Abstract
MicroRNAs (miRNAs) are a class of small, noncoding RNAs that function by regulating gene expression post-transcriptionally. Alterations in miRNA expression can strongly influence cellular physiology. Here we demonstrated cross-regulation between two components of the RNA interference (RNAi) machinery in human cells. Inhibition of exportin-5, the karyopherin responsible for pre-miRNA export, downregulated expression of Dicer, the RNase III required for pre-miRNA maturation. This effect was post-transcriptional and resulted from an increased nuclear localization of Dicer mRNA. In vitro assays and cellular RNA immunoprecipitation experiments showed that exportin-5 interacted directly with Dicer mRNA. Titration of exportin-5 by overexpression of either pre-miRNA or the adenoviral VA1 RNA resulted in loss of Dicer mRNA–exportin-5 interaction and reduction of Dicer level. This saturation also occurred during adenoviral infection and enhanced viral replication. Our study reveals an important cross-regulatory mechanism between pre-miRNA or viral small RNAs and Dicer through exportin-5.
miRNAs are single-stranded RNA of 19–24 nucleotides that are predicted to regulate up to 30% of protein-encoding genes. miRNA have been implicated in a vast array of cellular processes including cell differentiation, proliferation and apoptosis1. miRNA repertoires are highly cell type specific and change markedly during development or upon cell activation2. Changes in miRNA expression profile have been linked to human pathologies such as cancer and neurodegenerative diseases3. In the nucleus, primary RNA polymerase II transcripts (pri-miRNA) containing imperfect hairpin structures are cleaved by the microprocessor complex, composed of the ribonuclease (RNase) III Drosha and its RNA-binding partner DGCR8. The resulting processed stem-loop structure of ~65 nucleotides, called a pre-miRNA, is recognized by exportin-5 (XPO5)4-6 and transits to the cytoplasm, where it is then cleaved by the ATP-dependent RNase III Dicer. Dicer generates a small duplex of 19–24 bp containing mismatches, called miRNA/miRNA*. The guide miRNA strand is incorporated into the RNA-induced silencing complex (RISC) containing the Argonaute proteins (Ago1–Ago4) as core components, while the miRNA* is degraded. Ago-bound miRNA serves as a guide to specifically recognize cellular mRNA so as to either induce their degradation and/or inhibit their translation7.
Dicer is a key component of the miRNA pathway. Even though it is well known that Dicer is ubiquitously expressed among many cell types, deregulation of its expression may result in miRNA biogenesis defects in specific cellular contexts. Recently, several studies have reported a modulation of Dicer expression in cancer cells. This regulation can be transcriptional: upon melanocyte differentiation, Dicer expression is induced through the binding of master transcriptional regulator MITF on Dicer promoter8. Dicer regulation can also be post-transcriptional. Dicer mRNA can be targeted by specific miRNAs: in human lung cancers, let-7 miRNA is upregulated and targets Dicer 3′ UTR region9; in breast cancers, miR-103/107 family expression is associated with metastasis and a poor outcome in patients, as these miRNAs target and inhibit Dicer expression, causing a global miRNA downregulation10.
Here, we investigate the regulation of Dicer expression in human cells. We describe a new regulatory mechanism for Dicer expression involving competition for XPO5 binding between Dicer mRNA and short hairpin RNA such as pre-miRNA, shRNA or viral RNA.
RESULTS
Exportin-5 controls Dicer expression post-transcriptionally
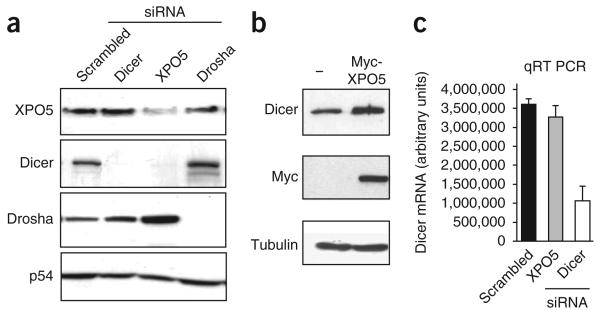
In an attempt to deplete the cellular miRNA content, we separately knocked down three key components of the miRNA pathway in HeLa cells—XPO5, Dicer and Drosha—using sequence-specific siRNAs. As expected, the targeted proteins were specifically inhibited by their respective siRNA (Fig. 1a); surprisingly, however, Dicer expression was also abolished upon transfection of siRNA targeting XPO5 (siXPO5; Fig. 1a). We ruled out an off-target effect because Dicer levels were similarly reduced after transfection of three additional siRNA designed against other regions of XPO5 messenger (Supplementary Fig. 1a). Moreover, this effect was specific to Dicer because siXPO5 had no effect on Drosha or on the P-body component DDX6; Fig. 1a). Conversely, XPO5 overexpression increased the level of cellular Dicer (Fig. 1b), consistent with previously reported enhancement of RNA interference in XPO5-overexpressing cells11. To assess whether XPO5 controls Dicer expression at the mRNA level, we analyzed transcript levels by quantitative RT-PCR (qRT-PCR; Fig. 1c) and northern blotting (Supplementary Fig. 1b) in the samples shown in Figure 1a. Dicer mRNA quantification was assessed using three sets of primers covering different regions of the messenger (Supplementary Fig. 1c). As expected, Dicer mRNA levels were reduced in cells treated with Dicer-specific siRNA. However, they remained unaffected upon siXPO5 treatment (Fig. 1c and Supplementary Fig. 1c). Together these results show that XPO5 regulates Dicer expression post-transcriptionally.
Figure 1.
Regulation of Dicer protein level by XPO5. (a) Expression of XPO5, Dicer, Drosha and DDX6 in HeLa cells transfected with control scrambled (Scr) or specific siRNA, as determined by western blotting. (b) Expression of Dicer, Myc-XPO5 and tubulin in HeLa cells transfected with either Myc-XPO5 or empty expression vectors, as determined by western blotting. (c) Dicer mRNA levels of samples described in a, as analyzed by qRT-PCR after total RNA extraction. Error bars, s.d.
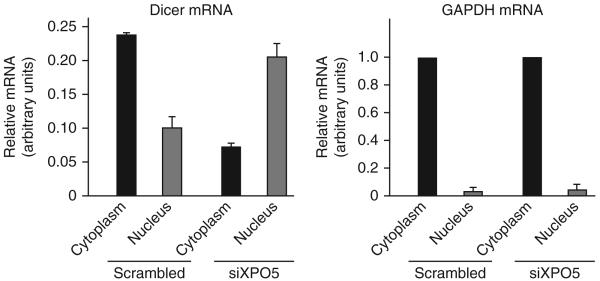
XPO5 is a member of the karyopherin family that is required for the transport from nucleus to cytoplasm of specific classes of small RNAs4,5,12-14 and double-stranded RNA–binding proteins15,16. XPO5 docks and translocates specific RNA cargoes through the nuclear pore complex, in an export process dependent on a RanGTP-RanGDP gradient across the nuclear membrane17. Only three classes of cellular RNA—tRNA, Y-RNA and miRNA precursors (pre-miRNAs)—are known to transit through XPO5. Small nuclear (sn) RNAs (U1–U5) and some cellular RNAs containing AU-rich elements depend on CRM1 for nuclear export or maturation, whereas tRNA is exported mainly through exportin-t (reviewed in refs. 17,18). In contrast, cellular mRNAs transit as ribonucleoprotein complexes through the TAP–p15 (also called NXF1–NXT1) complex in a RanGTP-independent manner. To test whether XPO5 knockdown compromises the nucleocytoplasmic distribution of Dicer mRNA, we extracted total mRNA from the cytoplasm or the nucleus19. We assayed levels of Dicer transcripts by qRT-PCR in siXPO5-treated cells and in control cells treated with scrambled siRNA. The analysis revealed that knockdown of XPO5 causes accumulation of Dicer mRNA in the nucleus (Fig. 2). By contrast, the subcellular distribution of GAPDH mRNA, which transits through the Tap-p15 pathway, remained unchanged.
Figure 2.
XPO5 knockdown results in accumulation of Dicer mRNA in the nucleus. Quantification of Dicer mRNA, U6 small nuclear RNA and GAPDH mRNA by qRT-PCR in nuclear and cytoplasmic fractionated RNA from HeLa cells transfected with XPO5-specific siRNA or control scrambled siRNA. Cytoplasmic RNAs were normalized to GAPDH mRNA and nuclear RNAs to U6 snRNA. Results are expressed in arbitrary units and are representative of three independent experiments. Error bars, s.d.
Dicer mRNA interacts with exportin-5 in vivo and in vitro
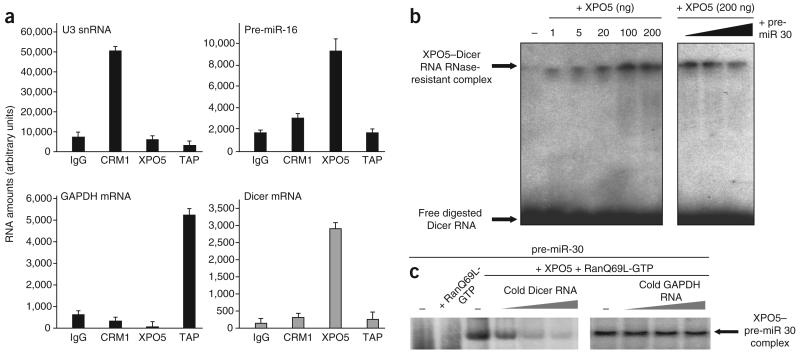
On the basis of these results, we hypothesized that XPO5 could be the cellular karyopherin responsible for Dicer mRNA nuclear export. To analyze whether Dicer mRNA interacts with XPO5 in vivo, we developed an immunoprecipitation assay to specifically recover RNA associated with each of the three major cellular export pathways (Supplementary Fig. 2). XPO5 interactions reported to date are RanGTP dependent14,20,21. Thus, in order to increase and stabilize specific interactions, we added nonhydrolyzable RanQ69L-GTP to the immunoprecipitation buffer. We then subjected selected RNAs associated with CRM1, XPO5 or TAP–p15 immunoprecipitates (Fig. 3a) to qRT-PCR analysis. As expected, U3, a small nucleolar (sno) RNA whose intranuclear transport to the nucleolus depends on CRM1 (ref. 22), was specifically immunoprecipitated with CRM1 (Fig. 3a, top left), pre-miR-16 with XPO5 (Fig. 3a, top right), and GAPDH mRNA with TAP–p15 (Fig. 3a, lower left). Dicer mRNA was specifically found in XPO5 immunoprecipitates (Fig. 3a, lower right), indicating that Dicer mRNA interacts with XPO5 in vivo. Electrophoretic mobility shift assays (EMSA) provided in vitro confirmation for a direct Dicer mRNA–XPO5 interaction20. We incubated in vitro–synthesized radiolabeled Dicer mRNA with increasing amounts of recombinant XPO5 and RanQ69L-GTP and followed this by treatment with RNaseT1 and RNase A. Analysis by nondenaturing acrylamide gel electrophoresis showed that Dicer mRNA was protected in a XPO5 dose–dependent manner (Fig. 3b). Notably, preincubation of XPO5 with increasing amounts of unlabeled pre-miR-30, a known substrate for XPO5, decreased the amount of protected Dicer mRNA in a dose-dependent manner (Fig. 3b), demonstrating the specificity of Dicer mRNA and XPO5 interaction. Conversely, preincubation of XPO5 with increasing amounts of unlabeled Dicer mRNA inhibited pre-miR-30–XPO5 complex formation in a dose-dependent manner (Fig. 3c). This effect was specific to Dicer mRNA, as the same amounts of GAPDH mRNA had no effect (Fig. 3c). As previously described for XPO5 partners, the interaction between Dicer mRNA and XPO5 depended on the presence of RanQ69L-GTP both in vivo (Supplementary Fig. 3a) and in vitro (Supplementary Fig. 3b). Taken together, these experiments strongly suggest that XPO5 is the karyopherin responsible for the nuclear export of Dicer mRNA.
Figure 3.
Dicer mRNA specifically interacts with XPO5 in vivo and in vitro. (a) HeLa cell extracts were subjected to immunoprecipitation using IgG (control) or antibodies to CRM1, XPO5 or TAP–p15 in the presence of RanQ69L-GTP. Fractions of the unbound (FT) material and immunoprecipitates (IPs) were analyzed by western blotting using specific antibodies (Supplementary Fig. 2), and the rest of the IPs were used for RNA extraction. Purified RNA were reverse transcribed and subjected to qRT-PCR using specific primers for U3 snoRNA, pre-miR-16, GAPDH mRNA and Dicer mRNA. (b) Electrophoretic mobility shift assay using a radiolabeled Dicer mRNA probe (see Online Methods). Complex was formed in the presence of increasing amounts of recombinant XPO5 and RanQ69L-GTP (lanes 1–7). Complexes were subjected to treatment with RNase and analyzed on nondenaturing 5% acrylamide/TBE gels. Increasing amounts of unlabeled pre-miR-30 (10, 50, 250 ng) were used as specific competitor. Both binding assays were carried out in the presence of RanQ69L-GTP to increase specificity (Supplementary Fig. 3). (c) Radiolabeled pre-miR-30 was incubated with 200 ng of XPO5 in the presence of RanQ69L-GTP as described in Online Methods. Complexes were formed in the absence (−) or in the presence of increasing amounts (0.5, 1 and 2 μg) of the competitor RNAs as indicated. Note that this EMSA is done in the absence of RNase treatment. Complexes were analyzed on nondenaturing 5% acrylamide/TBE gels.
Overexpression of small RNA decreases Dicer protein level
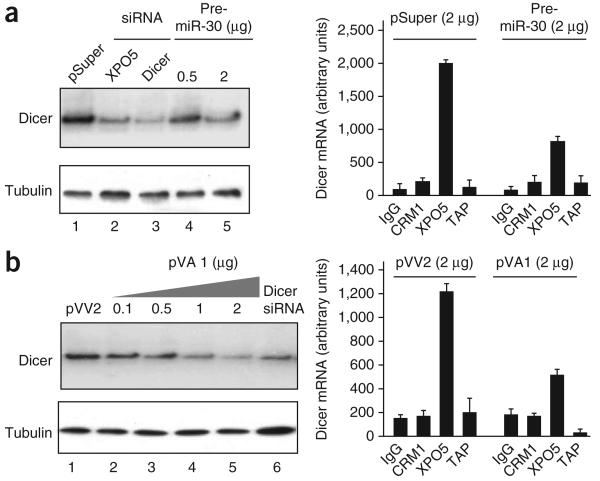
The above data show that Dicer mRNA binds specifically to XPO5, as previously reported for pre-miRNA23 and adenoviral VA RNA I (VA1)14. Because intracellular expression levels of XPO5 are limiting, it can be saturated by overexpression of its substrates in vivo24 and in vitro25. We therefore tested whether pre-miRNA or VA1 could outcompete Dicer mRNA for endogenous XPO5 amounts and, consequently, decrease Dicer protein levels. For this purpose, we transfected HeLa cells with increasing amounts of plasmids expressing either pre-miR-30 (Fig. 4a) or VA1 (Fig. 4b) or the corresponding empty plasmids (pSuper or pVV2, respectively) and analyzed Dicer expression by western blotting. Both pre-miR-30 (Fig. 4a, left) and VA1 (Fig. 4b, left) overexpression decreased Dicer levels in a dose-dependent manner. At the highest saturating dose of pre-miRNA or VA1, cell extracts were immunoprecipitated using antibodies to CRM1, XPO5 or TAP, or a control isotype IgG, and Dicer mRNA was quantified by qRT-PCR. Pre-miR-30 and VA1 overexpression decreased the levels of XPO5-bound Dicer mRNA and pre-miR-16 (Fig. 4a,b, right; Supplementary Fig. 4). However, overexpression of pre-miR-30 and VA1 had no effect on the amounts of U3 snoRNA and GAPDH mRNA recovered from CRM1 and TAP immunoprecipitates, respectively (Supplementary Fig. 4). Thus, saturation of XPO5 by pre-miRNA or VA1 diminished its association with Dicer mRNA, and consequently decreased Dicer protein levels in cells. Taken together, our data show that Dicer levels are tightly regulated by XPO5, through direct interaction between XPO5 karyopherin and Dicer mRNA.
Figure 4.
Overexpression of pre-miRNA or adenoviral VA1 RNA affects Dicer protein levels in cells. (a) HeLa cells were transfected with XPO5- or Dicer-specific siRNA, empty vector, or increasing amounts of a vector expressing pre-miR-30. Dicer and tubulin expression were analyzed by western blot (left), CRM1, XPO5 or TAP–p15 were immunoprecipitated from cell extracts and associated RNAs were extracted. Recovered RNAs were subjected to qRT-PCR to quantify U3 snoRNA, U6 snRNA, pre-miR-16 and GAPDH mRNA, as controls (Supplementary Fig. 4), as well as Dicer mRNA. (b) As in a except that cells were transfected with either empty vector (pVV2) or increasing amounts of plasmid expressing VA1 RNA (pVA1).
Exportin-5 inhibition enhances viral infection
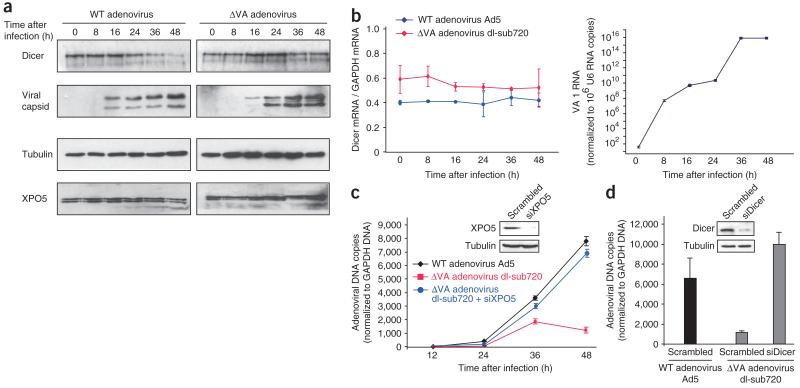
Modulating the available level of XPO5 offers a simple and efficient cross-regulatory mechanism in which high expression of pre-miRNA could outcompete Dicer mRNA, preventing its export and translation and thereby regulating miRNA levels in the cell. This feedback mechanism might thus contribute to balance out Dicer and pre-miRNA levels. We therefore assessed whether the level of Dicer protein would be affected in a pathologic situation such as adenoviral infection. During adenoviral replication, VA1 and VA2 RNA are produced in large amounts and accumulate in infected cells, reaching up to 20% of cellular RNA26. We thus compared the effect of adenovirus infection using a wild-type virus (Ad5) or a mutant from which both VA structures had been deleted (Ad720) on Dicer protein level (Fig. 5a). Ad5 mediates a reduction of Dicer protein level (Fig. 5a), without affecting Dicer mRNA levels (Fig. 5b, left). This downmodulation is correlated with increased amounts of VA RNA (Fig. 5b, right). In contrast, Ad720 had no effect on Dicer levels (Fig. 5a,b). Together, these results clearly show that viral associated RNAs, massively produced at the late stage of adenovirus infection, are able to knockdown Dicer expression by saturating XPO5 and preventing nuclear export of Dicer mRNA.
Figure 5.
XPO5 inhibition enhances adenovirus replication. (a,b) HeLa cells were infected with wild-type (Ad5) or Ad720 mutant (dlsub720) adenovirus. Infected cells were harvested at different times after infection. A fraction of each cell extract was analyzed for viral capsid, Dicer, XPO5 and tubulin by western blotting (a) and the remainder was subjected to RNA purification. Dicer and GAPDH mRNAs (b, left), U6 snRNA and VA1 RNA (b, right) were quantified by qRT-PCR. (c,d) Top panels show knockdown efficiency of XPO5 (c) and Dicer (d) in HeLa cells. siRNA-transfected HeLa cells were infected with Ad5 or Ad720 (0.1 particle per cell). Cells were harvested every 12 h up to 48 h after infection (c) or just at 48 h after infection (d). Bottom panels show levels of adenoviral DNA as quantified by qPCR using primers amplifying the DBP viral gene present in both viruses. Results are expressed after normalization with respect to GAPDH. Error bars, s.d.
In order to evaluate whether this function provides an advantage for the wild-type adenovirus, we also assessed the kinetics of replication for both viruses. After infection, we purified genomic DNA, quantified adenoviral DNA by qPCR, and normalized DNA levels to those of GAPDH DNA. As previously described27, the virus was deleted for both VA replicates slowly compared to wild-type virus (Fig. 5c). We therefore asked whether Ad720 replication could be rescued by reducing XPO5 expression. To determine this, we transfected HeLa cells with siXPO5 for 48 h and then infected them with Ad720. As a control, cells transfected with scrambled siRNA were infected with Ad720 or Ad5. Adenoviral replication was quantified after 12, 24, 36 and 48 h (Fig. 5c). XPO5 knockdown enhanced Ad720 replication to levels comparable to those for the wild-type virus. Inhibition of XPO5 could thus complement VA deficiency. Notably, knockdown of Dicer also complements Ad720 replication to wild-type levels (Fig. 5d). Taken together, these experiments show that adenovirus targets the regulation of Dicer expression by XPO5 for optimal replication.
DISCUSSION
In this study we describe a previously unknown cross-regulation between two essential mediators of small RNA–mediated silencing: pre-miRNA and Dicer. Three salient points emerge from our study. First, the observation that overexpression of pre-miRNA decreases Dicer level (Fig. 4a), suggests that this cross-regulatory mechanism may contribute to the homeostatic control of miRNA biogenesis, similarly to the recently described post-transcriptional cross-regulation between the two subunits of the microprocessor complex, Drosha and DGCR8 (refs. 28,29). These results suggest that a cross-regulatory mechanism exists between the key proteins involved in RNAi. In support of this, we observed that knockdown of Dicer leads to an increased level of Drosha (Fig. 1) by an unknown mechanism. Saturation of XPO5 has been described in vivo in mice treated with high doses of shRNA vector24. They show signs of severe toxicity and ultimately die within a month24. Morbidity was associated with downregulation of liver-derived miRNA. This observation was attributed to saturation of XPO5 resulting in reduced pre-miRNA export. We provide evidence that XPO5 saturation decreases not only pre-miRNA export as previously described4,24, but also Dicer expression. Second, regulation of Dicer mRNA by XPO5 may be exploited by viruses to modulate cellular miRNA expression and overcome RNA interference. Indeed, viruses that produce small, highly structured RNA able to bind and saturate XPO5 may inhibit Dicer expression, thus preventing the maturation of viral RNAs to small noncoding RNAs (miRNAs or siRNAs). Targeting of mRNA export pathways upon viral infection has been documented. Influenza A virus, through its NS1 viral protein, inhibits the TAP–p15 export pathway. The reduction of mRNA export leads to a higher permissivity of cells to influenza virus replication30. Third, the finding that XPO5 interacts with and regulates Dicer mRNA expression opens up the possibility that other cellular mRNAs could interact with XPO5. Although structural determinant responsible for the recognition of short hairpin RNAs by XPO5 have been precisely defined, XPO5 is able to mediate the nuclear export of some unspliced mRNA lacking such a structural motif31. This may uncover a new gene-regulatory mechanism involving mRNA export through the limiting XPO5 pathway.
ONLINE METHODS
Plasmids
Plasmids encoding Myc-XPO5, His-XPO5 and His-RanQ69L were provided by I. Macara12. pSuper and pSuper-premiR-30 were provided by B. Cullen4 (see Acknowledgments).
siRNA cell transfection
HeLa cells were cultured in DMEM medium supplemented with 10% (v/v) FCS. Cells are transfected with siRNA using INTERFERin following manufacturer’s instruction (Polyplus). siRNAs were synthesized by MWG with the following sequences. To target XPO5: siXPO5, 5′-UGUGAGGAGGCAUGCUUGU-3′; siXPO5#2, 5′-GCCCUCAAGUUUUG UGAGG-3′; siXPO5#3, 5′-CUCGAUUGGAGAAGGUGUA-3′; and siXPO5#4, 5′-GGAUAAUACAGACCAACUA-3′. To target Dicer: siDICER, 5′-UGCUUG AAGCAGCUCUGGA-3′. Expression levels of knockdown proteins were analyzed by western blotting 48 h after transfection. Cell extracts were resolved by SDS-PAGE. Proteins were transferred to PVDF membrane by semidry electroblotting and probed overnight at 4 °C with primary antibody (antibody to Dicer, XPO5 or TAP–p15 (Abcam) or antibody to CRM1 or p54 (Bethyl)), washed, and incubated with secondary antibody (Amersham) for 1 h. Proteins were visualized by chemiluminescence according to the manufacturer’s protocol (Pierce).
RNA immunoprecipitation and qRT-PCR
HeLa cells were grown in 60-mm dishes and transfected with the indicated plasmids using jetPEI (Polyplus). Cells were harvested 48 h after transfection, lysed for 15 min in 2 ml of RIP buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 2.5 mM MgCl2•6H2O, 250 mM sucrose, 0.05% (v/v) NP-40 and 0.5% (v/v) Triton X-100) containing 10 U ml−1 of RNasin (Promega) and 1 mM DTT, and centrifuged to pellet debris. Supernatants were split into four equal parts and incubated overnight with antibodies recognizing XPO5, TAP–p15 or CRM1 or an IgG control in the presence of 10 μg of recombinant RanQ69L-GTP at 4 °C, and then incubated for 1 h with protein G–Sepharose or protein A–Sepharose. Flowthroughs were recovered and analyzed at the protein and RNA level. Immunoprecipitates were washed five times with RIP buffer, and nucleic acids were extracted with phenol/chloroform/isoamyl alcohol, precipitated with isopropanol, washed with ethanol and resuspended in RNase-free water. Total RNA was treated with DNase I. After heat inactivation, RNA was reverse transcribed using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Reverse transcription products were PCR-amplified successively using primers specific for GAPDH (GAPDH forw, 5′-CTGGCGTCTTCACCACCATGG-3′; GAPDH rev, 5′-CATCACGCCACAGTTTCCCGG-3′); U3 (U3 forw, 5′-TTCT CTGAGCGTGTAGAGCACCGA-3′; U3 rev, 5′-GATCATCAATGGCTGACGG CAGTT-3′); VA (VA forw, 5′-GTCCGCCGTGATCCATGC-3′; VA rev, 5′-CGTT GTCTGACGTCGCAC-3′) or Dicer (DICER 190–292 forw, 5′-GCAGTAAGC TGTGCTAGAAC-3′; DICER 190–292 rev, 5′-ATTGGTGAGGAAGCAGGG-3′; DICER 5662–5811 forw, 5′-TGGAGACAGTCTGGCAGGTGTA-3′; DICER 5662–5811 rev, 5′-TCCCGTCGTAAGTTCTCTCAGC-3′; DICER 7460–7639 forw, 5′-TCCCATCAACATACCAGTAGAG-3′; DICER 7460 rev, 5′-CAGATAAA GCAGGAAGGACAG-3′). To detect pre-miRNA, an additional step was added in which RNA was polyadenylated with ATP by poly(A) polymerase at 37 °C for 1 h using the RNA tailing kit (Ambion) and reverse transcribed using 0.5 μg of poly(dT) adaptor primer (Invitrogen). RT products were amplified using U3 forward primer; pre-miR-16 forward primer (5′-GTCAGCAGTGCCTTAGCAGCAC-3′), pre-miR-30 forward (5′-GCGACUGUAAACAUCCUCGACUGGA-3′) and reverse primer (based on the adaptor sequence). Amplification was done using a LightCycler 480 (Roche).
Nuclear and cytoplasmic mRNA fractionation and quantification
Nuclear and cytoplasmic RNA were purified following a published protocol19. Briefly, cells were harvested, resuspended in 1 ml of buffer (10 mM Tris, pH 7.4; 10 mM NaCl; 3 mM MgCl2), incubated on ice for 3 min and centrifuged. The volume of swelled cell pellet was estimated and resuspended by slow pipetting with four times its volume of buffer A (10 mM Tris, pH 7.4; 10 mM NaCl; 3 mM MgCl2; 10% (v/v) glycerol, 0.5% (v/v) NP-40; 0.5 mM DTT; 100 U ml−1 RNasin). Nuclei were immediately collected by centrifugation at 4,500g for 3 min, and supernatant was saved as cytoplasm extract. Nuclei were further resuspended in buffer A supplemented with detergent (3.3% (w/v) sodium deoxycholate, 6.6% (v/v) Tween 20) and incubated on ice for 5 min. Nuclei were collected by centrifugation at 10,000g for 5 min. Nuclear integrity was monitored by microscopy after trypan blue staining and resuspension in buffer A. Cytoplasmic and nuclear RNA were extracted by Trizol following manufacturer’s instructions (Invitrogen). 2 μg of RNA was reverse transcribed and used as template for qPCR. Fractionation was assessed by checking nuclear U6 quantification.
Northern blot
RNAs were isolated by using mirVana Kit (Ambion) and analyzed following the NorthernMax-Gly Protocol. Briefly, RNAs were separated on 1% acrylamide-glyoxal gel and transferred for 3 h to nylon membrane. After UV cross-linking, membranes were prehybridized for 1 h at 68 °C in Ultrahyb (Ambion) and incubated overnight at 56 °C with [α-32P]UTP-radiolabeled RNA probe complementary to Dicer or actin RNA.
Electrophoretic mobility shift assay
Recombinant XPO5-his and RanQ69L-his were produced as previously described12. Purified proteins were further dialyzed against buffer B (PBS, pH 7.5, 14 mM β-mercaptoethanol and 10% (v/v) glycerol). Protein expression and structures were verified by western blotting and by XPO5’s ability to specifically bind VA1 RNA more efficiently in the presence of RanQ69L-GTP and not bind to the mutant VA Mut10 (data not shown)14. Dicer mRNA was radiolabeled in vitro by co-transcribing the complete ORF of 5.6 kb and the 3′ UTR of 4.27 kb in the presence of [α-32P]UTP using the T7 AmpliScribe kit (Epicentre). Pre-miR-30 was also radiolabeled in vitro using [α32-P]UTP. RNA-binding reactions were carried out in binding buffer (20 mM HEPES, pH7.9; 50 mM KCl, 5 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 10% (v/v) glycerol) for 20 min complemented with 50 nM of Mut10VA to reduce nonspecific binding and 10 ng of RanQ69L-GTP to increase specificity. In the case of Dicer mRNA, because of its length, 0.01 μl of RNase A/RNase T1 cocktail mix (Ambion) was added for 10 min at room temperature to degrade unprotected RNA. 1 μl of loading buffer (0.6 mg per ml heparin, 1 mg per ml bromophenol blue) was added to the samples for an additional 5 min. The complexes were resolved on a pre-run 5% nondenaturing acrylamide/TBE gel. Gels were dried and exposed by autoradiography.
Adenovirus infection
HeLa cells were infected with 100 particles per cell of wild-type Ad5 or dlsub720 (a derivative Ad5 strain deleted for VA RNAI and II, provided by G. Akusjärvi (see Acknowledgments)) at 4 °C for 4 h27. Cells were washed twice with PBS and incubated at 37 °C with DMEM/10% (v/v) FCS medium. Infection was followed for 48 h. Cells were harvested every 8 h, and protein, RNA and DNA were extracted. Proteins were analyzed by western blotting, RNAs were reverse transcribed, and Dicer, GAPDH and VA1 RNA were quantified by qRT-PCR. DNA was extracted by the Hirt method32. In another set of experiments, HeLa cells were transfected with a scrambled siRNA as control or siXPO5. 48 h later, cells were infected with 0.1 particles per cell of Ad5 or Ad720. Cells were harvested every 12 h up to 48 h after infection. DNA was extracted and adenoviral DNA was quantified by qRT-PCR using primers amplifying the DBP viral gene: forward, 5′-CAGGGACACGTTGCGATACT-3′; reverse, 5′-GCGGAGGCCCCAACTGCGACTTC-3′. Results were normalized by quantification of cellular GAPDH DNA. Amplification was done using a LightCycler 480 (Roche).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to R. Kiernan and members of the Laboratoire de Virologie Moléculaire for helpful discussions and for critically reading the manuscript, and to G. Akusjärvi (Uppsala University, Uppsala, Sweden), B. Cullen (Duke University, Durham, North Carolina, USA) and I. Macara (University of Virginia, Charlottesville, Virginia, USA) for providing reagents. Work in M.B.’s laboratory was supported by Agence Nationale de Recherche sur le SIDA, SIDACTION, Agence Nationale de la Recherche-BLAN-0040, European Research Council (ERC 250333) and the Fondation pour la Recherche Médicale Equipe labéllisée FRM.
Footnotes
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 6.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 8.Levy C, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 10.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calado A, Treichel N, Muller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gwizdek C, et al. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 2003;278:5505–5508. doi: 10.1074/jbc.C200668200. [DOI] [PubMed] [Google Scholar]
- 15.Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol. Cell. Biol. 2004;24:6608–6619. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwizdek C, et al. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J. Biol. Chem. 2004;279:884–891. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- 17.Macara IG. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39:356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Gwizdek C, et al. Terminal minihelix, a novel RNA motif that directs polymerase III transcripts to the cell cytoplasm. Terminal minihelix and RNA export. J. Biol. Chem. 2001;276:25910–25918. doi: 10.1074/jbc.M100493200. [DOI] [PubMed] [Google Scholar]
- 21.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulon S, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 25.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J. Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhat RA, Thimmappaya B. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 1984;12:7377–7388. doi: 10.1093/nar/12.19.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triboulet R, Chang HM, Lapierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satterly N, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 32.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.