL-MAML1), NOTCH1 assembles a multifunctional complex containing the transcription coactivator AF4p12, the PBAF nucleosome remodeling complex, and the histone demethylases LSD1 and PHF8 acting through their demethylase activity to promote epigenetic modifications at Notch-target genes. Remarkably, LSD1 functions as a corepressor when associated with CSL-repressor complex and as a NOTCH1 coactivator upon Notch activation. Our work provides new insights into the molecular mechanisms that govern Notch transcriptional activity and represents glimpse into NOTCH1 interaction landscape, which will help in deciphering mechanisms of NOTCH1 functions and regulation.
INTRODUCTION
Signal-transduction pathways are evolutionarily conserved cell communication mechanisms that underpin all aspects of metazoan development. Despite remarkable diversity in cell types and tissues, few signaling pathways are required to unfold most developmental programs. Signaling by Notch receptors determines cell fates throughout embryonic development and in adult tissues (Artavanis-Tsakonas et al., 1999). Notch pathway activation relies on ligand-induced proteolytic cleavage of the receptor that results in the release of the intracellular domain of Notch (ICN). ICN then travels to the nucleus and initiates highly diverse transcriptional programs that govern a vast array of cellular functions. Nuclear responses downstream of Notch activation are tightly regulated by various posttranslational modifications that affect ICN trafficking, half-life, and transcriptional activity, thereby contributing to signaling diversity (Andersson et al., 2011). Due to the prominent role of Notch signaling in dictating cell-fate decisions, dysfunctions in the pathway are associated with various human diseases including inherited genetic disorders and cancers. Human NOTCH1 was discovered in leukemic T cells harboring the t(7;9) translocation and was subsequently shown to be a master regulator of T cell development with a strong oncogenic potential in developing T cells (Aifantis et al., 2008). The central role of NOTCH1 in the pathogenesis of human T-ALL is supported by the presence of somatic mutations in more than 50% of T-ALL cases (Weng et al., 2004). In recent years, it has become increasingly evident that in addition to its oncogenic role in T-ALL, Notch signaling drives the growth of a wide range of hematopoietic malignancies and solid tumors (South et al., 2012). Paradoxically, mutations that inactivate the Notch pathway have also been described in human cancers indicating that, depending on the cellular context, Notch signaling can be either oncogenic or tumor suppressive (South et al., 2012).
Over the past decades important progress has been made in deciphering Notch signal transduction and identifying processes that are influenced by Notch (Kopan and Ilagan, 2009). The emerging picture posits that most Notch-dependent physiological and pathological processes rely on the ability of nuclear ICN to convert the DNA-binding protein CSL (also known as RBPJ) from a transcriptional repressor into an activator. This regulation involves the formation of a stable ternary complex composed of CSL, ICN, and Mastermind-like family of coactivators (MAML) (Nam et al., 2006; Wilson and Kovall, 2006). Although little is known about its physical partners, the ICN-CSL-MAML complex is thought to serve as a platform for recruitment of coactivators and subsequent transcriptional activation of Notch-target genes (Borggrefe and Oswald, 2009). Deciphering how the ICN-CSL-MAML ternary complex orchestrates transcriptional activation and how diversity in the transcriptional program is established depending on the cellular context is a major challenge in the field.
Using tandem affinity chromatography, followed by mass spectrometry, we purified the intracellular active form of NOTCH1 (ICN1) and identified its associated partners in human T-ALL cells. We found a large set of proteins associated with ICN1, including transcriptional regulators and protein modifiers. Moreover, we found that ICN1 associates with lineage-specific transcription factors and components of other signaling pathways that could cooperate with Notch to confer a specific program of gene expression. Importantly, biochemical and functional analysis led to the identification of several components of the Notch-activation complex and provide insights into the molecular mechanisms that govern Notch-mediated activation of its target genes.
RESULTS
Identification of NOTCH1-Associated Cofactors
Mutational activation of NOTCH1 leading to aberrant ICN1 production and translocation into the nucleus is a frequent event in T-ALL. Beside CSL and MAML1, nuclear partners that support NOTCH1 transcriptional program and tumorigenesis remain to be determined. To identify such factors, we purified ICN1 from SupT1 cells, a human NOTCH1-dependent T-ALL cell line. We generated stable SupT1 cells expressing human ICN1 tagged with both FLAG and HA epitopes (F/H-ICN1). Western blot (WB) assay showed that tagged ICN1 is expressed in the nucleus at a level comparable to that of endogenous ICN1 (Figure S1A). Importantly, F/H-ICN1 was able to restore Notch-responsive gene expression (Figure S1B) and Notch-dependent proliferation (Figure S1C) after inhibition of endogenous Notch using g-secretase inhibitor (GSI), indicating that F/H-ICN1 is functional.
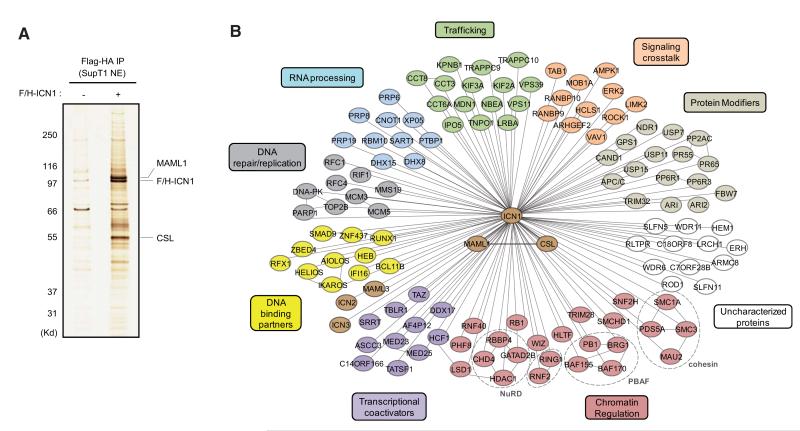
F/H-ICN1 and its associated partners were purified from nuclear extracts derived from SupT1 cells using tandem affinity chromatography (Figure 1A). Mass spectrometry (MS) analysis identified protein-partners of ICN1 (Figure 1B and Table S1). Major MS-identified ICN1 nuclear partners are the core components of the Notch-activation complex: MAML1 and CSL. The numbers of recovered peptides (Table S1) and the intensity of the corresponding silver-stained bands (Figure 1A) indicate that MAML1 and CSL are stoichiometric ICN1-partners. Other Notch pathway components such as MAML3, ICN2, and ICN3 were also recovered. Importantly, 27 out of 127 interacting proteins have been previously associated with Notch signaling (Table S2). Finally, some of ICN1-partners were tested and confirmed by WB (Figure S1D). Taken together, these results validate our ICN1-purification strategy.
Figure 1. Identification of Notch1-Associated Nuclear Factors.
(A) Purification of NOTCH1 intracellular domain (ICN1) and its interacting partners. Nuclear extracts (NEs) prepared from FLAG-HA epitope-tagged ICN1 (F/H-ICN1)-expressing SupT1 cells or mock-transduced SupT1 cells were subjected to sequential immunoprecipitation using anti-FLAG and anti-HA beads. Proteins were resolved by SDS-PAGE and visualized by silver staining.
(B) Interaction network of ICN1-associated proteins identified by Mass Spectrometry (MS). The false positive interactors were excluded by removing all proteins that were also detected in the control purification. See also Figure S1 and Tables S1, S2, and S3.
Interaction network analysis of ICN1 partners revealed several functional classes of proteins (Figure 1B). These include protein-modifying enzymes with a potential role in NOTCH1 regulation such as the tumor suppressor FBW7 known to target ICN1 for ubiquitination and degradation (Aifantis et al., 2008), as well as deubiquitinating enzymes, kinases, and phosphatases. ICN1 associates with several lineage-specific transcription factors that comprise major regulators of T cell development (BCL11B, HEB, and RUNX1) and IKAROS family members, including IKZF1, a tumor suppressor that represses Notch transcriptional responses. Moreover, interactions between ICN1 and components of other signaling pathways, such as the MAP kinase family member ERK2 and the TGFβ/BMP signaling mediator SMAD9, were also detected. Importantly, among ICN1-associated proteins we found numerous regulators of gene expression that act at various steps of transcriptional activation (Table S3). These encompass well-characterized coactivators, such as HCF1, ASCC3, and subunits of the Mediator complex, as well as proteins with putative functions in transcriptional regulation, for example the MLL-fusion partner AF4p12 (also known as FRYL), a poorly-characterized protein that exhibits transcriptional activation properties (Hayette et al., 2005). Chromatin-modifying enzymes, including the PBAF nucleosome-remodeling complex, RNF40 (a subunit of the E3 ubiquitin-ligase BRE1 that monoubiquitinates H2B) and the histone demethylases LSD1 and PHF8, were also recovered.
Thus, ICN1 interacts with a varied set of proteins in T-ALL cells that could reflect the diversity of Notch functions and regulation. In the present study, we focused on the characterization of ICN1-cofactors important for its transcriptional activity.
The PBAF Complex, LSD1, PHF8, and AF4p12 Associate with ICN1
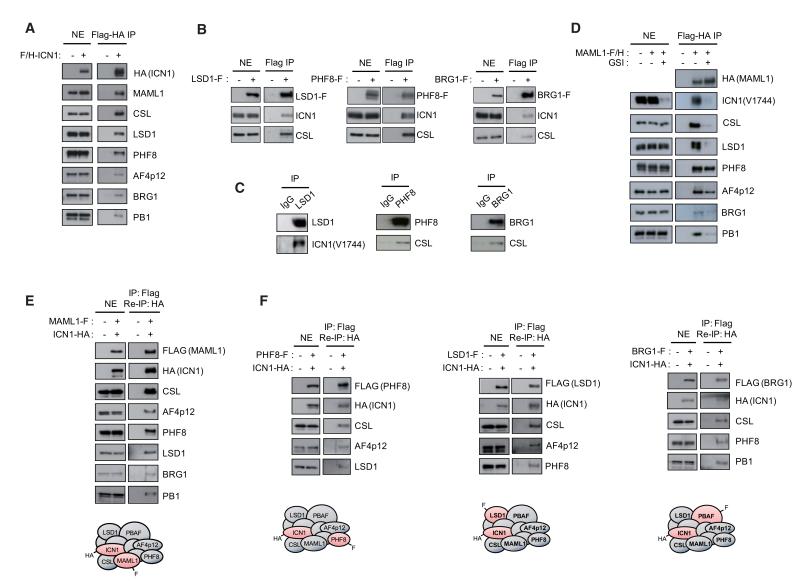
Western Blot analysis of ICN1 purified material confirmed the interaction with PBAF subunits BRG1 and PB1, the histone demethylases LSD1 and PHF8, and AF4p12 (Figure 2A). Importantly, endogenous ICN1 and CSL were immunoprecipitated with FLAG-tagged LSD1, PHF8 or BRG1 (Figure 2B). Moreover, immunoprecipitation of endogenous LSD1, PHF8, and BRG1 showed a specific interaction with components of the Notch-activation complex, as revealed by the presence of endogenous ICN1 or CSL (Figure 2C), suggesting that PBAF, LSD1, and PHF8 are part of the Notch-activation complex. In agreement with these observations, WB analysis of MAML1-interacting proteins purified from SupT1 nuclear extracts confirmed its association with the identified cofactors (Figure 2D). Interestingly, GSI treatment severely altered MAML1 association with CSL, LSD1, PB1, and BRG1, indicating that these interactions are Notch-dependent. However, binding of PHF8 and AF4p12 to MAML1 was only marginally reduced (Figure 2D). This suggests that MAML1 might recruit PHF8 and AF4p12 to the Notch-activation complex, while LSD1 and the PBAF complex could be ICN1 or CSL partners. Alternatively, recruitment of cofactors, including LSD1 and PBAF, may result from conformational changes induced by ICN1-CSL-MAML1 ternary complex formation.
Figure 2. PBAF, LSD1, PHF8, and AF4p12 Associate with ICN1-CSL-MAML1.
(A) FLAG and HA-immunopurified ICN1-associtated proteins from SupT1 nuclear extracts (NEs) were resolved on SDS-PAGE and the presence of partners identified by mass spectrometry was confirmed by Western blot (WB).
(B) NEs from SupT1 stably expressing LSD1-F (FLAG-tagged), PHF8-F or BRG1-F were subjected to immunoprecipitation (IP) using anti-FLAG beads. The presence of endogenous ICN1 and CSL in the purified material was revealed by WB. The anti-ICN1 antibody specifically recognizes the γ-secretase-cleaved active form of NOTCH1 (V1744).
(C) Interaction between endogenous LSD1, PHF8 or BRG1 with components of the Notch-activation complex. LSD1, PHF8, and BRG1 were purified from SupT1 NEs using specific antibodies and the presence of ICN1 or CSL in the purified material was revealed by WB.
(D) SupT1 cells stably expressing FLAG and HA-tagged MAML1 (MAML1-F/H) were treated with DMSO or GSI (500 nM, 8 hr). MAML1-associated proteins were FLAG-HA immunopurified from NEs and analyzed by WB using the indicated antibodies.
(E) BRG1, PB1, LSD1, PHF8, and AF4p12 are associated with the Notch-activation complex. NEs from SupT1 stably transduced with FLAG-MAML1 and HA-ICN1 were subjected to sequential IP using anti-FLAG and anti-HA beads. The presence of Notch cofactors in the purified material was analyzed by WB.
(F) Notch cofactors assemble into a single complex containing ICN1-CSL-MAML1. Reciprocal IPs with anti-FLAG and anti-HA beads were performed using NEs from SupT1 cells stably coexpressing HA-ICN1 and FLAG-PHF8, -LSD1 or -BRG1. Eluates were subjected to WB.
NOTCH1-Associated Factors Assemble into a Single Complex Containing ICN1-CSL-MAML1
To further confirm that Notch cofactors are integral components of the Notch-activation complex, nuclear extracts from SupT1 cells stably expressing FLAG-MAML1 and HA-ICN1 were subjected to sequential immunopurifications using anti-FLAG followed by anti-HA beads. Because MAML1 incorporation into the complex requires a contact with both ICN1 and CSL (Nam et al., 2006; Wilson and Kovall, 2006), this approach allowed us to isolate factors associated with the ICN1-CSL-MAML1 ternary complex. WB assay of the purified material revealed the presence of ICN1 cofactors (Figure 2E), indicating that BRG1, PB1, LSD1, PHF8, and AF4p12 are physical partners of the Notch-activation complex.
To determine whether these factors form one or distinct ICN1-CSL-MAML1-containing complexes, we stably coexpressed HA-ICN1 and FLAG-tagged PHF8, LSD1 or BRG1 in SupT1. Reciprocal immunoprecipitations detected the association of CSL with the purified PHF8-ICN1-, LSD1-ICN1-, and BRG1-ICN1-containing complexes (Figure 2F). Importantly, WB analysis also revealed the presence of LSD1 and AF4p12 in the PHF8-ICN1 purification, suggesting that these factors exist in a single complex. Furthermore, AF4p12 and PHF8 coimmunoprecipitated with LSD1-ICN1 complex, and both PHF8 and PB1 were detected in BRG1-ICN1 purification (Figure 2F). These results indicate that PBAF, LSD1, PHF8, and AF4p12 associate in a single multifunctional complex containing ICN1-CSL-MAML1. Of note, our analysis does not exclude that other coactivators including those identified by MS analysis (Table S3) are also components of this complex, and it remains possible that Notch might form different complexes with other transcriptional regulators.
NOTCH1 Cofactors Are Recruited to Notch-Target Genes
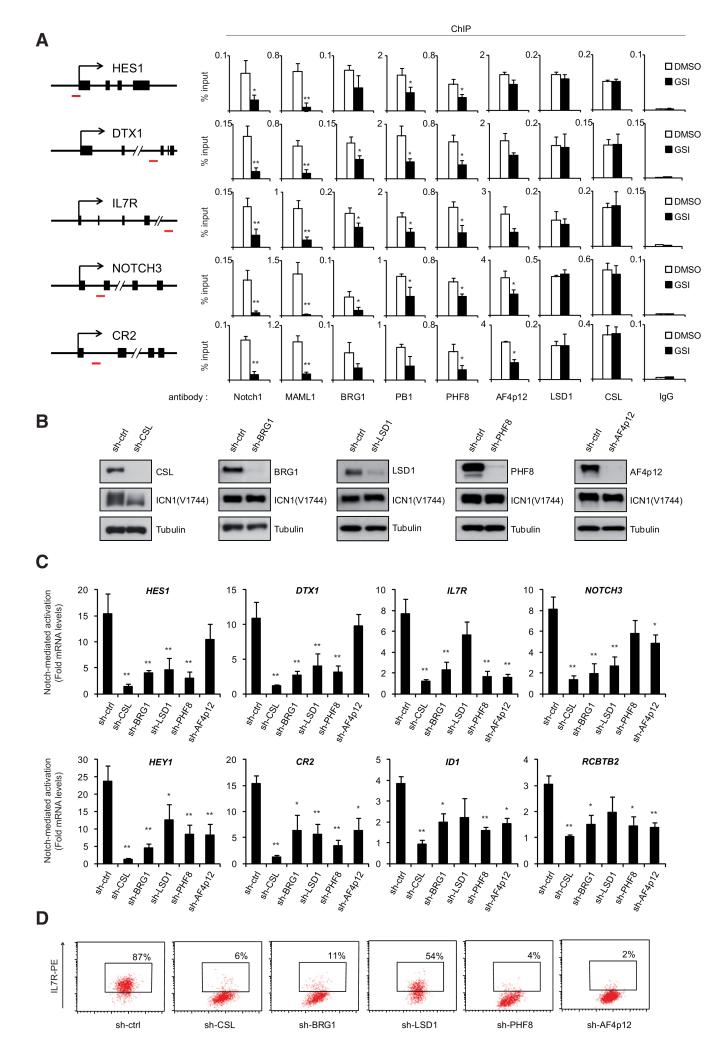
To investigate the functional relevance of ICN1-associated complex, we addressed its role in Notch-mediated transcriptional activation. First, we studied the recruitment of PBAF, LSD1, PHF8, and AF4p12 to known Notch-target genes in immature T cells (HES1, DTX1, IL7R, NOTCH3, and CR2). ICN1-binding sites were determined based on the recently published genome-wide map of NOTCH1 occupancy in T-ALL cell lines (Wang et al., 2011), and further validated by chromatin immunoprecipitation assay (ChIP) in SupT1 (Figure S2A). Chromatin prepared from SupT1 cells treated with DMSO or GSI was subjected to ChIP using the indicated antibodies. Consistent with the biochemical interactions (Figure 2), MAML1, CSL, the PBAF complex (as revealed by BRG1 and PB1 binding), PHF8, LSD1, and AF4p12 were found associated with ICN1-binding sites (Figure 3A). Importantly, GSI treatment, which released ICN1 and MAML1 from chromatin, reduced the recruitment of PBAF, PHF8, and AF4p12, but did not significantly affect CSL and LSD1 binding (Figure 3A). Accordingly, a link between LSD1 and CSL-mediated repression has been reported (Di Stefano et al., 2011; Mulligan et al., 2011; Wang et al., 2007), supporting the idea that LSD1 occupancy of Notch-target genes after GSI treatment might be mediated by its association with CSL. These results suggest that ICN1, MAML1, PBAF, PHF8, and AF4p12 are recruited to CSL-binding sites after Notch signaling activation and may associate with resident LSD1 to form a functional Notch-activation complex.
Figure 3. Notch-Associated Cofactors Are Required for Notch Transcriptional Responses.
(A) BRG1, PB1, PHF8, AF4p12, and LSD1 are recruited to Notch-target genes. Chromatin from SupT1 cells treated with DMSO or GSI (1 μM, 8 hr) was subjected to ChIP. The ICN1-binding regions (as determined in Figure S2A) were PCR amplified from the precipitated and input DNA. The position of the PCR amplicons is illustrated in the left panel (red dash). Results are expressed as percentage relative to input. Shown are means ± SD (n ≥ 3).
(B) SupT1 cells were transduced with CSL, BRG1, LSD1, PHF8, AF4p12 specific shRNAs or control shRNA (sh-ctrl). Knockdown efficiency and ICN1 expression were monitored by WB.
(C) BRG1, LSD1, PHF8, and AF4p12 regulate Notch-mediated activation of its target genes in T-ALL. SupT1 cells expressing specific shRNAs (from B) were further treated with DMSO or GSI (1 μM, 24 hr). Notch-induced expression of eight direct target genes (see also Figure S2B) was measured by quantitative RT-PCR. mRNA levels were normalized to GAPDH mRNAs and represented relative to their expression level in the absence of Notch (GSI-treated cells). Shown are means ± SD (n ≥ 3).
(D) Surface expression of IL7R on SupT1 cells 6 days after transduction with the indicated shRNA was analyzed by flow cytometry using anti-IL7RPE antibody. The x axis represents the forward scatter (cell size). The gate and percentage in the dot plots indicate IL7R-positive cells as determined relative to an irrelevant isotype-matched antibody (not shown). See also Figure S2C for IL7R expression in HPB-ALL, TALL1, and DND41. (All panels) *p < 0.05 and **p < 0.01.
PBAF, LSD1, PHF8, and AF4p12 Are Required for Notch Transcriptional Activity
We next asked whether Notch-cofactors are required for its transcriptional activity. Efficient depletion of BRG1, LSD1, PHF8, and AF4p12 in SupT1 cells did not affect ICN1 levels (Figure 3B). The eight Notch-responsive genes analyzed were all found to be directly bound by NOTCH1 and positively regulated by Notch signaling in several T-ALL cell lines (Wang et al., 2011), including SupT1 (Figure S2B). As expected, CSL depletion completely abolished Notch transcriptional activity as measured by quantitative RT-PCR (Figure 3C). Knockdown of the ATPase subunit of PBAF remodeling complex, BRG1, strongly reduced ICN1-mediated transcriptional activation of all tested genes. Moreover, knockdown of LSD1, PHF8 or AF4p12 altered the expression of Notch-responsive genes, although some targets showed a differential sensitivity to LSD1, PHF8 or AF4p12 depletion (Figure 3C). This selective requirement might depend on gene-specific features, including the chromatin context and other regulatory sequences, rather than differences in the ability of ICN1 to mediate their recruitment (Figure 3A).
IL7R expression at the cell surface plays a key role in Notch-induced T cell development and leukemia (Magri et al., 2009) (González-García et al., 2009). Thus, we analyzed several human Notch-dependent TALL cell lines that constitutively express IL7R at the cell surface (SupT1, HPB-ALL, TALL1, and DND41). We found that disruption of Notch signaling by CSL depletion severely impaired IL7R expression in SupT1, HPB-ALL, and TALL1, but not DND41 cells (Figures 3D and S2C). Consistently, knockdown of Notch-cofactors resulted in a downregulation of IL7R in SupT1, HPB-ALL, and TALL1, but did not affect Notch-independent expression of IL7R in DND41 cells (Figures 3D and S2C). This suggests that PHF8, LSD1, and AF4p12 are not general regulators of IL7R expression, but are specifically required for Notch-mediated regulation of IL7R. Overall, these results indicate that the identified cofactors of the Notch-activation complex control the expression of Notch-responsive genes.
AF4p12 Is a Notch Transcriptional Coactivator
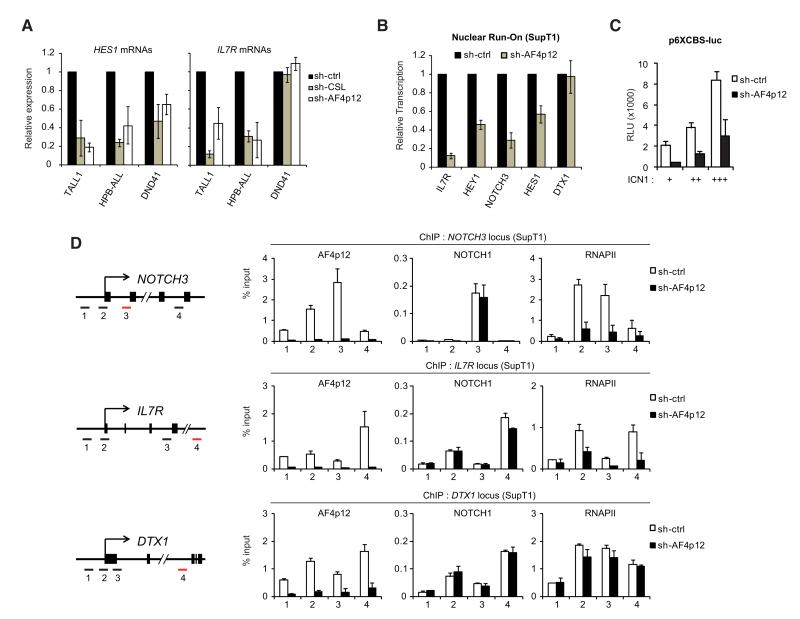
The function of AF4p12 in transcription remains largely uncharacterized (Hayette et al., 2005). Decreased expression of Notch-target genes, including HES1 and IL7R, was observed after depletion of AFp12 in several T-ALL cell lines (Figure 4A). To exclude any defect in cotranscriptional RNA processing or alteration in mRNA stability, we first measured the levels of nascent pre-mRNAs using intronic primers. AF4p12 knockdown caused a severe decrease in pre-mRNA levels of several Notch-responsive genes (Figure S3A), suggesting that AF4p12 affects the rate of transcription. To test this, we isolated nuclei from SupT1 cells expressing control or AF4p12 specific shRNA and performed nuclear run-on experiments. Analysis of transcripts generated during the run-on indicates that AF4p12 positively regulates the transcription of Notch-target genes (Figure 4B). Additionally, ICN1-mediated activation of a transiently transfected Notch-responsive luciferase reporter (p6XCBS-luc) was reduced by AF4p12 depletion (Figures 4C and S3C), supporting a role of AF4p12 in Notch transcriptional activity. Next, we determined the consequence of AF4p12 depletion on ICN1 and RNA polymerase II (RNAPII) recruitment to Notch-target genes using ChIP assay. While knockdown of AF4p12 had no effect on ICN1 recruitment, it reduced the recruitment of RNAPII to NOTCH3 and IL7R locus (Figure 4D). Consistent with the absence of a role for AF4p12 in ICN1-mediated transcription of DTX1 in SupT1 (Figure 4B), AF4p12 depletion did not affect RNAPII recruitment to the DTX locus (Figure 4D). Taken together, our results show that AF4p12 acts as a Notch coactivator and plays a role in transcriptional activation events subsequent to ICN1 recruitment that are required for RNAPII assembly at several Notch-responsive loci.
Figure 4. AF4p12 Is a Notch Transcriptional Coactivator.
(A) AF4p12 is required for Notch-target gene expression in T-ALL cell lines. TALL1, HPB-ALL, and DND41 were transduced with control, CSL or AF4p12 shRNA. Expression of HES1 and IL7R was measured by quantitative (Q-) RT-PCR (mean ± SD, n = 2). Knockdown efficiencies are shown in Figure S2D.
(B) AF4p12 affects the rate of transcription of Notch-target genes. Nuclear run-on assays (n = 3) were performed on isolated nuclei from SupT1 cells expressing control or AF4p12 shRNA. Transcripts generated during the run-on were purified using anti-BrdU beads and analyzed by Q-RT-PCR. See also Figure S3A for quantitative analysis of pre-mRNAs in AF4p12 knockdown cells.
(C) AF4p12 controls Notch transcriptional activity in transient reporter assay. HeLa cells expressing control or AF4p12 shRNA were transfected with a Notch-responsive luciferase reporter (p6XCBS-luc) and various amounts of ICN1 expression vector. The values are Relative Luciferase Units (RLU) represented as fold induction by ICN1 (mean ± SD, n = 2). Efficiency of AF4p12 knockdown is shown in Figure S3B.
(D) AF4p12 is required for RNA polymerase II (RNAPII) recruitment to Notch-target loci. ChIP assay were performed in SupT1 expressing control or AF4p12 shRNA. Results for AF4p12, NOTCH1, and RNAPII occupancy at the NOTCH3, IL7R, and DTX1 loci are shown as percentage relative to input (mean ± SD, n = 2). Numbers on the x axes represent PCR amplicons used for each locus, as depicted in the left panel.
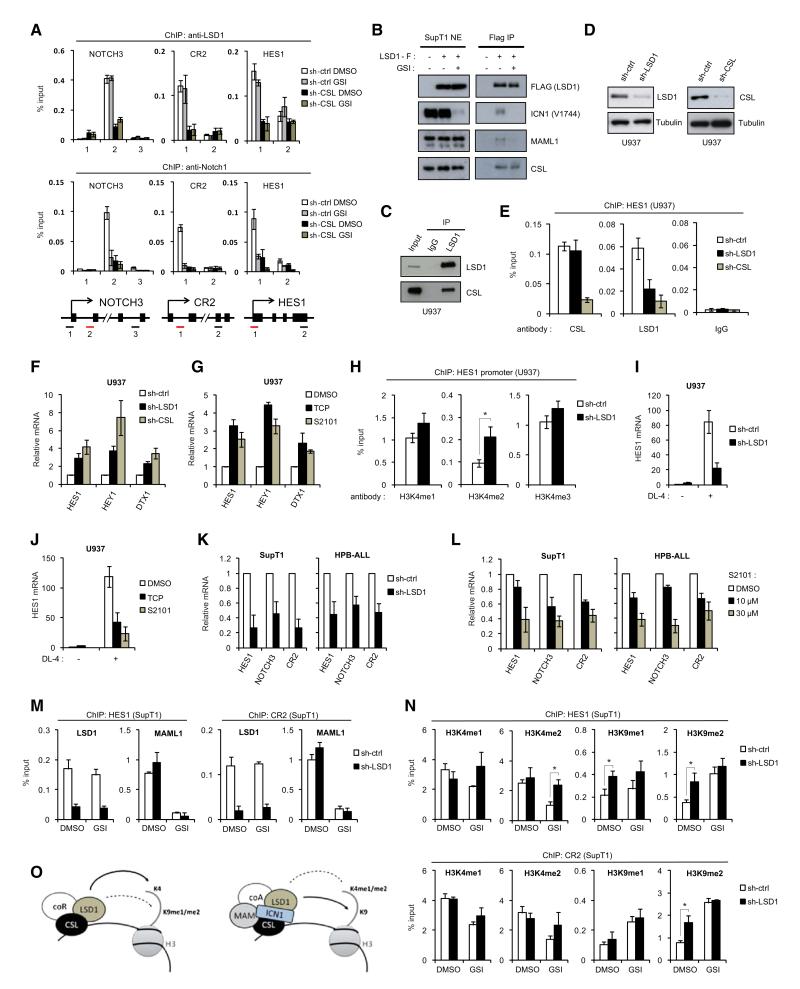
LSD1 Is a Component of the CSL-Repressor Complex and Notch-Activation Complex
In line with recent reports (Wang et al., 2007) (Mulligan et al., 2011), results shown in Figure 3A suggest that LSD1 associates with the CSL-repressor complex. Additionally, our results demonstrate that LSD1 is an integral component of the Notch-activation complex (Figure 2). We therefore reasoned that LSD1 might play a dual role in Notch signaling. First, we examined LSD1 binding to NOTCH3, CR2, and HES1 genes in SupT1 cells expressing control or CSL-specific shRNA. While Notch inactivation by GSI did not significantly affect LSD1 binding, knockdown of CSL reduced LSD1 occupancy regardless of Notch activation (Figure 5A). These results support a model in which LSD1 occupies Notch-target genes as part of the CSL-repressor complex and the Notch-activation complex. In both cases, LSD1 recruitment is CSL dependent. Next, we performed coimmunoprecipitation experiments in SupT1 cells treated with DMSO or GSI. While LSD1 binding to endogenous MAML1 and ICN1 was strongly reduced by GSI, CSL coimmunoprecipited with LSD1 under both conditions (Figures 5B and S4A). Thus, LSD1 can interact with CSL independently of ICN1. Consistently, endogenous LSD1 and CSL interact in U937 cells, a myeloid cell line that does not express a constitutively active NOTCH1 (Figure 5C). Moreover, CSL knockdown in U937 cells (Figure 5D) reduced LSD1 recruitment to the HES1 promoter (Figure 5E) and HEY1 enhancer (Figure S4B), indicating that LSD1 is a component of the DNA-bound CSL-repressor complex.
Figure 5. Opposing Role of LSD1 in the Regulation of Notch-Target Genes.
(A) LSD1 binding to Notch-target genes in the presence or absence of ICN1 requires CSL. ChIP assay were performed using chromatin prepared from SupT1 expressing control or CSL shRNA. Cells were treated with DMSO or GSI (1 μM, 24 hr). ChIP results for LSD1 and NOTCH1 at the NOTCH3, CR2, and HES1 loci are shown as percentage relative to input DNA (n = 2). Schematics of each locus and position of the amplicons are shown under the ChIP data.
(B) LSD1 is a component of both the Notch-activation complex and the CSL-repressor complex. Nuclear extracts (NEs) from FLAG-LSD1 expressing SupT1 cells treated with DMSO or GSI (500nM, 8 hr) were subjected to IP using anti-FLAG beads. The presence of ICN1, MAML1, and CSL in the IP elutions was revealed by WB. See also Figure S4A.
(C) Endogenous LSD1 was purified from U937 NEs and the presence of CSL in the eluates was revealed by WB. IP using irrelevant antibody (IgG) was used as a control.
(D) U937 cells were transduced with control, LSD1 or CSL specific shRNA. Efficiency of LSD1 and CSL knockdown was monitored by WB.
(E) U937 cells from (D) were subjected to ChIP using anti-CSL, anti-LSD1 or control (IgG) antibody. CSL and LSD1 occupancy of the HES1 promoter is shown as percentage relative to input DNA (n = 2). See also Figure S4B for HEY1 locus.
(F) LSD1 represses Notch-target genes in the absence of Notch signaling activation. Quantitative RT-PCR (Q-RT-PCR) analysis of HES1, HEY1, and DTX1 mRNA levels was performed in U937 expressing LSD1 or CSL shRNA (from D) (n = 3).
(G) U937 cells were treated with LSD1 inhibitors tranylcypromine (TCP, 1mM) and S2101 (30μM) for 8 hr. HES1, HEY1, and DTX1 mRNA levels were measured by Q-RT-PCR (n = 3). Results obtained in THP1 are shown in Figure S4C. See also Figures S4D and S4E for results in GSI-treated SupT1.
(H) H3K4 methylation at the HES1 promoter was assessed after LSD1 knockdown in U937 cells by ChIP. Data are presented as percentage of input (n = 3, *p < 0.05). See also Figure S4F for HEY1 and DTX1 loci.
(I) LSD1 is required for ligand-mediated HES1 activation. U937 cells expressing control or LSD1 shRNA were cultured on precoated DL-4 Notch ligand (5μg/mL) for 1 hr. HES1 expression was analyzed by Q-RT-PCR (n = 3). WB analyses are shown in Figure S4G. See also Figures S4H–S4J for LSD1 effects on a Notch-responsive reporter.
(J) Analysis of HES1 expression in U937 cells treated with LSD1 inhibitors (1mM TCP or 30μM S2101, for 8 hr) then cultured on DL-4 ligand for 1 hr (n = 3).
(K) LSD1 controls Notch-target gene expression in NOTCH1-dependent T-ALL cell lines. SupT1 and HPB-ALL were transduced with control or LSD1 shRNA. Expression of HES1, NOTCH3, and CR2 was measured by Q-RT-PCR (n = 3). Results obtained in TALL1 cells are shown in Figure S4K.
(L) Q-RT-PCR analysis of HES1, NOTCH3, and CR2 expression in SupT1 and HPB-ALL treated for 4 hr with DMSO or S2101 (n = 3).
(M) SupT1 cells expressing control or LSD1 shRNA were treated with DMSO or GSI and subjected to ChIP assays. Results for LSD1 and MAML1 occupancy at HES1 promoter (left panel) and CR2 enhancer (right panel) are shown as percentage of input DNA (n = 2). See also Figure S4L for CSL binding.
(N) H3K4me1/me2 and H3K9me1/me2 levels at HES1 promoter (upper panel) and CR2 enhancer (lower panel) were analyzed by ChIP using SupT1 cells described in (M) (n = 3, *p < 0.05).
(O) Model for LSD1 functions in Notch-target gene regulation. (All panels) All Q-RT-PCR were normalized to GAPDH mRNAs and are represented as mean ± SD.
LSD1 Is Required for CSL-Mediated Repression of Notch-Target Genes
We next determined the role of LSD1 in CSL-mediated repression. Similar to CSL knockdown, depletion of LSD1 in U937 cells resulted in a significant increase of HES1, HEY1, and DTX1 mRNA levels (Figure 5F). Moreover, treatment of U937 cells with LSD1 inhibitors, tranylcypromine (TCP) and S2101, also increased HES1, HEY1, and DTX1 expression (Figure 5G). A similar result was obtained in the monocytic cell line THP1 that does not express a constitutively active NOTCH1 (Figure S4C). Importantly, inhibition of LSD1 by either shRNA (Figure S4D) or S2101 treatment (Figure S4E) in GSI-treated SupT1 cells resulted in a derepression of Notch-responsive genes, while the expression of the control S14 gene was not affected. LSD1 represses transcription through H3K4me1/me2 demethylation (Shi et al., 2004). Therefore, we tested whether LSD1 depletion is associated with an increase of H3K4 methylation at Notch-target genes. While H3K4me1 and H3K4me3 were not significantly affected, we observed a significant increase in H3K4me2 levels at Notch-target loci after LSD1 depletion in U937 cells (Figures 5H and S4F). These results indicate that in the context of the CSL-repressor complex, LSD1 contributes to Notch-target gene repression by removing the activating H3K4me2 mark.
A Functional Switch of LSD1 Activity Controls Notch-Target Genes Activation
LSD1 can either suppress or promote transcription depending on its substrate (Metzger et al., 2005). Our results suggest a functional switch of LSD1 activity after Notch activation. To test this hypothesis, we cultured U937 cells expressing control or LSD1 specific shRNA in the presence of recombinant Delta-like 4 (DL-4) Notch ligand. LSD1 knockdown reduced Notch-induced HES1 transcription (Figure 5I), without affecting ligand-mediated NOTCH1 cleavage or stability (Figure S4G), suggesting a transcriptional inhibition downstream of ICN1 release. Consistently, both TCP and S2101 reduced HES1 activation (Figure 5J), indicating that the demethylase activity of LSD1 is required for HES1 induction by Notch. Knockdown of LSD1 (Figures S4H and S4I) and TCP treatment (Figure S4J) impaired ICN1-induced activation of the Notch-responsive reporter (p6XCBS-luc) in HeLa cells. These data imply that LSD1, which acts as a transcription repressor in the absence of Notch, also functions as a Notch coactivator. In support of this conclusion, knockdown of LSD1 in human NOTCH1-dependent T-ALL cell lines impaired the expression of the several Notch-dependent genes, including HES1, NOTCH3, and CR2 (Figures 5K and S4K). Importantly, inhibition of LSD1 activity by S2101 in T-ALL cells resulted in a dose-dependent repression of Notch-target genes (Figure 5L).
We next explored the mechanism underlying the functional requirement of LSD1 demethylase activity in Notch-dependent transcription. Knockdown of LSD1 in SupT1 reduced its binding to HES1 and CR2, but did not alter CSL binding or Notch-dependent MAML1 recruitment (Figures 5M and S4L), indicating a block downstream ICN1-CSL-MAML1 binding. LSD1 is reported to function as a coactivator for several transcription factors through demethylation of H3K9me1/me2 repressive marks (Metzger et al., 2005). Accordingly, LSD1 depletion significantly increased H3K9me2 levels, but not H3K4 methylation, at HES1 and CR2 gene in DMSO treated SupT1 cells (Figure 5N). In contrast, in GSI treated cells, knockdown of LSD1 did not significantly affect H3K9 methylation, but resulted in a specific increase in H3K4me2 levels (Figure 5N). Taken together, our results suggest that a critical function of LSD1 in Notch-dependent transcription is to trigger H3K9me2 demethylation. However, in the absence of Notch, LSD1 acts preferentially on H3K4me2 and contributes to CSL-mediated repression (Figure 5O).
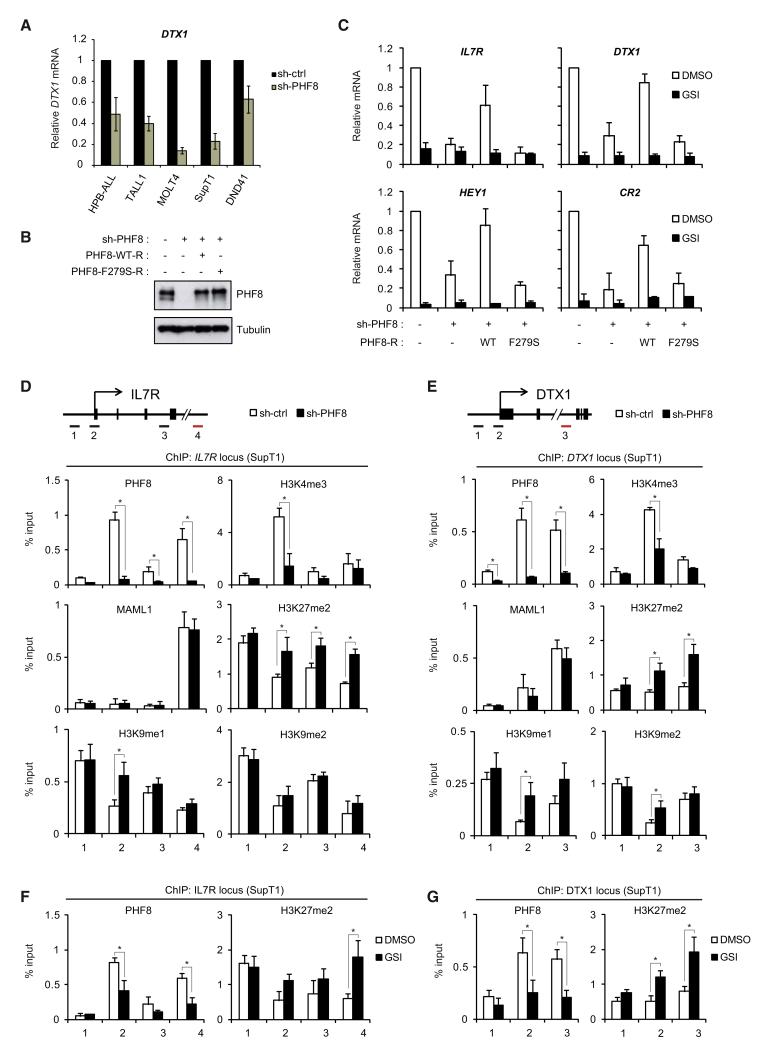
PHF8 Demethylase Activity Is Required for Notch-Mediated Activation of Its Target Genes
Consistent with the results obtained in SupT1 cells (Figure 3), knockdown of PHF8 in several T-ALL cell lines decreased the expression of Notch-responsive genes (Figures 6A and S5A). Moreover, depletion of PHF8 in HeLa cells impaired ICN1-mediated activation of the p6XCBS-luc reporter (Figure S5B), indicating that PHF8 acts as a transcriptional coactivator of Notch.
Figure 6. PHF8 Demethylase Activity Is Required for Notch-Mediated Activation of Its Target Genes.
(A) PHF8 is required for Notch-responsive gene expression in T-ALL cell lines. HPB-ALL, TALL1, MOLT4, SupT1 and DND41 were transduced with control or PHF8 shRNA. Expression of DTX1 was measured by quantitative RT-PCR (Q-RT-PCR) and normalized to GAPDH. See also Figure S5A for other Notch-responsive genes.
(B) SupT1 expressing PHF8 shRNA were transduced with shRNA-resistant wide-type (WT) or catalytic inactive (F279S) PHF8 mutant. Endogenous and ectopically expressed PHF8 levels were determined by WB.
(C) mRNA levels of IL7R, DTX1, HEY1, and CR2 were measured by Q-RT-PCR in SupT1 cells coexpressing PHF8 shRNA and the indicated PHF8 construct (from B). Shown are means ± SD (n = 3).
(D and E) ChIP assays for PHF8, MAML1, H3K4me3, H3K27me2, H3K9me1, and H3K9me2 at the IL7R locus (C) and DTX1 locus (D) were performed in SupT1 cells expressing control or PHF8 shRNA (shown as percentage relative to input DNA, n = 3, *p < 0.05). The upper panel depicts the locus structure and positions of the PCR amplicons. See also Figure S5C for H4K20me1 levels. H3K27me2/me3 ChIP results at other Notch-target loci are shown in Figures S5D and S5E.
(F and G) ChIP assays were performed in SupT1 cells treated with DMSO or GSI. Results for PHF8 and H3K27me2 at the IL7R locus (E) and DTX1 locus (F) are shown. ChIP primers described in (D) and (E) were used (n = 3, *p < 0.05). See also Figure S5B for PHF8 effects on a Notch-responsive reporter.
PHF8 removes multiple transcriptional repressive marks, including H3K9me1/me2, H4K20me1, and H3K27me2 (Horton et al., 2010; Liu et al., 2010b; Loenarz et al., 2010; Qi et al., 2010). To determine whether PHF8 demethylase activity is required for Notch-target gene activation, we performed rescue experiments in SupT1 by expressing shRNA-resistant PHF8 or the catalytically inactive F279S mutant (Figure 6B). As shown in Figure 6C, the expression of Notch-responsive genes was restored by wide-type PHF8 but not by the inactive mutant. These data indicate that PHF8 controls Notch transcriptional responses through its demethylase activity.
To investigate histone marks regulated by PHF8 at Notch target loci, we performed ChIP experiments in SupT1. Analysis of PHF8-binding at the IL7R locus (Figure 6D) and DTX1 locus (Figure 6E) showed that PHF8 peaks at ICN1-containing enhancers but also at the transcription start site (TSS), consistent with its ability to bind H3K4me3. In agreement with its function as a transcriptional coactivator, PHF8 depletion reduced H3K4me3 levels at the IL7R and DTX1 TSS. An increase of H3K9me1 levels, and to a lesser extent H3K9me2 levels, was observed only at the TSS-region (primer 2 for IL7R and DTX1), but not at ICN1-binding region (primer 4 for IL7R and primer 3 for DTX1). Moreover, loss of PHF8 did not lead to any detectable increase in H4K20me1 levels (Figure S5C). In contrast, while PHF8 depletion did not affect total H3 level (data not shown), it caused an accumulation of the repressive H3K27me2 mark at the two tested loci (Figures 6D and 6E), suggesting that it is actively removing this mark. Consistently, upon knockdown of PHF8, we observed a robust increase of H3K27me2 levels, but not H3K27me3, at other Notch-target genes (including HES1, CR2, and NOTCH3) (Figures S5D and S5E). These results suggest that in the context of the Notch-activation complex, PHF8 may control Notch responses by removing H3K27me2. In agreement with this model, reduction in PHF8 recruitment after Notch signaling inhibition by GSI was accompanied by an increase of H3K27me2 mark at both IL7R (Figure 6F) and DTX1 loci (Figure 6G).
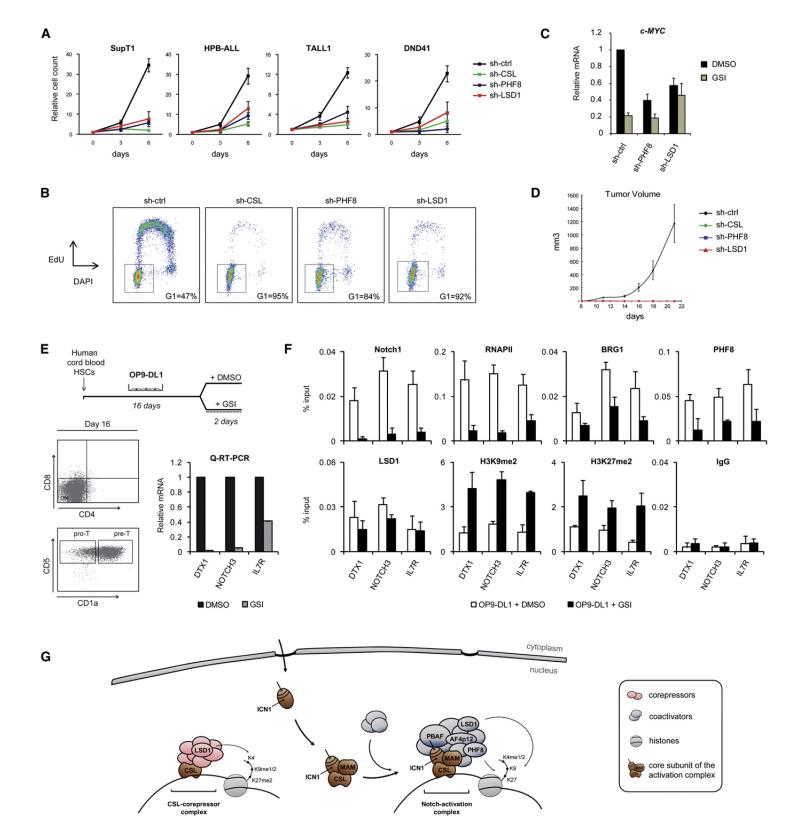
LSD1 and PHF8 Are Required for NOTCH1-Dependent T-ALL Cell Proliferation and Tumor Growth
Previous studies have shown that inhibition of the Notch pathway induces cell-cycle arrest and alters growth capacities of NOTCH1-dependent leukemia cells (Weng et al., 2004). These observations prompted us to assess the role of LSD1 and PHF8 in NOTCH1 oncogenic functions. Abrogation of Notch signaling by depletion of CSL, PHF8, or LSD1 in a panel of T-ALL cell lines (SupT1, HPB-ALL, TALL1, and DND41) resulted in a marked reduction in their proliferation (Figure 7A). Importantly, knockdown of CSL, PHF8, or LSD1 induced a G0/G1 cell-cycle arrest in all NOTCH1-dependent T-ALL cells tested (Figure 7B and S6A), but did not significantly affect cell-cycle progression in MOLT4 and H9 (Figure S6A), which are Notch-independent T-ALL cell lines (Weng et al., 2004). Depletion of PHF8 and LSD1 suppressed Notch-mediated expression of c-MYC (Figure 7C), a gene that has a key role in tumorigenesis induced by NOTCH1, which further support their role in the oncogenic activity of Notch signaling. Consistent with this, T-ALL cells expressing CSL, PHF8 or LSD1 shRNA did not establish tumors in a mouse xenograft model (Figures 7D and S6B). These results indicate that LSD1 and PHF8 are key regulators of NOTCH1 oncogenic functions in T-ALL cells and support the idea that targeted therapies interfering with PHF8 or LSD1 activities could be useful in the treatment of cancers that have activated NOTCH1 alleles.
Figure 7. Functional Relevance of Notch Cofactors in T-ALL Proliferation and Gene Expression during T Cell Development.
(A) LSD1 and PHF8 are required for NOTCH1-dependent T-ALL cells growth. SupT1, HPB-ALL, TALL1, and DND41 cells were transduced with control, CSL, PHF8 or LSD1 specific shRNA. One week posttransduction, cell count proliferation assays were performed (n = 3). The observed effects of CSL, PHF8, and LSD1 depletion is significant (p < 0.05).
(B) Effects of PHF8 and LSD1 depletion on cell cyle progression. SupT1 cells expressing control, CSL, PHF8 or LSD1 shRNA were pulse labeled with EdU for 2 hr. DNA synthesis (EdU incorporation, y axis) and total DNA content (DAPI staining, x axis) was measured by flow cytometry. The gate and percentage indicate cells in G0/G1 phase. See also Figure S6A for CSL, LSD1, and PHF8 depletion effects on cell cycle in a panel of NOTCH1-dependent or independent T-ALL cell lines.
(C) Notch-mediated expression of c-MYC in T-ALL cells requires PHF8 and LSD1. Quantitative RT-PCR analysis of c-MYC expression was performed in TALL1 expressing control, PHF8 or LSD1 shRNA and treated with DMSO or GSI (n = 3).
(D) Depletion of PHF8 and LSD1 impairs T-ALL progression in vivo. SupT1 expressing control, CSL, LSD1 or PHF8 shRNA were implanted subcutaneously in SCID mice (n = 5). Xenograft tumor volume was monitored over 21 days. See also Figure S6B.
(E) Hematopoietic stem cells (HSCs) from human cord blood were induced to differentiate to the T cell lineage in OP9-DL1 cocultures. T cell precursors (pre-T) were further treated with DMSO or GSI (upper panel). Cell surface expression of CD4, CD8, CD5, and CD1a at day 16 was analyzed by flow cytometry (Dot plots, lower panel). Expression of DTX1, NOTCH3, and IL7R was analyzed by quantitative RT-PCR on total RNA extracted from DMSO or GSI-treated pre-T (Graphs, lower panel).
(F) Notch cofactors are recruited to target genes during T cell development. ChIP experiments were performed in pre-T from (E) using the indicated antibodies. ICN1-binding region at the DTX1, NOTCH3, and IL7R locus was PCR amplified from the precipitated and input DNA. An irrelevant antibody (IgG) was used as a control. Shown are means ± SD (n = 2).
(G) Model for the regulation of Notch-target genes.
Notch Cofactors Are Recruited to Notch-Target Genes during T Cell Development
Notch signaling pathway plays a key role during T cell development. To investigate whether Notch cofactors identified in leukemic T cells are recruited to Notch-target genes during physiological T lymphopoiesis, we performed ChIP experiments using an in vitro T cell differentiation system that allows analysis of early stages (Schmitt and Zúñiga-Pflücker, 2002). Hematopoietic stem cells (HSCs) from human umbilical cord blood were cultured for 16 days on OP9-DL1 cells, a bone-marrow stromal cell line that ectopically expresses the Notch ligand DL1, then DMSO or GSI was added for 2 additional days (Figure 7E). The coculture induced the expression of several Notch-target genes (including DTX1, NOTCH3, and IL7R) that was accompanied by differentiation and proliferation of the progenitors (not shown). At day 16 of culture, most precursors were committed to the T cell lineage, as indicated by the acquisition of CD1a (Figure 7E). ChIP assays confirmed the binding of ICN1 to the DTX1, NOTCH3, and IL7R enhancers in T cell precursors (Figure 7F). Consistent with the Q-RT-PCR analysis (Figure 7E), GSI treatment impaired RNAPII recruitment to Notch-target genes (Figure 7F). Importantly, several components of the Notch-activation complex, including BRG1, PHF8, and LSD1 were associated with ICN1-containing enhancers. As in T-ALL cells, binding of BRG1 and PHF8, but not LSD1, requires activation of Notch signaling (Figure 7F). Furthermore, we detected an accumulation of the repressive marks H3K9me2 and H3K27me2 after turning off Notch signaling (Figure 7F). This is consistent with LSD1 and PHF8 demethylase activities. These results suggest that ICN1 might trigger the formation of a similar activation complex in developing T cell and their malignant counterpart.
DISCUSSION
Notch signaling is involved in virtually all developmental processes and implicated in many human diseases including T cell lymphoblastic leukemia. Here we identified nuclear ICN1- partners in human T cell leukemia providing a framework to elucidate ICN1 regulation and mechanisms of action.
Insights into the Molecular Mechanisms Involved in Notch-Dependent Transcription
Upon Notch activation, ICN1 directs the formation of the ternary ICN1-CSL-MAML1 complex required for Notch transcriptional responses. Here we expand our understanding of Notch-mediated transcription by identifying components of the Notch-activation complex. An important finding from this study is that Notch activation leads to the assembly of a large multisubunit complex containing ICN1-CSL-MAML1 and several classes of transcriptional regulators that could act at different steps of the transcriptional activation process. The control of gene expression through chromatin requires proteins that enzymatically regulate nucleosomal structure and histone modifications. The SWI/SNF remodeling complex PBAF was found here to interact with the core Notch-activation complex. Importantly, the catalytic subunit of PBAF, BRG1, is required for endogenous Notch-target gene expression in T-ALL cells. This finding is consistent with the previously reported role of BRG1 in Notch signaling during mouse embryonic development (Takeuchi et al., 2007).
Our data also uncovers an important function for AF4p12 in Notch-mediated transcriptional activation. AF4p12 is a poorly characterized protein that was first identified as one of the MLL translocation partners in leukemia (Hayette et al., 2005). Importantly, its C-terminal part displays transcription activation properties when fused to GAL4 DNA-binding domain (Hayette et al., 2005). While the precise role of AF4p12 awaits further investigation, our results indicate that it is required for RNAPII recruitment at several Notch-target genes. AF4p12 therefore acts as a transcriptional coactivator of ICN1.
Epigenetic Regulation of Notch-Target Genes
Our results provide strong evidence indicating that histone modifications play a central role in Notch-target gene regulation. Several histone modifiers, including the BRE1 subunit RNF40, and the demethylases LSD1 and PHF8, were detected by MS. Interestingly, the homolog of BRE1 is required for Notch activity in Drosophila (Bray et al., 2005), suggesting that H2B monoubiquitination might play a conserved function in Notch signaling. In the current study, we focused on the role of LSD1 and PHF8 in modulating Notch responses.
Previous studies in various species have linked LSD1 to the repression of Notch-target genes (Di Stefano et al., 2011; Mulligan et al., 2011; Wang et al., 2007). In agreement with these observations, we found that LSD1 is bound to chromatin as part of the CSL-repressor complex and prevents transcription by maintaining low levels of H3K4me2. However, our study reveals a transcriptional coactivator function for LSD1 in the context of the Notch-activation complex. Indeed, we show that LSD1 is required for Notch-mediated activation of its target genes by insuring efficient H3K9me2 demethylation. Consistently, mutant alleles of Drosophila LSD1 suppressed gain-of-function phenotypes of Notch (Di Stefano et al., 2011), suggesting a conserved role of LSD1 in the activation of Notch signaling. What are the mechanisms that govern alteration of LSD1 activity after Notch activation? First, LSD1 substrate specificity could be modulated as a result of CSL complex remodeling. Second, histone tail modifications may play a role. Accordingly, in vitro demethylation of H3K4 by LSD1 is completely blocked by Ser10 phosphorylation (Forneris et al., 2005), a mark that is strongly increased after ICN1 recruitment (Fryer et al., 2004). Thus, as a consequence of Notch activation, a sequence of events that remains to be determined, including remodeling of protein complexes and histone marks, might modify LSD1 catalytic activity.
Similarly to LSD1, the PHD finger- and JmjC-containing histone demethylase, PHF8, exhibits differential substrate specificities in different contexts. For example, PHD-mediated binding to adjacent H3K4me3 is required for H3K9me1/2 but not H3K27me2 demethylation (Horton et al., 2010; Liu et al., 2010b). In the context of Notch responses, PHF8 recruitment to ICN1-containing enhancers is associated with a robust demethylation of H3K27me2. Our results also reveal an additional activity of PHF8 toward lysine 9 at the TSS region, probably mediated through its interaction with H3K4me3. Because most Notch-responsive genes lack ICN1 binding sites at the promoter region and are regulated through enhancers (containing low levels of H3K4me3) (Wang et al., 2011), the major PHF8 substrate is likely H3K27me2. Thus, ICN1 recruitment to Notch-responsive enhancers is associated with at least two demethylase activities: H3K9me1/me2 demethylation by LSD1 and H3K27me2 by PHF8. However, in some cases, both LSD1 and PHF8 might contribute to Notch-induced demethylation of H3K9me1/2. Additionally, PHF8 and LSD1 may function by targeting other substrates, including nonhistone proteins.
Additional Partners Involved in Notch Transcriptional Activity?
Other ICN1-interacting proteins identified in this study may also associate with the Notch-activation complex and play nonredundant functions in Notch-target gene activation (see Table S3). Some of these factors, such as the corepressor/coactivator exchange factor TBLR1 or the poorly characterized nuclear protein ERH, have been described as positive regulators of Notch signaling (see Table S2). Thus, further characterization of additional partners will undoubtedly help to complete our understanding of Notch functions. Moreover, factors involved in transcriptional repression, including the NuRD complex and subunits of the polycomb repressive complex 1 (PRC1), copurified with ICN1 (Table S1). Since the primary function of ICN1 is to activate transcription, investigations on the biological significance of these interactions might reveal important aspects of Notch signaling.
Shaping Notch Transcriptional Responses
Our study indicates that ICN1 interacts with other transcription factors (TFs) and signaling pathways in T-ALL cells. Notably, most of the identified ICN1-associated TFs (such as BCL11B, HEB, RUNX1, and IKAROS) are lineage-specific master regulators of T cell differentiation (Rothenberg and Taghon, 2005) and were reported to co-operate with Notch signaling (Table S2). We therefore speculate that co-operativity between ICN1 and master TFs promotes the expression of a cell-type specific subset of Notch-target genes. Conversely, combinatorial interactions between ICN1 and transcriptional repressors (such as IKAROS and BCL11B) could restrict the repertoire of Notch-activated genes in T cells by selectively inhibiting the expression of co-occupied enhancers.
Strikingly, several TFs found associated with ICN1 have tumor suppressive functions in T cells. For instance, defects in Ikaros, Runx1 or Bcl11b lead to the development of T cell leukemia in mice (Kundu et al., 2005; Wakabayashi et al., 2003). Moreover, mutations in IKAROS, RUNX1, and BCL11B have been identified in human T-ALL (Zhang et al., 2012). Therefore, these interactions might trigger a Notch transcriptional program supporting differentiation rather than cell growth.
ICN1 is able to form homodimers and the dimerization-defective NOTCH1 mutant fails to induce leukemia (Liu et al., 2010a). Because residues that mediate dimerization are conserved in all paralogues, the presence of NOTCH2 and NOTCH3 peptides in the ICN1-purified material may result from direct interactions between paralogues. Accordingly, knockdown of NOTCH3 expression has been recently reported to alter the growth of NOTCH1-dependent T-ALL cells, including SupT1 cells (Xiang et al., 2012), allowing one to speculate that heterodimerization may play a role in Notch-induced tumorigenesis.
Regulation of NOTCH1 by Posttranslational Modifications
Posttranscriptional modifications (PTMs) regulate several aspects of Notch biology including ICN1 trafficking and transcriptional activity (Andersson et al., 2011). Moreover, the signal strength and duration are tightly regulated by various PTMs that contribute to fine-tuning ICN1 half-life. For instance, phosphorylation of the PEST domain by various kinases such as CDK8 (Fryer et al., 2004) creates a recognition site for FBW7 E3-ligase that targets ICN1 for ubiquitination and proteosomal degradation. Remarkably, various kinases, phosphatases, and E3-ligases including FBW7 were recovered in ICN1 purified material (Table S1). Several kinases identified, such as ERK2, ROCK1, and AMPK, may link Notch to other signaling or metabolic pathways. It will be important to determine if ICN1 is the substrate and how these phosphorylations impact Notch signaling, including ICN1 turnover.
The most common mechanisms for deregulation of Notch pathway in human cancers affect proteasome-mediated degradation of ICN1. These include deletion of the PEST domain and inactivation of FBW7 (Aifantis et al., 2008; South et al., 2012). Beside FBW7, ICN1 may also be the substrate for other E3 ligases or deubiquitinating enzymes identified in this study (Table S1). An important question is whether deregulation of these interactions may contribute to ICN1 stabilization and tumorigenesis. Further characterization of these interactions will likely uncover mechanisms of fine-tuning Notch signaling and may open therapeutic opportunities.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures can be found in supplemental information.
ChIP Assays
Chromatin prepared from cross-linked cells was sonicated and subjected to ChIP using specific antibodies prebound to magnetic beads. Antibodies and primers are described in the Supplemental Information.
Nuclear Run-On
5×105 nuclei isolated from SupT1 cells were incubated with ATP, CTP, GTP, and br-UTP (500μM each) in the presence of 0,5% sarkosyl. The reaction was performed as previously described (Core et al., 2008).
Xenograft Tumor Model of T-ALL
Female SCID mice were subcutaneously inoculated with human T-ALL cells. 5×106 SupT1 cells expressing ctrl, CSL, PHF8 or LSD1 shRNA were resuspended in 100μL DMEM: 50% Matrigel and injected to each mouse by subcutaneous route (n = 5 per group). Tumor volume was evaluated three times a week during the follow-up period (23 days).
In Vitro T Cell Differentiation
CD34+ cells from human umbilical cord blood samples were sorted (Stemcell technologies) (≥95% purity) and cultured on OP9-DL1 stromal cells in αMEM media containing FLT3L (5ng/ml), and IL7 (5 ng/ml) for 16 days. At day 16, most progenitors (~90%) were engaged to the T cell lineage (pre-T cells) as determined by the expression of CD5 and CD1a. pre-T cells were cultured for 2 additional days on OP9-DL1 cells in presence or absence of GSI (5 μM).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Molecular Virology Lab for critical reading of the manuscript and E. Six, A.B. Lasser, J.C. Aster, A. Israel, T.M. Kristie, Y. Shi, and H. Qi for sharing reagents. This work was supported by grants from the ERC (250333), ANR-BLAN-0040, and FRM “équipe labéllisée” to M.B. A.Y. was supported by MESR, École de l’INSERM-Liliane Bettencourt, and FRM fellowships. S.L.C. was supported by ERC.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures, three tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2012.08.022.
REFERENCES
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev. Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Walker JA, Burgio G, Corona DF, Mulligan P, Näär AM, Dyson NJ. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011;25:17–28. doi: 10.1101/gad.1983711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- González-García S, García-Peydró M, Martín-Gayo E, Ballestar E, Esteller M, Bornstein R, de la Pompa JL, Ferrando AA, Toribio ML. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7Ralpha gene expression in early human thymopoiesis and leukemia. J. Exp. Med. 2009;206:779–791. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayette S, Cornillet-Lefebvre P, Tigaud I, Struski S, Forissier S, Berchet A, Doll D, Gillot L, Brahim W, Delabesse E, et al. AF4p12, a human homologue to the furry gene of Drosophila, as a novel MLL fusion partner. Cancer Res. 2005;65:6521–6525. doi: 10.1158/0008-5472.CAN-05-1325. [DOI] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Compton S, Garrett-Beal L, Stacy T, Starost MF, Eckhaus M, Speck NA, Liu PP. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood. 2005;106:3621–3624. doi: 10.1182/blood-2005-04-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chi AW, Arnett KL, Chiang MY, Xu L, Shestova O, Wang H, Li YM, Bhandoola A, Aster JC, et al. Notch dimerization is required for leukemogenesis and T-cell development. Genes Dev. 2010a;24:2395–2407. doi: 10.1101/gad.1975210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010b;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum. Mol. Genet. 2010;19:217–222. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri M, Yatim A, Benne C, Balbo M, Henry A, Serraf A, Sakano S, Gazzolo L, Lévy Y, Lelièvre JD. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur. J. Immunol. 2009;39:1231–1240. doi: 10.1002/eji.200838765. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell. 2011;42:689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu. Rev. Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin. Cell Dev. Biol. 2012;23:458–464. doi: 10.1016/j.semcdb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc. Natl. Acad. Sci. USA. 2007;104:846–851. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, Mishima Y, Niwa O, Kominami R. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem. Biophys. Res. Commun. 2003;301:598–603. doi: 10.1016/s0006-291x(02)03069-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Xiang J, Ouyang Y, Cui Y, Lin F, Ren J, Long M, Chen X, Wei J, Zhang H. Silencing of Notch3 Using shRNA driven by survivin promoter inhibits growth and promotes apoptosis of human T-cell acute lymphoblastic leukemia cells. Clin Lymphoma Myeloma Leuk. 2012;12:59–65. doi: 10.1016/j.clml.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.