SUMMARY
Transcription elongation is increasingly recognized as an important mechanism of gene regulation. Here, we show that microprocessor controls gene expression in an RNAi-independent manner. Microprocessor orchestrates the recruitment of termination factors Setx and Xrn2, and the 3′–5′ exoribonuclease, Rrp6, to initiate RNAPII pausing and premature termination at the HIV-1 promoter through cleavage of the stem-loop RNA, TAR. Rrp6 further processes the cleavage product, which generates a small RNA that is required to mediate potent transcriptional repression and chromatin remodeling at the HIV-1 promoter. Using chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq), we identified cellular gene targets whose transcription is modulated by microprocessor. Our study reveals RNAPII pausing and premature termination mediated by the co-operative activity of ribonucleases, Drosha/Dgcr8, Xrn2, and Rrp6, as a regulatory mechanism of RNAPII-dependent transcription elongation.
INTRODUCTION
Many cellular genes, in particular highly inducible genes, undergo transcriptional initiation but are regulated at the level of transcriptional elongation (Guenther et al., 2007). Indeed, genome-wide mapping studies have shown that transcription initiation from cellular genes is extremely pervasive, confirming promoter-proximal RNA polymerase II (RNAPII) pausing as an important mechanism of transcriptional control (Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009; Core and Lis, 2008; Core et al., 2008; Seila et al., 2008). Nucleosome architecture profiles have been correlated with the tendency for a promoter to undergo pausing (Gilchrist et al., 2010). However, the mechanisms that control RNAPII pausing are poorly understood. Transcription factors that contribute to pausing have been clearly identified. Furthermore, whereas a regulatory role for the small promoter-associated RNA products of such abortive transcription has been speculated (Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009; Core and Lis, 2008; Core et al., 2008; Seila et al., 2008), the mechanisms controlling their processing and function remain to be discovered.
The HIV type 1 (HIV-1) promoter is a well-defined, convenient, and thus widely used model, which has provided considerable insight into transcriptional elongation control. In the absence of the viral transactivator Tat, transcription from the long terminal repeat (LTR) leads to RNAPII pausing and premature termination after synthesis of a short stem-loop RNA, the transactivation response element (TAR) (Brès et al., 2008). HIV-1 Tat, together with the positive transcription elongation factor PTEF-b, binds a bulge-loop within TAR, allowing CDK9 to phosphorylate RNAPII CTD at serine 2 and NTEFs (Negative Transcription Elongation factors), licensing RNAPII for productive elongation (Brès et al., 2008). The molecular mechanisms involved in RNAPII pausing and premature termination at the HIV-1 promoter are unknown.
The microprocessor complex that consists of at least two subunits, the RNase III Drosha and the dsRNA-binding protein Dgcr8, is required for the regulation of mature miRNA abundance (Han et al., 2009; Kim et al., 2009). Microprocessor is essential for the first processing step characterized by recognition of the canonical stem-loop structure of the miRNA by Dgcr8. Drosha cleaves both strands of the primary transcript (pri-miRNA) at sites near the base of the stem loop that liberates the precursor miRNA (pre-miRNA), which is further processed in the cytoplasm by Dicer (Newman and Hammond, 2010; Seitz and Zamore, 2006). The mature miRNA mediates posttranscriptional gene silencing (PTGS) through translational inhibition and destabilization of the target mRNA (Han et al., 2009). Interestingly, PTGS-independent regulation of gene expression in mammalian cells by the microprocessor has been suggested by Han et al. (2009). However, the molecular mechanism for this function of the microprocessor is unknown.
Transcriptional termination of several classes of RNA in yeast occurs via a complex containing the RNA/DNA helicase, Sen1 (Steinmetz et al., 2006; Ursic et al., 1997). The Sen1 termination complex associates with RNAPII near promoters and appears to be most important for termination within 500 bp of transcription start sites (TSSs) (Kim et al., 2010; Steinmetz et al., 2006). Sen1-mediated termination is potentiated by a 5′–3′ exoribonuclease, Rat1p/Xrn2, that, following cleavage, degrades the uncapped nascent transcript to promote the release of RNAPII from its template (Kawauchi et al., 2008). The human homolog of Sen1, Senataxin (Setx), was recently shown to promote Xrn2-dependent transcriptional termination at the 3′ end of the B-actin gene (Skourti-Stathaki et al., 2011). The Sen1 complex was also recently shown to antagonize transcriptional elongation and promote premature transcription termination of the yeast FKS2 gene (Kim and Levin, 2011). Rrp6 is the catalytic subunit of nuclear exosome, a highly conserved complex possessing 3′–5′ exoribonuclease activity that exerts an indispensable role in RNA processing and quality control (Houseley et al., 2006). Rrp6 is a member of the DEDD family of 3′ → 5′ exonucleases that act on nucleic acid by 3′ hydrolysis (Moser et al., 1997) and possesses intrinsic distributive exoribonuclease activity in vitro.
Here, we describe a mechanism of premature termination and RNA-dependent transcriptional gene silencing (TGS) at the HIV-1 promoter. A PTGS-independent function of the microprocessor complex, in cooperation with termination factors, Setx and Xrn2, regulates RNAPII pausing and premature termination, whereas additional processing of the cleavage product by Rrp6 generates a small RNA that represses transcription. Modifications to the local chromatin architecture restrict access of RNAPII to the promoter, thereby limiting transcriptional output.
RESULTS
Microprocessor Regulates HIV-1 Transcription Independently of the RNAi Pathway
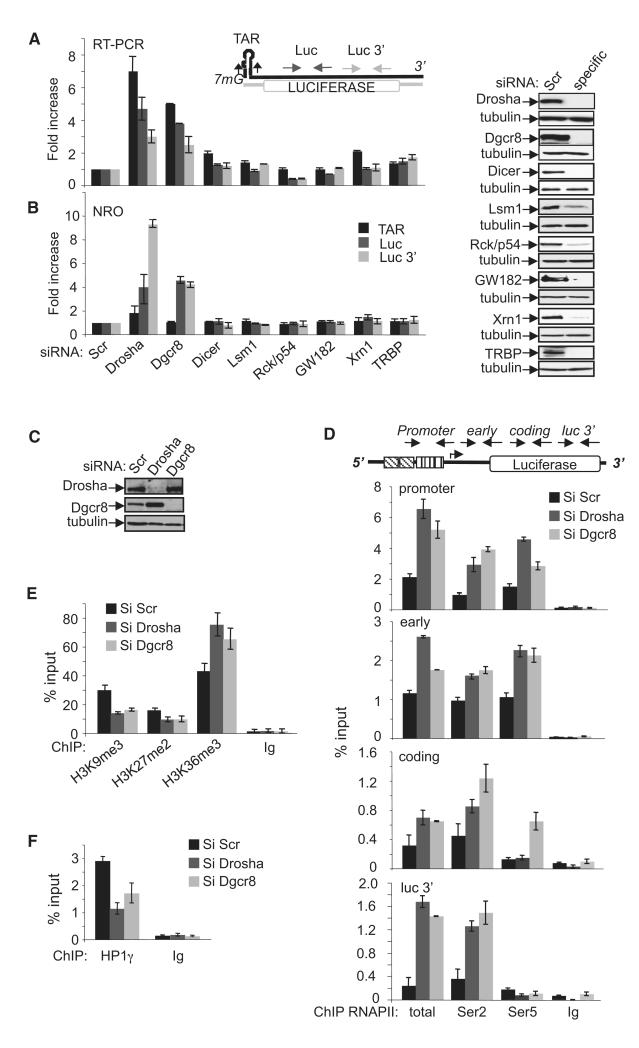
Given the resemblance between TAR RNA and the stem-loop structure of miRNA, we hypothesized that microprocessor may play a role in transcriptional repression and premature termination at the HIV-1 promoter. To determine whether microprocessor can regulate transcription independently of PTGS, RNAi directed against components of PTGS was performed in HeLa cells containing a stably integrated LTR linked to a luciferase reporter gene (HeLa-LTR-luc; du Chéné et al., 2007; Lassot et al., 2007). Knockdown of either Drosha or Dgcr8 increased the abundance of transcripts emanating from an integrated LTR, as measured by real-time PCR, and corresponding luciferase activity (Figure 1A; see Figure S1A available online). Importantly, the effect of Drosha and Dgcr8 is independent of small RNA-mediated PTGS because invalidation of this pathway using small interfering RNA (siRNA) to Dicer, Lsm1, Rck/p54, GW132, TRBP, Xrn1, Ago1, or Ago2 had no effect (Figures 1A, S1A, and S1B). To determine whether microprocessor regulates the level of LTR transcription independently of PTGS, Nuclear Run-On transcription assay (NRO) was performed in cells knocked down for RNAi factors. The rate of transcription from the HIV-1 LTR was increased after knockdown of Drosha and Dgcr8, but not other PTGS factors as compared to control cells (Figure 1B). Enhanced basal LTR-luc activity after Drosha knockdown was rescued by expression of a Drosha siRNA-resistant mutant (Figure S1C). To determine whether microprocessor also modulated basal transcription of full-length HIV-1, cells were infected with HIV-1 that lacks the transactivator protein, Tat, and subjected to RNAi against microprocessor or Rck/p54. Nascent transcription was analyzed by NRO using primers in the Gag region of HIV-1. Knockdown of microprocessor, but not Rck/p54, enhanced transcription of full-length HIV-1 (Figure S1D). Taken together, these experiments point to a PTGS-independent function for Drosha and Dgcr8 acting as transcriptional repressors of the HIV-1 LTR.
Figure 1. Drosha and Dgcr8 Are Transcriptional Repressors of the Integrated HIV-1 Promoter.
(A and B) RNA isolated from HeLa LTR-Luc cells transfected with the indicated siRNAs was analyzed by reverse-transcription q-PCR and NRO using the primers indicated on the schematic above the graphs. Values were normalized to that of GAPDH in the same samples. The result for Scrtreated cells was attributed a value of 1. The knockdown of specific factors was validated by immunoblot (right).
(C) Validation of RNAi knockdowns by immunoblot using the indicated antibodies.
(D) ChIP assay was performed using the indicated antibodies and chromatin prepared from HeLa LTR-Luc cells transfected with control (Si Scr), Drosha, or Dgcr8 siRNAs as indicated. Locations of primers used are indicated on the schematic above the graphs. The amount of immunoprecipitated material for each PCR was normalized to the input DNA.
(E) Native ChIP was performed using the indicated antibodies and chromatin from cells transfected with the indicated siRNAs. The promoter region was amplified by q-PCR. The amount of immunoprecipitated material was normalized to the input DNA.
(F) ChIP was performed using anti-HP1g or a control IgG and chromatin prepared from HeLa LTR-Luc cells transfected with the indicated siRNAs. The promoter region was amplified by q-PCR. The amount of immunoprecipitated material was normalized to the input DNA.
All graphs show mean ± SE from three independent experiments. See also Figure S1.
Microprocessor Regulates RNAPII Processivity and Chromatin Organization at the HIV-1 Promoter
To further characterize the involvement of Drosha and Dgcr8 in HIV-1 LTR transcriptional repression, we analyzed the consequence of their knockdown on the recruitment and modification of RNAPII at the LTR using chromatin immunoprecipitation (ChIP) assay. Knockdown of Drosha or Dgcr8 (Figure 1C) enhanced RNAPII occupancy at the viral promoter and across the gene body (Figure 1D). Importantly, phosphorylation of RNAPII Ser5 and Ser2 was increased when Drosha or Dgcr8 levels were reduced. This experiment suggests that the enhanced recruitment of RNAPII after Drosha and Dgcr8 knockdown is accompanied by modifications required for promoter clearance and processive transcription elongation at the HIV-1 promoter.
Chromatin organization, particularly nucleosome1 (Nuc1) localized at approximately 100 nt after the TSS of the HIV-1 promoter, is known to play a role in regulating transcription from the integrated LTR (Bisgrove et al., 2005). Interestingly, knockdown of Drosha/Dgcr8 had no effect when HIV-1 LTR-luc was transiently transfected in HeLa cells, suggesting that microprocessor-mediated repression may depend on a properly chromatinized environment (data not shown). Thus, we analyzed the consequence of Drosha and Dgcr8 knockdown on chromatin marks associated with Nuc1. As previously shown by du Chéné et al. (2007), transcriptionally inactive HIV-1 LTR is associated with histone H3 repressive marks, H3K9me3 and H3K27me3. Knockdown of Drosha and Dgcr8 diminished these repressive marks and significantly increased H3K36me3 that is associated with transcription elongation (Figure 1E). Consistent with loss of H3K9me3, association of Hp1γ with the repressed LTR was also reduced after Drosha and Dgcr8 knockdown (Figure 1F). These experiments suggest that Drosha and Dgcr8 maintain the chromatin at the HIV-1 LTR in a repressed state. Their loss promoted the establishment of a transcriptionally competent chromatin environment.
Microprocessor Is Recruited to the HIV-1 LTR through TAR RNA
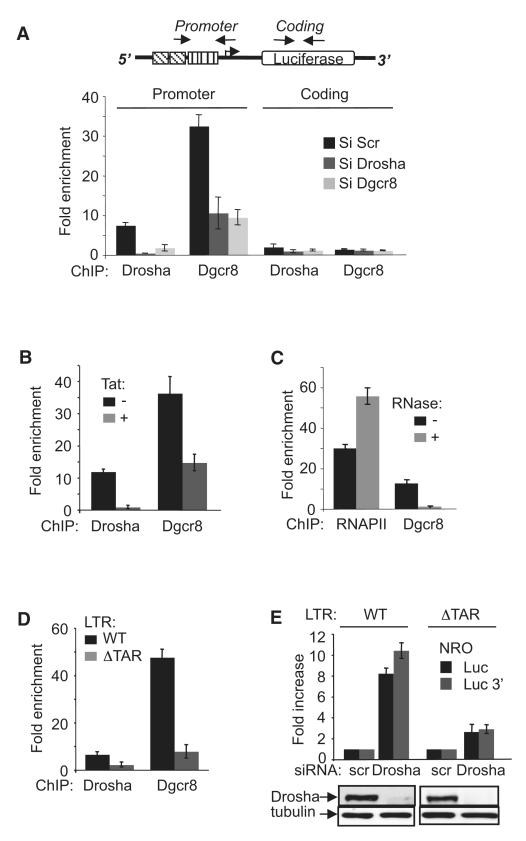
Next, we asked whether Drosha and Dgcr8 are directly associated with the HIV-1 LTR. ChIP using Drosha- and Dgcr8-specific antibodies immunoprecipitated the HIV-1 promoter-proximal region (Figure 2A). Association of Drosha and Dgcr8 with the LTR is specific because the signal was significantly reduced in siRNA-treated cells. Interestingly, knockdown of Drosha led to loss of Dgcr8, and vice versa, suggesting that association of Drosha and Dgcr8 with the HIV-1 LTR requires the presence of the two subunits (Figure 2A). The presence of Drosha and Dgcr8 at the HIV-1 LTR promoter region but not at the coding region suggests that they act at an early step of transcription (Figure 2A). Importantly, transcriptional activation of the HIV-1 LTR by Tat diminished the association of Drosha and Dgcr8 with the promoter as shown by ChIP (Figure 2B), suggesting that Tat overcomes Drosha- and Dgcr8-mediated transcriptional repression by inducing their release from the promoter.
Figure 2. HIV-1 TAR RNA Is Required for Drosha and Dgcr8-Mediated Transcriptional Repression of the HIV-1 Promoter.
(A) ChIP assay was performed as in Figure 1 using the indicated antibodies and chromatin prepared from HeLa LTR-Luc cells transfected with the indicated siRNAs. Results are defined as enrichment over that of a mock precipitation with an unrelated IgG antiserum. Locations of primers used to amplify promoter-proximal and coding region sequences are indicated on the schematic above the graph.
(B) ChIP was performed using the indicated antibodies and chromatin prepared from HeLa LTR-Luc cells treated with Tat or mock treated as indicated. The presence of HIV-1 promoter region in immunoprecipitated materials was determined by q-PCR. Results are presented as fold enrichment over that of a mock precipitation using an unrelated IgG antiserum.
(C) ChIP was performed using the indicated antibodies and chromatin prepared from HeLa LTR-Luc cells that had been pretreated with RNase or mock treated as indicated. The presence of HIV-1 promoter-proximal region in immunoprecipitated materials was determined by q-PCR. Results are presented as fold enrichment over that of a mock precipitation using an unrelated IgG antiserum.
(D) ChIP assay was performed using the indicated antibodies and chromatin prepared from HeLa LTR-Luc (WT) and HeLa LTRΔTAR-Luc (ΔTAR) cells. The presence of HIV-1 promoter-proximal region in immunoprecipitated materials was determined by q-PCR. Results are presented as fold enrichment over that of a mock precipitation using an unrelated IgG antiserum.
(E) Nuclei isolated from HeLa LTR-Luc (WT) or HeLa LTRΔTAR-Luc (ΔTAR) cells transfected with the indicated siRNAs were analyzed by NRO using the primers indicated. Values were normalized to that of GAPDH in the same samples. The result for scr-treated cells was attributed a value of 1. The knockdown of Drosha was validated by immunoblot (bottom).
All graphs show mean ± SE from three independent experiments. See also Figure S2.
Drosha and Dgcr8 are known to bind the canonical stem-loop structure of the miRNA within the primary miRNA transcript (Seitz and Zamore, 2006). Because HIV-1 produces a stem-loop RNA, TAR, we hypothesized that TAR RNA may contribute to the recruitment of Drosha and Dgcr8 to the viral promoter. In support of this hypothesis, pretreatment of chromatin with RNase abolished the association of the RNA-binding component of microprocessor, Dgcr8, but not RNAPII, with the HIV-1 promoter region showing that microprocessor association is RNA dependent (Figure 2C). We next generated HeLa cells containing an integrated TAR-deleted LTR-luciferase construct (LTR-ΔTAR-luc). It is important to note that both wild-type (WT) LTR-luc and LTR-ΔTAR-luc constructs were inserted individually at the same position in the genome to avoid an integration position effect (du Chéné et al., 2007; Tréand et al., 2006). Deletion of TAR reduced the association of Drosha and Dgcr8 with the HIV-1 promoter region as shown by ChIP (Figure 2D). Comparing basal WT LTR activity to LTRΔTAR, we observed up to a 9-fold increase of luciferase activity, suggesting that deletion of TAR results in transcriptional derepression of the LTR (Figure S2). Both WT and TAR-deleted constructs respond similarly to activation by PMA/Ionomycin when normalized to their own basal activity excluding the possibility that TAR deletion may affect the general responsiveness of the promoter (Figure S2). To determine the importance of TAR sequence in microprocessor-mediated repression, NRO was performed in HeLa LTR-luc containing either WT or TAR-deleted LTR, and transfected with control or Drosha-specific siRNA (Figure 2D). Consistent with increased basal luciferase activity in HeLa LTRΔTAR-luc cells, run-on transcripts were approximately 2-fold higher in TAR-deleted cells compared to cells containing WT LTR when transfected with Scr siRNA (data not shown). Importantly, LTR derepression after Drosha knockdown was significantly diminished in TAR-deleted cells compared to controls (Figure 2E). Taken together, these experiments show that Drosha and Dgcr8-mediated transcriptional repression of the HIV-1 LTR requires the presence of the TAR RNA.
Microprocessor Induces Premature Transcription Termination at the HIV-1 LTR through the Recruitment of Setx, Xrn2, and Rrp6
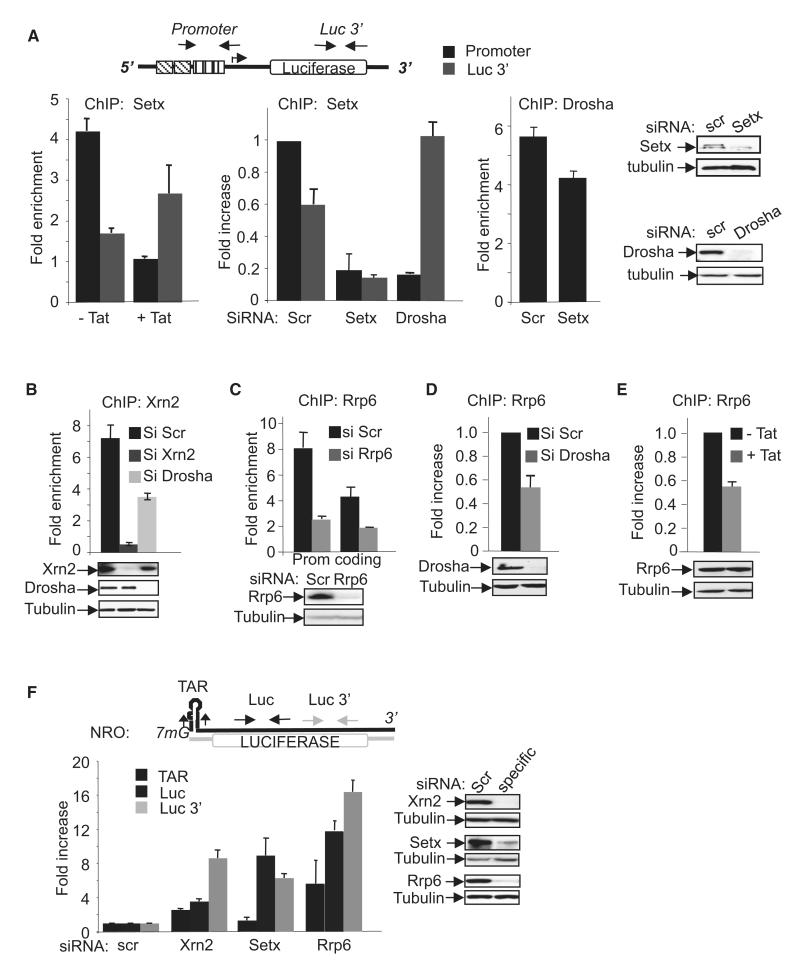
Based on the aforementioned, we built a working model in which synthesis of TAR RNA leads to recruitment of Drosha and Dgcr8 to the HIV-1 promoter, leading to endoribonucleolytic cleavage of the nascent TAR-containing transcript by Drosha that initiates premature termination of transcription. In support of this model, we first observed that Drosha-mediated repression of the HIV-1 LTR is dependent on its endonuclease activity because overexpression of a catalytic mutant resulted in enhanced HIV-1 LTR expression (Figure S3). To further investigate premature termination at the LTR, we analyzed the involvement of the Sen1 RNA/DNA helicase, Setx, which participates in termination of transcription, particularly of short transcripts (Kim et al., 2010; Steinmetz et al., 2006). Under basal transcription conditions, Setx was associated predominantly with the promoter-proximal region (Figure 3A, –Tat). Activation of transcription by Tat resulted in a shift of Setx to the 3′ end of the gene, consistent with a role in termination of short and long transcripts, respectively. Ablation of Setx by RNAi led to loss of Setx from both the promoter and 3′ end of luciferase (Figure 3A, siSetx, ChIP Setx). Interestingly, ablation of Drosha also resulted in a significant shift of Setx toward the 3′ end (Figure 3A, siDrosha, ChIP Setx). However, ablation of Setx only modestly affected Drosha association (Figure 3A, siSetx, ChIP Drosha). Sen1/Setx cooperates with the 5′–3′ exonuclease, Xrn2, which is known to play a role in transcription termination by RNAPII (Ballarino et al., 2009; Gromak et al., 2006) through subsequent degradation of the nascent transcript leading to cessation of RNA synthesis and termination by RNAPII-DNA dissociation. Association of Xrn2 with the HIV-1 promoter region was detected in control cells and was abolished in Xrn2 knockdown cells (Figure 3B). In support of our model, recruitment of Xrn2 to the promoter-proximal region was dependent on the presence of microprocessor because knockdown of Drosha significantly reduced Xrn2 recruitment (Figure 3B).
Figure 3. Drosha-Dependent Recruitment of Setx, Xrn2, and Rrp6 to the HIV-1 Promoter-Proximal Region.
(A) ChIP assay was performed using the indicated antibody and chromatin from Tat-treated or siRNA-treated HeLa LTR-Luc cells, as indicated. Locations of primers used to amplify the promoter-proximal and luc 3′ regions are indicated on the schematic above the graphs. Results are presented as fold enrichment over that of a mock precipitation using an unrelated IgG antiserum. Knockdown efficiency of the siRNA was assessed by immunoblotting using the indicated antibodies.
(B–E) ChIP assay was performed using the indicated antibody and chromatin from siRNA-treated or Tat-treated HeLa LTR-Luc cells. Unless indicated otherwise, the promoter-proximal region was amplified by q-PCR. Results are presented as fold enrichment over that of a mock precipitation using an unrelated IgG antiserum or fold increase relative to the control sample (siScr), which was attributed a value of 1. Knockdown efficiencies of the siRNAs were assessed by immunoblotting using the indicated antibodies.
(F) NRO performed using nuclei prepared from HeLa LTR-Luc cells transfected with the indicated siRNAs. Regions amplified by PCR are indicated above the graph. Values were normalized to the amount of GAPDH RNA in the same samples. The result for Scr-treated cells was attributed a value of 1. Knockdown efficiencies of the siRNAs were assessed by immunoblotting using the indicated antibodies.
All graphs show mean ± SE from three independent experiments. See also Figure S3.
In addition to providing a substrate for the 5′–3′ exonuclease, Xrn2, endonucleolytic cleavage of TAR RNA by Drosha will generate a free 3′ end that could serve as a recruitment signal for a 3′–5′ RNase such as Rrp6, which can carry out 3′–5′ RNA processing or degradation. We hypothesized that microprocessor-dependent cleavage of HIV-1 TAR might signal Rrp6 recruitment to the LTR. Thus, we tested whether Rrp6 physically associates with HIV-1 chromatin, and whether its association is dependent on the microprocessor. ChIP analysis showed that Rrp6 was enriched at both the promoter and coding regions (Figure 3C). Consistent with the hypothesis that Rrp6 is recruited following transcript cleavage, knockdown of Drosha diminished Rrp6 recruitment to the HIV-1 promoter (Figure 3D). Because the HIV-1 transactivator, Tat, displaces Drosha from the LTR (Figure 2B), we might expect Tat to affect Rrp6 recruitment. Association of Rrp6 with the LTR was reduced by Tat accordingly (Figure 3E). Thus, Rrp6 is recruited to HIV-1 chromatin in a manner that depends at least partly on the microprocessor.
To determine whether Setx/Xrn2 termination pathway and Rrp6 regulate basal transcription from the LTR, NROs were performed in cells in which this pathway was invalidated. Run-on transcripts were increased following knockdown of Xrn2, Setx, or Rrp6 (Figure 3F). Interestingly, no significant stabilization of TAR RNA was detected in Xrn2 or Setx knockdown cells compared to controls, whereas transcript corresponding to the coding region was increased (Figure 3F). The lack of TAR RNA accumulation suggests that the substrate of Xrn2 may be a transcript that is cleaved down stream of TAR. Furthermore, neither Setx nor Xrn2 knockdown increased luciferase activity significantly (data not shown), suggesting that the transcripts induced are not competent for protein synthesis.
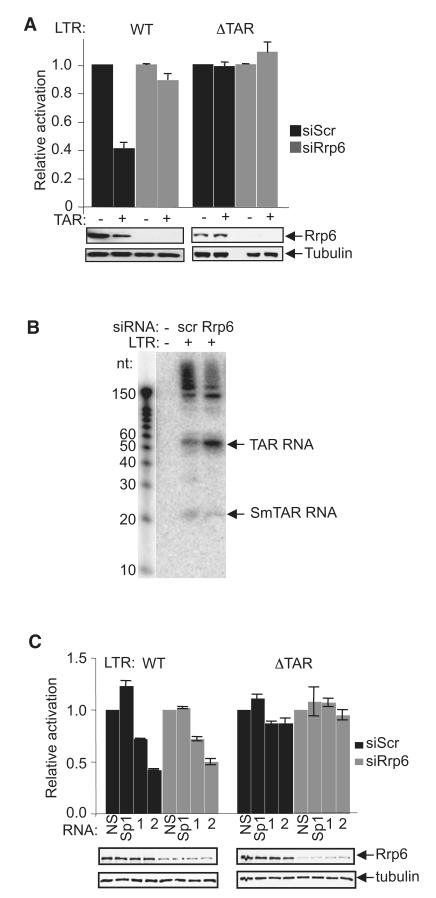
Interestingly, an increase of TAR RNA was observed after Rrp6 knockdown (Figure 3F). Thus, in considering how Rrp6 might contribute to transcriptional repression, we asked whether it further processes the product of microprocessor cleavage, the promoter-proximal transcript, TAR, which may be required for transcriptional repression. In support of this, TAR-containing RNA, when transfected into control HeLa-LTR-luc cells, further repressed basal LTR activity (Figure 4A), as described previously by Klase et al. (2007). However, TAR RNA-mediated repression was abolished in cells depleted of Rrp6, suggesting that further processing of TAR occurs in vivo that necessitates Rrp6. To better understand how TAR RNA can inhibit the LTR, we transfected TAR RNA into HeLa cells carrying a TAR-deleted LTR. TAR did not inhibit the LTR that lacks the corresponding DNA sequence (Figure 4A). These results show that TAR RNA is repressive toward the LTR in cells that express WT levels of Rrp6 and contain the corresponding TAR DNA sequence.
Figure 4. Small TAR-Derived RNAs Repress LTR Activity.
(A) HeLa LTR-Luc (WT) or HeLa LTRΔTAR-Luc (ΔTAR) cells transfected with control (scr) or Rrp6 siRNA, with or without TAR RNA (1 μg), as indicated, were harvested for luciferase assay (top) and immunoblotting using the antibodies indicated (bottom). For each condition, values were normalized to the control sample that was mock transfected with TAR RNA, which was assigned a value of 1.
(B) Northern blot analysis of TAR and TAR-derived RNAs obtained from HIV-1-infected cells transfected with the indicated siRNAs. RNA decade marker run in parallel is shown at left.
(C) HeLa LTR-Luc (WT) or HeLa LTRΔTAR-Luc (ΔTAR) cells transfected with control (scr) or Rrp6 siRNA and the indicated RNA oligonucleotides were harvested for luciferase assay (top) and immunoblotting using the antibodies indicated (bottom). For each condition, values were normalized to the control sample, which was assigned a value of 1.
All graphs represent mean ± SE obtained from at least three independent experiments. See also Figure S4.
Rrp6-Dependent Biogenesis of Small TAR RNAs that Repress HIV-1 LTR Activity
To determine whether small RNAs derived from TAR can be detected in vivo, HIV-1-infected cells were analyzed by small RNA-sequencing (RNA-seq) (Schopman et al., 2012). Sequences corresponding to TAR were identified among the reads (Figure S4). To determine whether Rrp6 may be implicated in the biogenesis of the TAR-derived RNAs, northern blotting was performed in cells transfected with control or Rrp6-specific siRNA (Figure 4B). Knockdown of Rrp6 diminished the abundance of small TAR RNAs and increased the abundance of unprocessed TAR RNA (Figure 4B). To determine whether TAR-derived oligonucleotides mediate repression, oligonucleotides corresponding to TAR-derived small RNAs identified in vivo (TAR1 nt 11–28 and TAR2 nt 40–58) were transfected into HeLa-LTR-luc cells (Figure 4C). The most abundant TAR-derived small RNA identified in vivo, TAR2 (nt 40–58) (Figure S4), further repressed LTR activity when compared to a nonsense oligo (NS) or a sequence within the nontranscribed Sp1 sites of the LTR, whereas the less abundant TAR1 nt 11–28 led to a modest reduction. A TAR-derived sequence that was not represented among the reads (TAR3 nt 15–35) did not repress LTR activity (data not shown). In contrast to full-length TAR RNA, TAR oligonucleotides repressed the LTR in a manner that was independent of Rrp6. Furthermore, small TAR oligonucleotides were not repressive in cells carrying a TAR-deleted LTR (Figure 4C). These results show that small RNAs derived from TAR, whose biogenesis depends on Rrp6, mediate repression of the LTR through a mechanism that may involve hybrid formation with TAR DNA sequences.
Microprocessor Regulates Transcription from the HERV Endogenous Retrovirus and Subset of Cellular Genes
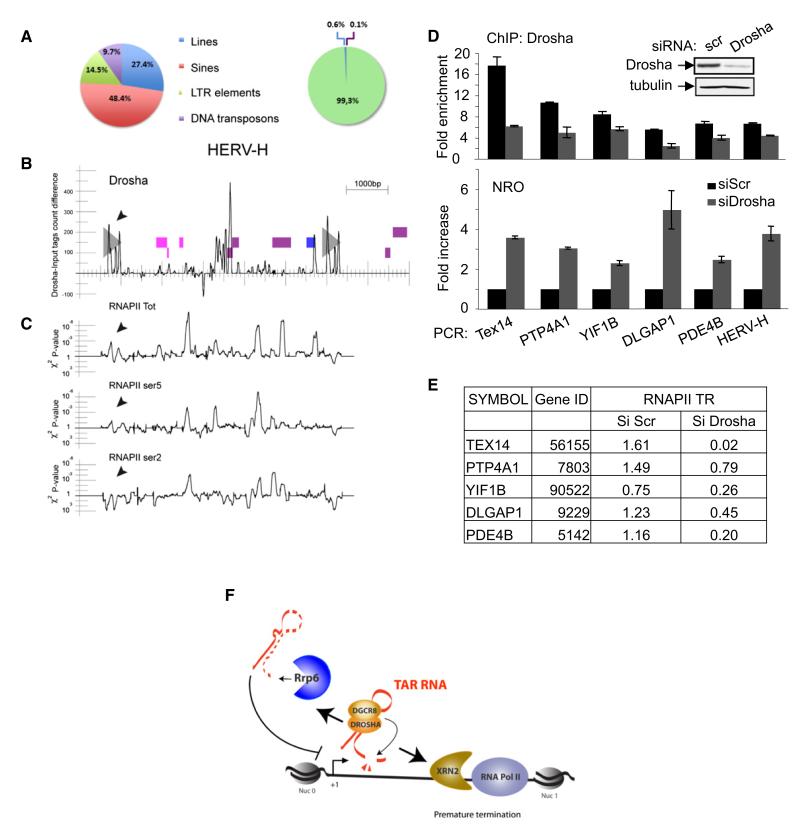
Genome-wide mapping studies revealed promoter-proximal pausing, shortly after initiation, of RNAPII at 30% of human genes, establishing postinitiation events as a hallmark of gene regulation (Core and Lis, 2008; Guenther et al., 2007). Thus, we asked whether the identified function for the microprocessor complex in regulating transcription by RNAPII is restricted to the HIV-1 promoter or if it also occurs at cellular genes. For this purpose we performed ChIP-sequencing (ChIP-seq) analysis using chromatin prepared from HeLa cells and antibody recognizing Drosha. First, the reads were filtered for repetitive sequences, and revealed a large number of tags with significant enrichment over input tags, corresponding to annotated GGAAT satellite sequence and LTR retroelements (Table S1). Interestingly, we noticed that the human endogenous retrovirus (HERV) families were highly represented among the Drosha-bound repetitive sequences (Figure 5A). Our analysis does not allow the determination of whether Drosha binds to each of the approximate 1,000 copies of HERV-H (de Parseval et al., 2001, 2003), or only a subset of them. However, the presence of sequences highly similar to env HERV-H genes among the reads suggests that Drosha binds preferentially to nearly full-length copies. Three major regions within the HERV-H sequence were found enriched in Drosha: 5′ and 3′ LTRs and regions within the middle of the genome (Figure 5B). The presence of Drosha at HERV-H was confirmed by ChIP using specific oligonucleotides (Figure 5D, top). Interestingly, knockdown of Drosha results in enhanced recruitment of RNAPII, in an elongation-competent form, to the HERV-H locus (Figure 5C) and an increase in nascent transcription as measured by NRO (Figure 5D, bottom). These data suggest that microprocessor-mediated transcriptional repression by RNAPII may be an ancient mechanism of regulation controlling the replication of endogenous retroviruses. Next, reads were filtered for human genes. Our analysis revealed that Drosha associates with 461 genes, none of which is annotated as miRNA-encoding genes (Table S2). Specific association of Drosha was confirmed by ChIP for five randomly selected genes in control and Drosha knockdown cells (Figure 5D, top). Nuclear run-on transcription analysis showed that ablation of Drosha increased transcription from these genes (Figure 5D, bottom). Furthermore, knockdown of Dicer or GW182 had no significant effect on their expression as measured by reverse-transcription quantitative PCR (q-PCR) (data not shown), suggesting that microprocessor-mediated regulation of these genes is RNAi independent. These results suggest that the microprocessor is physically associated with a subset of cellular genes and is implicated in their transcriptional regulation. To determine whether microprocessor affects RNAPII distribution across these genes, the traveling ratio (TR) of RNAPII in control and siDrosha samples was calculated as described previously by Rahl et al. (2010). In control cells the TR was 1 at most genes analyzed, suggesting that they show signs of RNAPII pausing (Figure 5E). In Drosha knockdown cells the TR was 1, suggesting that the genes were actively transcribing.
Figure 5. ChIP-seq Analysis Reveals the Endogenous Loci that Are Targets for Drosha.
(A) Pie charts summarizing the relative abundance of LINES, SINES, LTR elements, and DNA transposons in the human genome (left) and those bound by Drosha (right). The observed and expected numbers of tags are significantly different (chi-square test, p < 10−15). See also Table S1.
(B) Chr6:21,360,000–21,368,000 region containing a HERV-H endogenous retrovirus. Gray triangles represent the two LTRs. Pink, violet, and blue boxes represent genomic fragments whose translations have similarities with Gag, Pol, and Env retroviral proteins. ChIP-seq tags density difference between Drosha siRNA and control conditions along the sequence. Positive values indicate an excess of mapped tags in the Drosha condition.
(C) Experiment was performed as in (B) except that RNAPII, RNAPII ser5, and RNAPII ser2 antibodies were used for ChIP-seq.
(D) Analysis of genes targeted by Drosha. HeLa LTR-Luc cells transfected with the indicated siRNA were analyzed by ChIP assay using antibody against Drosha (top) or by NRO (bottom). Regions amplified by q-PCR using specific oligonucleotides are indicated. ChIP results are presented as fold enrichment over that of a mock precipitation using an unrelated IgG antiserum. For NRO, values were normalized to the amount of GAPDH RNA in the same samples. The result for Scrtreated cells was attributed a value of 1. All graphs show mean ± SE from at least three independent experiments. See also Table S2.
(E) TR of RNAPII is modified by Drosha. ChIP-seq data using anti-RNAPII performed on chromatin from HeLa LTR-Luc cells transfected with the indicated siRNA were analyzed, and the TR of RNAPII was determined.
(F) A proposed model for premature termination and transcriptional repression at the HIV-1 promoter. See text for details.
DISCUSSION
Our data demonstrate a mechanism of RNA-dependent TGS that depends on a PTGS-independent function for the microprocessor complex. In cooperation with the termination factors, Setx and Xrn2, and Rrp6, microprocessor regulates RNAPII pausing and premature termination at the HIV-1 promoter. Based on data presented, we propose a model for the establishment of a repressive cycle of transcription (Figure 5F). Recruitment of the microprocessor to the nascent TAR RNA leads to cleavage of the TSS transcript by Drosha, which generates nonadenylated TAR on the 5′ side of the cut, and an uncapped transcript 3′ to the cut. The uncapped RNA serves as a signal for recruitment of the termination factor, Xrn2, which degrades the ongoing transcript, leading to termination of transcription, whereas the free 3′ end of TAR signals recruitment of Rrp6. By further processing the cleavage product, an RNA species is generated that represses transcription. Access of RNAPII to the promoter becomes severely restricted, leading to transcriptional repression. In support of this model, (1) knockdown of Drosha/Dgcr8 diminishes recruitment of termination factors, Setx and Xrn2, and Rrp6 to the promoter-proximal region, and (2) biogenesis of a repressive small TAR-derived RNA depends on Rrp6. Consequently, microprocessor facilitates the establishment of transcriptionally repressive chromatin. Thus, in agreement with reports showing that small RNA-mediated chromatin remodeling and TGS can be induced in mammalian cells (Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009; Benhamed et al., 2012; Cernilogar et al., 2011; Guang et al., 2010; Rahl et al., 2010; Seila et al., 2008), we propose that the microprocessor, in concert with Setx, Xrn2, and Rrp6, establishes TGS at the viral promoter. This mechanism of repression is dynamic because it depends on the availability and recruitment of the different factors involved. Furthermore, it is dependent on a low level of ongoing transcription to provide the RNA component, which may form R loop or RNA:DNA hybrids that have been linked to the recruitment of repressive enzymatic complexes (Schmitz et al., 2010). Consistently, small TAR RNA-mediated repression requires the presence of the corresponding DNA sequence. Additionally, low-level transcription is important to prevent nucleosome assembly over the TSS that could result in more profound transcriptional repression (Gilchrist et al., 2010).
Genome-wide studies have firmly established that a significant proportion of genes undergo promoter-proximal pausing of RNAPII following the synthesis of a short transcript. However, it is currently unclear whether such polymerases remain in a paused state until their eventual release into productive elongation or whether they enter a termination pathway, thus clearing the way for new rounds of initiation (Nechaev and Adelman, 2011). Our data suggest that RNAPII pausing at the HIV-1 promoter under nonactivating conditions leads to premature termination of transcription that is initiated by the endonuclease activity of microprocessor toward TAR RNA. ChIP-seq data further indicate that a similar mechanism may operate at a subset of cellular genes. Consistent with this idea, Xrn2 and mRNA-decapping factors were recently identified at the 5′ end of a large number of paused genes, and loss of these factors correlated with enhanced elongation (Brannan et al., 2012). Thus, it appears likely that promoter-proximal RNAPII detected on a genome-wide scale represents a pastiche of both paused and terminated polymerases. Further studies will be required to discriminate between these outcomes at specific genes.
The finding that Drosha and DGCR8 are associated with a subset of cellular genes that are not known to encode miRNA supports the idea that these factors have additional functions in transcription. A highly abundant class of noncoding transcripts corresponds to small RNAs that appear to arise from polymerases that have stalled shortly after initiation (Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009; Core et al., 2008; Seila et al., 2008), and which have been shown to downregulate myc gene expression (Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009). We show that repression of the LTR can be enhanced by transfection of small RNAs, in an Rrp6-dependent manner. Thus, in agreement with reports showing that small RNA-mediated chromatin remodeling and TGS can be induced in mammalian cells (Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project, 2009; Guang et al., 2010; Rahl et al., 2010; Seila et al., 2008), we propose that the microprocessor, together with Setx, Xrn2, and Rrp6, initiates premature termination of transcription and TGS at the HIV-1 promoter and a subset of cellular genes.
Antiretroviral treatment (ART) potently controls HIV replication in infected individuals but does not eradicate the virus due, in part, to the establishment of a reservoir of stably integrated provirus that is transcriptionally silent. Establishment of the silent reservoir has been shown to depend on chromatin repression and a block to transcription elongation (Siliciano and Greene, 2011). Microprocessor-mediated TGS at the HIV promoter may have important implications in the establishment of a transcriptionally silent viral reservoir. Consistently, we have previously shown that knockdown of Drosha in PBMCs isolated from patients with HIV under suppressive therapy results in virus reactivation through RNAi-dependent and -independent mechanisms (Triboulet et al., 2007). Here, we show that microprocessor modulates transcription from silent HIV-1 provirus. Further work is required to determine whether microprocessor in concert with the Setx, Xrn2, and Rrp6 plays a role in HIV transcriptional silencing leading to the establishment and/or the maintenance of the transcriptionally latent viral reservoir.
EXPERIMENTAL PROCEDURES
Antibodies and Plasmids
Rabbit anti-Dgcr8, mouse anti-Dicer, Rabbit anti-Rrp6, mouse mAb anti-RNA Polymerase II CTD (clone 4H8), and rabbit anti-H3K36me3 were purchased from Abcam (Cambridge). Rabbit anti-Drosha (Up07717), mouse anti-Hp1γ (Up05690), and rabbit anti-H3K27me3 (Up07449) were purchased from Millipore. Rabbit anti-RNA Polymerase II CTD Phospho S2, rabbit anti-RNA Polymerase II CTD Phospho S5, rabbit anti-Xrn1, rabbit anti-Rck/p54, and rabbit anti-Setx were purchased from Bethyl (Montgomery, TX, USA). Mouse anti-Flag (M2), mouse anti-tubulin (clone DM1A), rabbit anti-Drosha, rabbit anti-Lsm1, and normal mouse IgA and IgG were purchased from Sigma-Aldrich (St. Louis). Rabbit anti-Xrn2 was purchased from ProteinTech Group. Rabbit anti-GW182 was purchased from Novus Biologicals (Cambridge). Rabbit anti-TRBP1 was obtained from Dr. K.-T. Jeang and has been described previously by Bennasser et al. (2006). Plasmids encoding WT Drosha and the mutant lacking the endonucleolytic activity (E1045Q/E1222Q) have been described previously by Han et al. (2006).
Cell Culture, Transfection, and Treatment
HeLa LTR-Luc and HeLa LTRΔTAR-Luc cells (Tréand et al., 2006) were obtained from Dr. Stéphane Emiliani (Institut Cochin, Paris) and were propagated in Dulbecco’s modified Eagle’s medium (DMEM; Lonza, Basel) supplemented with 10% fetal bovine serum (FBS; AbCys, Paris) and antibiotics. For transfection of siRNAs, HeLa cells harboring a stably integrated pNL4-3 (Tat minus) provirus or HeLa-LTR-luc cells were transfected with siRNAs (10 or 30 nM final concentration) using oligofectamine (Invitrogen, Paisley, UK) or Interferin (PolyPlus Transfection, Illkirch, France) according to the manufacturers’ instructions. Plasmid transfection was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) or Jet PEI (PolyPlus Transfection) according to the manufacturers’ instructions. Where indicated, cells were treated with phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) overnight. Treatment with GST and GST-Tat was carried out as previously described by du Chéné et al. (2007).
For transfection of RNAs, HeLa-LTR-luc cells were transfected with a first round of siRNA (20 nM final concentration) using oligofectamine according to the manufacturers’ instructions. Twenty-four hours later, the cells were transfected a second time with the same concentration of siRNA supplemented with 1 μg of the indicated RNA oligonucleotide, unless otherwise stated. Cells were harvested at approximately 60 hr for luciferase activity and western blot.
siRNAs and q-PCR Oligonucleotides and RNA Oligonucleotides
Double-stranded RNA oligonucleotides used for RNAi were purchased from Eurofins MWG Operon (Ebersberg, Germany). Target sequences are shown in Extended Experimental Procedures. PCR primer sequences to amplify TAR, early, luc (coding region), luc 3′, and GAPDH-Q have been described elsewhere (Nakamura et al., 2012). Sequences of additional oligonucleotide pairs used are shown in Extended Experimental Procedures. TAR RNA was purchased as a purified RNA oligonucleotide from Thermo Scientific. NS, Sp1 and TAR 1, TAR2 and TAR3 RNA oligonucleotides were purchased from MWG Operon. The sequences are shown in Extended Experimental Procedures.
Luciferase Assays and Immunoblot
Luciferase activity was measured according to the manufacturer’s protocol (Promega, Madison, WI, USA) and was normalized to cell protein concentration. Immunoblot was performed as described previously (Kiernan et al., 2001).
Quantitative RT-PCR
Total RNA was extracted from cells using TRIzol (Invitrogen) according to the manufacturer’s instructions and reverse transcribed using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). Reverse-transcription products were amplified by real-time PCR (LightCycler; Roche) using QuantiTect SYBR Green (QIAGEN, Germantown, MD, USA) with the indicated oligonucleotides. q-PCR cycling conditions are available on request. Unless otherwise stated, samples were analyzed by reverse-transcription q-PCR using the specific oligonucleotide pairs indicated, and GAPDH. The amount of the indicated mRNA was normalized to the amount of GAPDH mRNA in each sample, and the values were normalized to those for the control transfection (Scr), which was attributed a value of one (1).
ChIP
Cells were transfected as indicated in the figures. Following 64 hr incubation, cells were washed and harvested for native ChIP, performed as described previously (Wagschal et al., 2007), or crosslink ChIP, which was performed as described previously (Nakamura et al., 2012) except that chromatin was precleared at 4°C for 2 hr with 20 μl protein A or G magnetic beads (Invitrogen) that had been preblocked with 1% BSA. Antibodies (2 μg) were incubated for 4 hr with 20 ml of blocked magnetic beads before the addition of sonicated chromatin overnight at 4°C. An aliquot was amplified by real-time PCR as described previously by Lassot et al. (2007) using specific oligonucleotide primers indicated. An aliquot of chromatin was amplified in parallel, and values obtained for immunoprecipitates were normalized to values for chromatin (percent [%] input DNA).
RNase treatment of chromatin was performed as described previously by Abruzzi et al. (2004) except that cells were prefixed for 2.5 min prior to the addition of RNase cocktail (Ambion). ChIP was then performed as described above.
Nuclear Run-On Transcription
Run-on transcription was performed as described previously by Core et al. (2008). Run-on transcripts were reverse transcribed and quantified by PCR using the oligonucleotide pairs indicated. Results were normalized to the amount of GAPDH run-on transcript in the same sample.
Small RNA-seq and Northern Blotting
Small RNA-seq and northern blotting were performed as described previously by Schopman et al. (2012). Full-length TAR RNA was used as a probe in northern blotting.
ChIP-seq
Sample preparation was performed using the ChIP-seq sample preparation kit from Illumina (ref. IP-102-1001) according to the manufacturer’s instructions. Briefly, 10 ng of sonicated chromatin was repaired using a mix of T4 DNA Polymerase, Klenow DNA Polymerase, and T4 Polynucleotide kinase. The resulting fragments were A tailed using Klenow DNA polymerase (3′–5′ exo minus). Illumina’s adapters were ligated to the DNA fragment using T4 DNA ligase. The libraries were size selected at 200 bp (± 25 bp) on a 2% agarose gel. Once extracted from agarose, 18 cycles of PCR were performed on the libraries using Illumina’s PCR primers. Each library was diluted to 10 nM, denatured, and diluted again to 8 pM. A total of 100 μl of the diluted library was hybridized on a lane of an Illumina’s Flow Cell. Clustering and 36 cycle sequencing were performed according to Illumina’s instructions.
Reads that aligned to unique positions in the genome were processed using peak-calling MACS (Zhang et al., 2008) with mfod ≥10 and p = 10−5 for the binomial distribution. Peaks with false discovery rate (FDR) ≤1% were conserved. Target genes (503) were identified when at least 1 nt overlaps between gene and peak positions. Calculation of RNAPII TR that compares the ratio between Pol II density in the promoter and in the gene region was performed as described previously by Rahl et al. (2010).
Reads that aligned to multiple genomic locations were aligned on the human Repbase. Differentially expressed repeat tags were identified by a Fisher’s exact test with Benjamini-Hochberg method to compute the FDR and a cutoff p value of 0.01 as implemented in the SAGE Genie resource (http://cgap.nci.nih.gov/SAGE).
All Drosha ChIP-seq tags filtered against Repbase were aligned on all human genomic regions known to contain a copy of an endogenous retrovirus member of the HERV-H family. Only tags that fully and exactly aligned were considered. Because the HERV-H family has several hundred copies in the human genome, we visually checked the tag density over the hundred copies on which the number of aligned tags was maximal without noticing any qualitative difference. The HERV-H copy on the Chr6:21,360,000–21,368,000 region was then selected for further analysis because it is one of the copies that has a high number of aligned tags.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Molecular Virology and Gene Regulation labs for critical reading of the manuscript, N. Kim for Drosha WT and mutant plasmids, and S. Emiliani for HeLa-LTR-luc. ChIP-seq experiments were performed using MGX-Montpellier facilities. This work was supported by grants from the ERC (250333), ANR-BLAN-0040, Sidaction, ANRS, and FRM “équipe labéllisée” to M.B. and ANRS and Sidaction to R.K. A.W. was supported by ANR and ERC; E.R. and M.N. by Sidaction; P.B. by ANRS; S.L.-C. by ERC; X. Chen by CNRS and FRM; K.Z. by CNRS; O.M. by RTRS grand Sud; and B.B. by The Netherlands Organization for Scientific Research (NWO-CW, Top grant).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.08.004.
REFERENCES
- Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix ENCODE Transcriptome Project. Cold Spring Harbor Laboratory ENCODE Transcriptome Project Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- Bisgrove D, Lewinski M, Bushman F, Verdin E. Molecular mechanisms of HIV-1 proviral latency. Expert Rev. Anti Infect. Ther. 2005;3:805–814. doi: 10.1586/14787210.3.5.805. [DOI] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley DL. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoterproximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval N, Casella J, Gressin L, Heidmann T. Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology. 2001;279:558–569. doi: 10.1006/viro.2000.0737. [DOI] [PubMed] [Google Scholar]
- de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan RE, Emiliani S, Nakayama K, Castro A, Labbé JC, Lorca T, Nakayama Ki K, Benkirane M. Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 2001;21:7956–7970. doi: 10.1128/MCB.21.23.7956-7970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, Chable-Bessia C, Coux O, Benkirane M, Kiernan RE. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Basavarajaiah P, Rousset E, Beraud C, Latreille D, Henaoui IS, Lassot I, Mari B, Kiernan R. Spt6 levels are modulated by PAAF1 and proteasome to regulate the HIV-1 LTR. Retrovirology. 2012;9:13. doi: 10.1186/1742-4690-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, Haasnoot J. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 2012;40:414–427. doi: 10.1093/nar/gkr719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Zamore PD. Rethinking the microprocessor. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. HIV Latency. Cold Spring Harb. Perspect. Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Tréand C, du Chéné I, Brès V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Delaval K, Pannetier M, Arnaud P, Feil R. Chromatin immunoprecipitation (ChIP) on unfixed chromatin from cells and tissues to analyze histone modifications. CSH Protoc. 2007 doi: 10.1101/pdb.prot4767. Published online June 1, 2007. 10.1101/pdb.prot4767. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.