SUMMARY
HIV-1 transactivator Tat has greatly contributed to our understanding of transcription elongation by RNAPII. We purified HIV-1 Tat-associated factors from HeLa nuclear extract and show that Tat forms two distinct and stable complexes. Tatcom1 consists of the core active P-TEFb, MLL-fusion partners involved in leukemia (AF9, AFF4, AFF1, ENL, and ELL), and PAF1 complex. Importantly, Tatcom1 formation relies on P-TEFb while optimal CDK9 CTD-kinase activity is AF9 dependent. MLL-fusion partners and PAF1 are required for Tat transactivation. Tatcom2 is composed of CDK9, CycT1, and 7SK snRNP lacking HEXIM. Tat remodels 7SK snRNP by interacting directly with 7SK RNA, leading to the formation of a stress-resistant 7SK snRNP particle. Besides the identification of factors required for Tat transactivation and important for P-TEFb function, our data show a coordinated control of RNAPII elongation by different classes of transcription elongation factors associated in a single complex and acting at the same promoter.
INTRODUCTION
The field of eukaryotic transcription has seen fundamental conceptual changes in recent years (Buratowski, 2008). Studies revealing promoter-proximal pausing of RNA polymerase II (RNAPII) at 30% of human genes established postinitiation events as an important step in gene regulation (Core and Lis, 2008; Guenther et al., 2007). The carboxy-terminal domain (CTD) of RBP1, the largest subunit of RNAPII, has been proposed to serve as a binding scaffold for factors that ensure proper maturation of the RNAPII complex through the transcription cycle and link mRNA biogenesis to processing and export. Transition through these steps is tightly coupled to phosphorylation of RNAPII CTD. From a simplistic view, RNAPII phosphorylated at Ser5 mostly occupies promoter regions, and productive elongation of nascent RNA requires the action of the positive transcription elongation factor b (P-TEFb) (Peterlin and Price, 2006). In vivo, two P-TEFb complexes predominate in equilibrium. Core active P-TEFb is composed of one molecule of the regulatory subunit Cyclin (Cyc) T1 (or the less abundant T2 or K) and one molecule of the catalytic subunit CDK9. Inactive P-TEFb (7SK snRNP) is composed of 7SK RNA (Nguyen et al., 2001; Yang et al., 2001), 7SK-capping enzyme MEPCE, LARP7, HEXIM1 and/or HEXIM2, SART3, CDK9, and CycT1 or T2 (Jeronimo et al., 2007; Krueger et al., 2008; Markert et al., 2008). Depending on the cellular demand, following various stress signals or proliferation for instance, the CDK9/CycT1 heterodimer is released from 7SK snRNP, leading to enhanced active P-TEFb that will stimulate transcription elongation from target genes. At mammalian genes, P-TEFb is often recruited through its binding to BRD4, forming a stoichiometric complex (Jang et al., 2005; Yang et al., 2005). Targeting of P-TEFb to a specific promoter leads to phosphorylation of the RNAPII-CTD at Ser2 that plays an important role in transcriptional regulation and cotranscriptional mRNA processing (Egloff and Murphy, 2008). In addition to the RNAPII CTD, P-TEFb/CDK9 also phosphorylates the negative transcription elongation factor NELF and the DRB-sensitivity-inducing factor DSIF/Spt4:Spt5 to overcome their negative action (Brès et al., 2008).
The HIV-1 promoter is a well-defined model that has provided considerable insight into P-TEFb-mediated control of transcriptional elongation. Transcription from the long terminal repeat (LTR) leads to RNAPII pausing after synthesis of a short RNA, the transactivation response element (TAR). The HIV-1 transactivator protein, Tat, together with CycT1, binds a bulge-loop within TAR (Wei et al., 1998), allowing CDK9 to phosphorylate RNAPII CTD and NTEFs, licensing RNAPII for productive elongation.
To gain more insight into P-TEFb function, we purified Tat and associated factors from HeLa nuclear extract. We show that Tat forms two stable and distinct complexes. Biochemical and functional analysis of Tatcom1 led to the identification of factors potentiating P-TEFb kinase activity and required for Tat/P-TEFb-mediated transactivation of the HIV-1 promoter. Those are MLL-fusion partners involved in leukemia (AF9, AFF4, AFF1, ENL, and ELL) and the PAF1 complex. Importantly, endogenous P-TEFb interacts with MLL-fusion partners and PAF1 in the absence of Tat. However, Tat increases the pool of P-TEFb associated with MLL-fusion partners and the PAF1 complex. Thus, Tat reassembles a stable transcription elongation complex containing distinct modules known to regulate different aspects of transcription elongation by RNAPII. Tatcom2 is composed of P-TEFb and 7SK snRNP containing LARP7 and MEPCE but lacking HEXIM1 and SART3. Remodeling of 7SK snRNP by Tat leads to formation of a stress-resistant 7SK snRNP particle, adding insight into Tat-mediated regulation of P-TEFb equilibrium.
RESULTS
HIV-1 Tat Binds P-TEFb, MLL-Fusion Partners, the PAF1 Complex, and Subunits of the Kinase-Inactive P-TEFb Complex, Including 7SK RNA
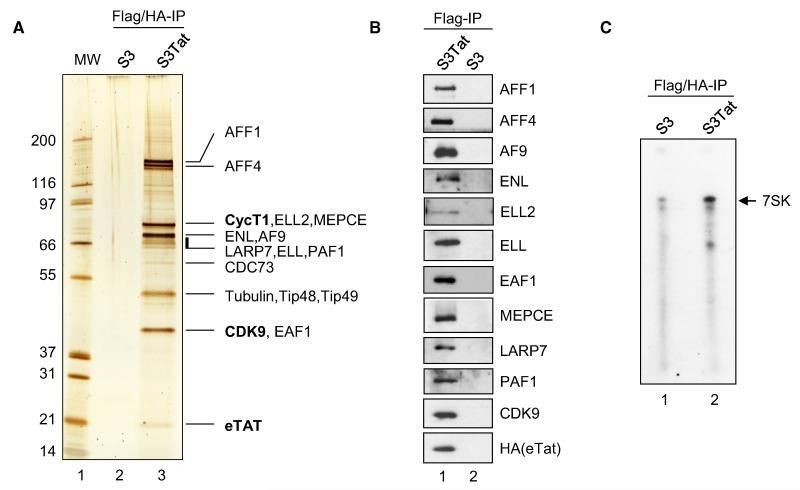
A myriad of proteins have been reported to interact with HIV-1 Tat, but as of today, CDK9 and CycT1 are the only known stoichiometric binding partners required for its transactivation function (Wei et al., 1998). To gain insight into Tat-mediated transcriptional regulation, we purified Tat and its associated partners using a previously established method (Nakatani and Ogryzko, 2003). HeLa S3 (S3) cells were stably transduced with full-length HIV-1 Tat (S3Tat) composed of 101 amino acids and fused to Flag and HA epitope-tags (eTat). Expression of eTat in S3Tat was comparable to that in Jurkat cells infected with HIV-1 (Tréand et al., 2006). eTat was purified from Dignam nuclear extracts derived from S3Tat cells using tandem affinity chromatography. At least 16 proteins that specifically associate with Tat were identified by tandem mass spectrometry. The percent protein coverage of all major eTat interactors (Figure S1A and Table S1), together with the intensity of the corresponding stained band (Figure 1A), gives an approximate estimate of the relative abundance of eTat-associated factors as compared to P-TEFb. As previously reported, CDK9 and CycT1 were found to be stoichiometric Tat-binding partners, suggesting a role for associated complexes in Tat/P-TEFb-mediated transcriptional regulation. Western blot (WB) assay of Flag-purified eTat with antibodies against the identified proteins confirmed the interactions (Figure 1B). Moreover, TatC22G mutant unable to bind CycT1 (Wei et al., 1998) failed to interact with Tat-associated factors (Figure S1B). Surprisingly, substantial interaction of eTat with LARP7 and MEPCE, members of the 7SK snRNP, was detected, suggesting the presence of a Tat-containing 7SK/P-TEFb particle. In support of this hypothesis, an RNA protection assay using a full-length 7SK-specific probe showed the presence of intact 7SK RNA in the double-purified eluate (Figure 1C). Of note, HEXIM1 and SART3, major constituents of the endogenous 7SK/P-TEFb complex, were not detected.
Figure 1. Purification of HIV-1 Tat Complexes.
(A) HIV-1 Tat was purified from Dignam nuclear extracts prepared from Flag-HA epitope-tagged Tat (eTat)-expressing HeLa S3 cells (S3Tat) or nontransduced S3. eTat was sequentially purified on anti-Flag and anti-HA antibody-conjugated agarose beads. Proteins were resolved by SDS-PAGE and visualized by silver staining. The identity of eTat-associated proteins was determined by mass spectrometry (see also Figure S1 and Table S1).
(B) Flag IP from samples shown in (A) were resolved on SDS-PAGE, and the presence of eTat-associated proteins identified was confirmed by WB.
(C) 7SK RNA copurifies with eTat. Total RNA was extracted from samples used in (A). The presence of 7SKRNA was assessed by RNase protection assay using a full-length 7SK probe.
Analysis of Tat-associated proteins also revealed the presence of PAF1, a well-established player in transcriptional elongation. With the exception of CDC73, other subunits of the PAF1 complex (Ctr9 and Leo1) were not identified by mass spectrometry. However, when assessed by WB using specific antibodies, both Ctr9 and Leo1 were detected in eTat-purified material as compared to PAF1 immunoprecipitates (Figure S1C). Interestingly, Leo1 immunoprecipitated AFF4, CDK9, and eTat from S3Tat nuclear extracts (Figure S1D). Together, these experiments suggest that the PAF1 complex is associated with eTat.
Interestingly, many common MLL-fusion partners (AFF1, AFF4, AF9, ENL, and ELL) involved in Acute Leukemia (Meyer et al., 2009) that have previously been linked to transcriptional regulation (Bitoun et al., 2007; Mueller et al., 2007; Schulze et al., 2009) copurified with eTat. ELL and ELL2 are well-characterized elongation factors both in vitro and in vivo (Shilatifard, 1998). We confirmed these interactions in HIV-1 target cells by purifying eTat from a stably transduced Jurkat T cell line (Figure S1E).
HIV-1 Tat Forms at Least Two Distinct P-TEFb-Containing Complexes
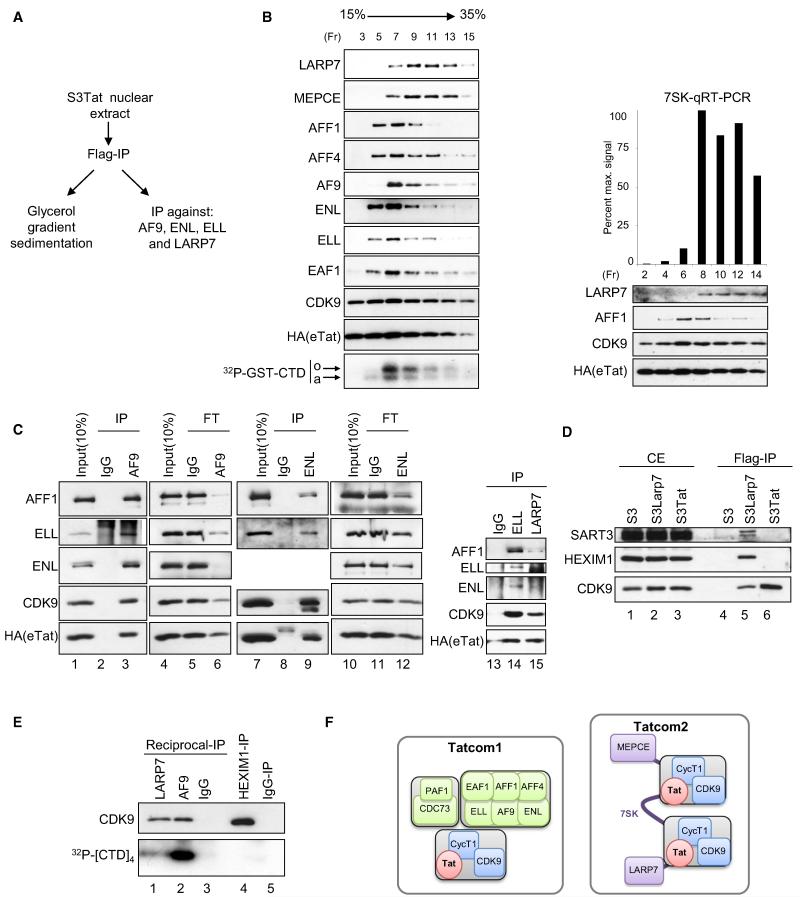
To test if the above-identified proteins form one or several complexes, glycerol gradient sedimentation of Flag-purified eTat was performed (Figure 2B), followed by WB analysis of collected fractions. eTat and CDK9 showed a near-identical distribution ranging from fraction 3 to 15. AF9, ENL, AFF1, AFF4, ELL and its associated factor EAF1, and PAF1 peaked between fractions 5 and 9 (Figures 2B, left panels, and S2). In contrast, MEPCE and LARP7 were most abundant in fractions 9–13. Quantitative RT-PCR analysis using 7SK-specific primers showed comigration of 7SK with MEPCE and LARP7 (Figure 2B, right panel). Interestingly, in vitro CTD-kinase reactions show that fraction 7, enriched in AFF1, AFF4, ENL, AF9, ELL, and EAF1, possesses stronger kinase activity as compared to the other fractions (Figure 2B, bottom left).
Figure 2. HIV-1 Tat Forms Two Distinct Complexes: Tatcom1, Containing Active P-TEFb, and Tatcom2, Containing 7SK, LARP7, and MEPCE.
(A) Experimental scheme for (B) and (C).
(B) Flag-purified eTat-associated complexes were separated by centrifugation through a 15%–35% glycerol gradient. Odd numbered fractions (Fr) were analyzed by WB and assayed for in vitro CTD-kinase activity using GST-CTD and [γ-32P]ATP. Reactions were separated on SDS-PAGE and visualized by autoradiography (bottom panel). Even-numbered fractions were probed with LARP7, AFF1, CDK9, and HA antibodies (right panel). RNA was extracted from these fractions and subjected to qRT-PCR analysis using 7SK-specific primers (top right panel). The hypophosphorylated (a) and hyperphosphorylated (o) forms of GST-CTD are indicated.
(C) Flag-purified eTat (input) was subjected to IP using AF9-, ENL-, ELL-, or LARP7-specific antibodies or irrelevant IgG. Input, IPs, as well as flowthrough (FT) from AF9 and ENL IPs were probed with the indicated antibodies (lanes 1–12). The presence of Tatcom1 subunits AFF1 and ENL in reciprocal IPs using ELL- or LARP7-specific antibodies, normalized for eTat levels, was assessed by WB (lanes 13–15).
(D) HEXIM1 and SART3 do not associate with eTat. Whole-cell extracts derived from S3 cells stably transduced with Flag- and HA-tagged LARP7, eLARP (S3LARP7), and eTat (S3Tat) were subjected to Flag IP. Cell extracts (CE) and IPs were analyzed for the presence of HEXIM1, SART3, and CDK9 by WB.
(E) Reciprocal IP with AF9 and LARP7 antibodies, normalized to amounts of CDK9 (top panel), as well as HEXIM1 IPs were assayed for in vitro kinase activity using [γ-32P]ATP and a tetrarepeat of the CTD consensus as substrate. Reactions were separated on SDS-PAGE and visualized by autoradiography (lower panel).
(F) Schematic representation of eTat-associated complexes. See also Figure S2.
Our data suggest that eTat/CDK9/PAF1/AF9/ENL/AFF1/AFF4/ELL/EAF1, referred to as Tatcom1, and eTat/CDK9/7SK/LARP7/MEPCE, or Tatcom2, may represent distinct biochemical entities. To further test this hypothesis, we performed reciprocal immunoprecipitations (IPs). Flag-purified eTat was subjected to IP with antibodies specific to members of Tatcom1 and Tatcom2. Anti-AF9 antibodies coimmunoprecipitated AFF1, ELL, and ENL together with CDK9 and eTat (Figure 2C, left panel). Furthermore, analysis of the flowthrough after AF9 IP shows specific codepletion of AFF1, ELL, and ENL and, to a lesser extent, of CDK9 and eTat (lanes 6 and 5). Similar results were obtained with ENL-specific antibody that immunoprecipitated AFF1, ELL, CDK9, and eTat (lanes 9 and 8) and depleted AFF1, ELL, and ENL more efficiently from eTat-associated complexes than eTat or CDK9 (Figure 2C, lanes 11 and 12). These results indicate the presence of a complex composed of eTat, CDK9, AF9, ENL, ELL, AFF1, and yet another eTat/P-TEFb-containing complex, since neither AF9 nor ENL reciprocal IPs depleted eTat and CDK9.
Reciprocal IP with ELL- and LARP7-specific antibodies were not depleting, but were normalized to the amount of coimmunoprecipitated eTat. Consistent with AF9 and ENL reciprocal coIPs, ELL IP copurified ENL, AFF1, CDK9, and eTat (Figure 2C, lane 14). Furthermore, reciprocal IP using LARP7-specific antibody copurified eTat and CDK9 but failed to copurify ENL and ELL, and only a weak interaction was observed for AFF1 (lanes 14 and 15).
Given the similar subunit composition between Tatcom2 and the endogenous 7SK snRNP, and since no HEXIM1 was detected by mass spectrometry, we asked whether eTat substitutes for HEXIM1 during Tatcom2 formation. S3 cells stably expressing Flag- and HA-tagged LARP7 (S3LARP7) or S3Tat were used to Flag purify eLARP7 and eTat, respectively. When normalized to the amount of copurified CDK9, neither SART3 nor HEXIM1 were detected in Flag-purified eTat as compared to Flag-purified eLARP7 (Figure 2D, lanes 5 and 6). The fact that eTat and eLARP7 were purified under the same conditions excludes the possibility that the absence of HEXIM1 and SART3 may be due to loss of 7SK RNA during the purification procedure. This, together with the results from Figure 1, infers that Tat remodels the cellular 7SK snRNP, possibly by replacing HEXIM1, resulting in Tatcom2. To further characterize Tatcom1 and Tatcom2, Flag-purified eTat was subjected to reciprocal IP using either AF9- or LARP7-specific antibodies. In vitro CTD-kinase activity was performed after normalization to CDK9 levels (Figure 2E). Tatcom2 displays lower CTD-kinase activity than Tatcom1 (lower panel, lanes 1 and 2). Significantly, Tatcom2 shows stronger activity than HEXIM1-bound P-TEFb (lanes 1–4).
Taken together, the glycerol gradient velocity analysis and reciprocal IPs, both using Flag-purified eTat as input material, strongly suggest the existence of two major biochemically and functionally distinct eTat-associated complexes (Figure 2F). Tatcom1, composed of at least P-TEFb, AF9, ENL, ELL, AFF1, AFF4, and PAF1, presenting strong CTD-kinase activity, is thus likely to be directly involved in transcriptional elongation. Tatcom2, a second P-TEFb-containing complex, binds Tatcom1 subunits less efficiently and is primarily composed of 7SK, LARP7, and MEPCE. Since no HEXIM1 was found associated with Tatcom2 and endogenous 7SK snRNP contains two molecules of HEXIM1, Tatcom2 is represented with two molecules of Tat/P-TEFb.
7SK Is an Integral Component of Tatcom2
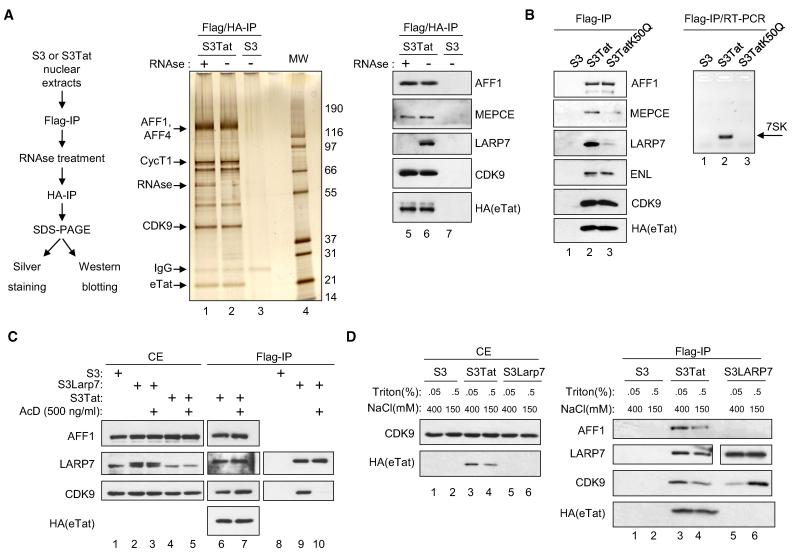
To assess the importance of 7SK in the integrity of Tatcom2, Flag-purified eTat was subjected to RNase digestion and further purified using anti-HA antibody. Immunoprecipitated material was visualized on a silver stained gel, essentially showing no readily observable differences between mock-treated and RNase-digested complexes (Figure 3A, middle panel). This infers that the integrity of eTat-associated complexes does not require 7SK. Thus, the architecture of Tatcom2 may be different from 7SK snRNP, which is disrupted upon RNase treatment. WB analysis of the same samples revealed a 7SK-dependent interaction with LARP7, since LARP7 was absent in RNase-treated Flag/HA-purified eTat (lanes 6 and 5). LARP7 comigrates with other proteins present in Flag-/HA-purified eTat, and its disappearance upon RNase treatment is difficult to appreciate from the silver stained gel. As observed for endogenous 7SK snRNP, the MEPCE/P-TEFb interaction was also RNase resistant in the context of Tatcom2 (lanes 5 and 6).
Figure 3. 7SK Is an Integral Component of Tatcom2.
(A) Left panel: Experimental scheme. Dignam nuclear extracts were prepared from S3Tat or S3 and subjected to Flag affinity chromatography. Flag-peptide eluates were treated with RNase A and RNase V1 or mock treated and subsequently HA purified and HA peptide eluted. The resulting material was separated on SDS-PAGE and visualized either by silver staining (middle panel) or WB (right panel).
(B) Whole-cell extracts from S3, S3Tat, or S3 cells stably expressing a TAR-binding-defective eTat mutant carrying a Lys50 to glutamine substitution (S3TatK50Q) were subjected to Flag IP. Eluates were either analyzed by WB (left panel) or the presence of 7SK monitored by RT-PCR using specific oligonucleotides (right panel, see also Figure S3).
(C) S3, S3Tat, or S3LARP7 cells were exposed to actD (500 ng/ml) for 1 hr or untreated prior to whole-cell extraction and Flag IP. Cell extracts (CE) and IPs were analyzed by WB (see also Figures S3E and S3F).
(D) S3, S3Tat, and S3LARP7 cells were lysed using buffer containing 400 mM NaCl and 0.05% Triton X-100 or 150 mM NaCl and 0.5% Triton X-100. The resulting extracts were subjected to Flag IP. Cell extracts (left panel) and IPs (right panel) were analyzed by WB. See also Figure S3.
Previous reports demonstrate that kinase-active P-TEFb is released from the 7SK snRNP. Therefore, a central question is whether Tatcom1 assembly depends upon Tatcom2 formation. In vitro, Tat can interact with 7SK through its TAR-binding motif (Sedore et al., 2007; Yik et al., 2004). We therefore compared subunit composition of wild-type eTat-associated complexes with a TAR-binding defective, transcriptionally inactive eTat mutant in which Lys50 is substituted by glutamine (eTatK50Q) but readily interacts with CycT1 (Brès et al., 2002). Flag IPs from S3Tat or S3TatK50Q were analyzed for the presence of eTat cofactors (Figure 3B, left panel). Tatcom1 subunits AFF1 and ENL bound eTatK50Q as efficiently as eTat. In contrast, binding to LARP7 and MEPCE, components of Tatcom2, was strongly reduced in eTatK50Q IPs (lanes 2 and 3). RT-PCR with 7SK-specific primers using RNA prepared from the same IPs showed reduced binding of 7SK to eTatK50Q as compared to eTat (right panel). These data imply that Tatcom2 formation is 7SK dependent but that formation of this complex is not a prerequisite for Tatcom1 assembly. In support of this conclusion, knockdown of LARP7 has no effect on Tatcom1 formation (Figure S3A). Furthermore, maintenance of most protein-protein interactions, except for LARP7, was RNase resistant and thus 7SK independent. Additionally, both eTat and eTatK50Q compete with HEXIM1 for CycT1 binding with the same efficiency when overexpressed (Figure S3B), indicating that competition between Tat and HEXIM1 for P-TEFb binding does not depend on their ability to interact with 7SK.
Lastly, we investigated whether Tatcom2 formation is part of an adaptation process due to stable expression of Tat. Whole-cell extracts (WCEs) from S3Tat cells kept in culture for 1 month and newly eTat-transduced S3 cells were Flag immunoprecipitated. The presence of AFF1, LARP7, and CDK9 was assessed by WB (Figure S3D). Since no significant differences were observed, we excluded Tatcom2 as an artifact due to long-term exposure of cells to Tat.
HIV-1 Tat-Associated Complexes Are Stress Resistant
Given the dynamic nature of P-TEFb shuttling between kinase-active and 7SK snRNP states, we tested the sensitivity of Tat-associated P-TEFb complexes to various forms of stress known to affect the P-TEFb equilibrium. WCEs derived from S3Tat, S3LARP7, or S3 cells, mock treated or treated with actinomycinD (ActD), were immunoprecipitated with anti-Flag antibody and analyzed by WB. As previously reported, LARP7 binding to P-TEFb, as revealed by the presence of CDK9 in eLARP7 IPs, was dramatically reduced upon drug treatment (Figure 3C, lanes 9 and 10). However, P-TEFb binding to Tat was unperturbed (lanes 6 and 7). Furthermore, the integrity of Tatcom1 and Tatcom2, as measured by copurified AFF1 and LARP7, respectively, was also unaffected (lanes 6 and 7). Similar results were obtained following UV or hexamethylene bisacetamide (HMBA) (Figures S3E and S3F) treatment, known to disrupt the 7SK snRNP.
To further investigate the idea that Tat-associated complexes may not behave like endogenous P-TEFb and may differ in architecture, we tested their susceptibility toward the ionic strength of buffers. BRD4 as well as HEXIM1 and LARP7 have all been shown to dissociate from P-TEFb at 400 mM NaCl (Yang et al., 2005). As expected, LARP7 dissociated from P-TEFb upon high-salt treatment as shown by a reduction of copurified CDK9 (Figure 3D, right panel, lanes 5 and 6). eTat, however, was less extractable under low-salt conditions (left panel), reflected by reduced eTat and associated proteins in Flag eluates (right panel). Importantly, interactions between eTat, CDK9, LARP7, and AFF1 were readily observed under high-salt conditions, indicating that Tatcom1 and Tatcom2 are resistant to high ionic strength.
HIV-1 Tat Increases the Pool of P-TEFb Bound to MLL-Fusion Partners and PAF1
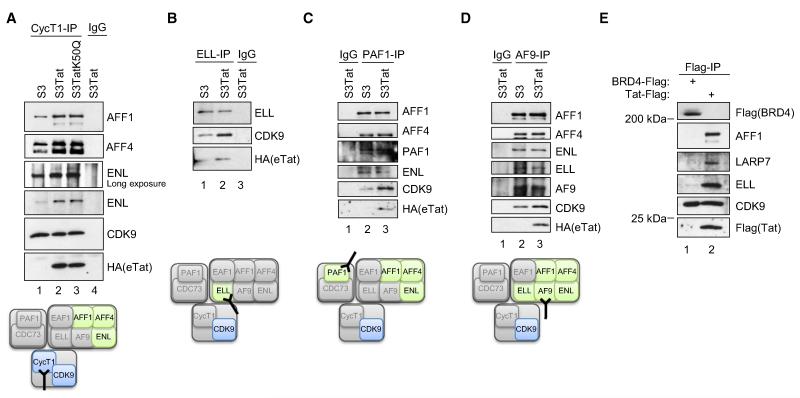
Given the subunit composition of Tatcom1 (Figure 2F) and the relative stoichiometry of the associated proteins in respect to CDK9 and CycT1 (Figure 1A), we asked whether the same repertoire of proteins bound P-TEFb in the absence of Tat. Thus, we analyzed the interaction between endogenous CDK9, AFF1, AFF4, AF9, ELL, ENL, and PAF1 in both S3 and S3Tat cells (Figure 4). Endogenous CycT1 immunopurified from S3 cells showed physical interactions with AFF1, AFF4, and ENL as compared to control IP (Figure 4A). However, these interactions were substantially increased in the presence of eTat. Of note, CycT1 immunoprecipitated equal amounts of CDK9 from S3 and S3Tat cells, suggesting that the increase of AFF1, AFF4, and ENL copurified with CycT1 from S3Tat is not due to IP of more P-TEFb. Consistent with results shown in Figure 3B, suggesting that Tat binding to 7SK snRNP is not required for Tatcom1 formation, IP of CycT1 from S3Tat and S3TatK50Q coimmunoprecipitated equivalent amounts of eTat together with AFF1, AFF4, and ENL (Figure 4A). These data confirm that Tat interaction with 7SK-bound P-TEFb is not a prerequisite for formation of Tatcom1, since a 7SK interaction-deficient mutant copurifies AFF1, AFF4, and ENL. Furthermore, eTat also increased ELL-bound P-TEFb (Figure 4B). Interestingly, PAF-1- and AF9-specific antibodies immunoprecipitated AFF1, AFF4, ENL, and ELL from S3 and S3Tat extracts with similar efficiency (Figures 4C and 4D). However, the amount of CDK9 associated with PAF-1 (Figure 4C) and AF9 (Figure 4D) was enhanced when Tat was present. Of note, endogenous CycT1, ELL, PAF-1, and AF9 coimmunoprecipitated eTat (Figure 4). These results indicate that a complex containing P-TEFb, AFF1, AFF4, AF9, ELL, ENL, and PAF1 exists under physiological conditions in vivo and that Tat enhances either its formation or stability.
Figure 4. HIV-Tat Increases the Pool of P-TEFb Bound to MLL-Fusion Partners and PAF1.
(A) Whole-cell extracts prepared from S3, S3Tat, and S3TatK50Q were subjected to IP using anti-CycT1 antibody or irrelevant IgG. IPs were analyzed by WB.
(B-D) Endogenous ELL (B), PAF1 (C), and AF9 (D) were immunopurified from S3 and S3Tat extracts using specific antibody or irrelevant IgG. The presence of Tatcom1 subunits in the IPs was analyzed by WB.
(E) Unlike Tat, BRD4 expression does not result in increased association of P-TEFb with ELL or AFF1. 293T cells were transfected with Flag-BRD4 or Flag-Tat. Cell extracts were subjected to Flag IP. IPs were normalized to amounts of copurified CDK9. The presence of AFF1, ELL, and LARP7 was analyzed by WB.
We next tested whether BRD4 expression induces P-TEFb binding to MLL-fusion proteins as robustly as Tat. 293T cells were transfected with Flag-tagged BRD4 or Tat, followed by Flag IP and WB against subunits of Tatcom1 (AFF1, ELL) and Tatcom2 (LARP7) (Figure 4E). When normalized to amounts of copurified CDK9, AFF1, ELL, and LARP7 were present in Tat-Flag IP but absent in Flag-BRD4 IP. These results are consistent with previous reports demonstrating no substantial interaction with leukemia proteins for BRD4 (Jang et al., 2005; Yang et al., 2005). However, we do not exclude that under certain conditions, such as specific cell cycle stage, such a complex may be formed (Mochizuki et al., 2008; Yang et al., 2008).
Tatcom1 Formation Relies on P-TEFb while Optimal CDK9 CTD-Kinase Activity Depends on the Presence of AF9
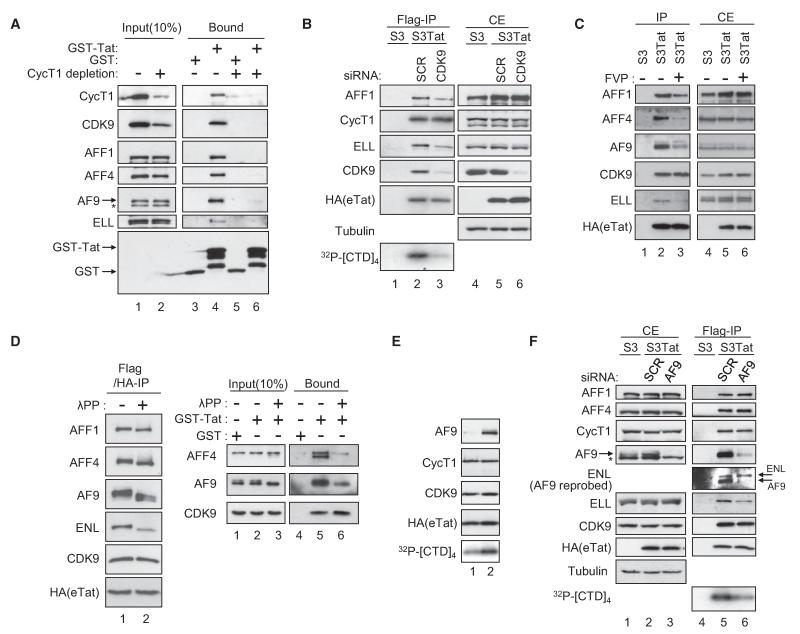
CoIP of endogenous proteins revealed specific interactions between P-TEFb and AFF1, AFF4, AF9, ELL, ENL, and PAF1 that were enhanced in the presence of Tat. Since Tat interacts directly with P-TEFb, we asked whether P-TEFb may play a role in the formation of Tatcom1. Thus, we analyzed the interaction between Tat and its associated factors in HeLa nuclear extracts immunodepleted for CycT1 (Figure 5A). While CycT1 and CDK9 were dramatically reduced in CycT1-depleted nuclear extracts, AFF1, AFF4, AF9, and ELL levels were not significantly affected. Gst-Tat, but not Gst, associates with all proteins of Tatcom1 tested when incubated with mock-depleted nuclear extracts. Interestingly, depletion of CycT1 resulted in dramatic loss of Gst-Tat association with AFF4, AFF1, AF9, ELL, and ENL. This shows the requirement for CycT1 in Tatcom1 formation. These results are consistent with the inability of TatC22G to interact with Tatcom1- and Tatcom2-associated factors (Figure S1B). Next, we assessed the contribution of P-TEFb to the formation of Tatcom1 using RNAi against CDK9 in S3Tat (Figure 5B). Knockdown of CDK9 reduced the amount of ELL and AFF1 associated with eTat, while it had no effect on Tat-CycT1 interaction. Thus, CDK9 has a role in the assembly or stability of Tatcom1. Importantly, CTD-kinase activity was dramatically reduced in eTat purified from CDK9-siRNA-transfected cells (lanes 2 and 3), indicating that CDK9 is the major CTD-kinase associated with eTat. This led us to ask whether P-TEFb kinase activity may affect the integrity of Tatcom1. eTat was purified from nuclear extracts prepared from mock- or flavopiridol-treated S3Tat cells. As shown in Figure 5D, flavopiridol treatment results in reduced association of AFF1, AFF4, AF9, and ELL with Tat, while the amount of CDK9 was unchanged (Figure 5C). Additionally, phosphatase treatment of immunoprecipitated eTat shows that AFF1, AFF4, AF9, and ENL are phosphorylated within Tatcom1 since they show a faster migration (Figure 5D, left panel). To assess whether phosphorylation of Tatcom1 subunits play a role in the assembly of the complex, HeLa nuclear extracts were mock- or phosphatase-treated prior to incubation with Gst or Gst-Tat. Phosphatase treatment reduced AFF4 and AF9 associated with Gst-Tat (right panel). Taken together, experiments in Figures 5A-5D indicate that Tatcom1 formation is dependent on Tat-P-TEFb interaction and CDK9 activity and may involve phosphorylation of AFF1, AFF4, AF9, and ENL.
Figure 5. Assembly and Activity of HIV-1 Tat Complexes.
(A) CycT1 is required for Tatcom1 formation. Gst and Gst-Tat were incubated with mock- or CycT1-depleted HeLa nuclear extracts for 2 hr at 4 °C. Input and bound materials were analyzed by WB.
(B) CDK9 is required for Tatcom1 formation. Extracts of S3 or S3Tat that had been transfected with siRNA against CDK9 or a control (SCR) were subjected to Flag affinity chromatography. The presence of AFF1, ELL, CycT1, CDK9, and eTat in Flag IPs was analyzed by WB. CTD-kinase activity associated with Flag peptide eluates was assessed as in Figure 2E.
(C) S3Tat cells were exposed to 100 nM flavopiridol (FVP) for 20 hr or left untreated prior to whole-cell extraction and Flag IP. Eluates (IP) and cell extracts (CE) were analyzed by WB.
(D) Flag- and HA-purified eTat was either mock treated or incubated with 800 units lambda phosphatase (λPP) for 30 min and probed with the indicated antibodies (left panel). λPP-treated HeLa nuclear extracts were subjected to Gst pull-down using Gst-Tat or Gst and analyzed by WB.
(E) Fractions 5 and 7 from the glycerol gradient shown in Figure S2 were analyzed by WB. CTD-kinase activity associated with these fractions was assessed as in (C).
(F) AF9 potentiates CDK9 kinase activity. Extracts of S3Tat treated with AF9 or scrambled siRNA were subjected to Flag IP. Cell extracts (CE) and Flag-purified eTat were analyzed by WB, and eTat-associated kinase activity was tested as in (C).
To explore the role of the identified Tat/P-TEFb-associated factors in P-TEFb function, we compared CTD-kinase activity associated with eTat in fractions 5 and 7 obtained from glycerol gradient (Figure S2), which were normalized to the amount of CDK9. For equal CDK9 levels, AFF4, AFF1, AF9, ELL, ENL, and PAF1, while present in fraction 7, were barely detectable in fraction 5 (Figures S2 and 5E). Interestingly, enhanced CTD-kinase activity was observed in fraction 7 as compared to fraction 5 (Figure 5E). To confirm this observation, eTat was purified from control (SCR) or AF9 knocked-down S3Tat extracts (Figure 5F). While knockdown of AF9 had no significant effect on the amount of AFF1, AFF4, ENL, CycT1, and CDK9 associated with eTat, the amount of ELL was reduced (lanes 5 and 6). Knockdown of AF9 led to low CTD-kinase activity associated with eTat (lanes 5 and 6), suggesting a requirement for the presence of AF9 and/or ELL for optimal CDK9 CTD-kinase activity.
The MLL-Fusion Partners and PAF1 Are Required for Tat-Mediated Transactivation of the HIV-1 Promoter
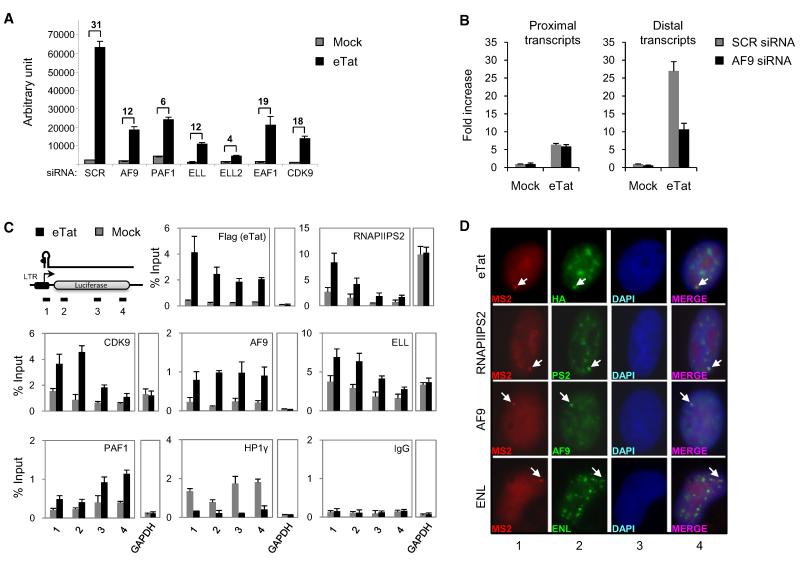
To investigate the biological significance of Tatcom1-associated factors, we asked whether they play a role in Tat-mediated transactivation of the HIV-1 LTR (Figure 6A). HeLa cells containing a single copy of an integrated LTR-luciferase reporter gene (HeLaLTR-luc) were transfected with siRNA specific for AF9, PAF1, ELL, ELL2, or EAF1. Knockdown of AF9, ELL, ELL2, EAF1, and CDK9 reduced both basal and Tat-mediated activation of the LTR. PAF1 knockdown modestly increased basal LTR activity (2-fold) and dramatically reduced Tat-mediated transactivation (5-fold). Knockdown of LARP7 had no effect (Figures S4A and S4B). Knockdown efficiency of targeted genes was assessed by WB (Figure S4C). Since P-TEFb is critical for NF-kB to activate elongation of transcription (Barboric et al., 2001), we asked whether Tatcom1 subunits may also play a role. Knockdown of Tatcom1 subunits affected TNF-α-mediated activation of the LTR (Figure S4D), suggesting their role in NF-κB transcriptional activity. In vivo evidence for the involvement of Tatcom1 in Tat-mediated transcription elongation of the viral promoter was assessed using a siRNA approach. Since AF9 is required for optimal CTD-kinase activity of P-TEFb, the effect of AF9 knockdown on promoter-proximal (initiation) and -distal (elongation) transcripts was monitored by qRT-PCR of mRNA transcribed from the LTR-luciferase reporter (Figure 6B). While knockdown of AF9 had no effect on promoter-proximal transcripts, it reduced the levels of distal transcripts, demonstrating a role for AF9 in Tat-mediated transcriptional elongation.
Figure 6. Tatcom1 Factors Are Required for Tat Transactivation and Are Recruited to Promoter and Coding Regions at the HIV-1 LTR.
(A) AF9, PAF1, ELL, ELL2, and EAF1 are required for Tat-mediated transactivation of the HIV-1 LTR. HeLa LTR-luciferase cells were transfected with specific or control siRNA (SCR). siRNA-transfected cells were transduced with a retrovirus encoding for eTat or empty vector. Luciferase was measured 24 hr after transduction. Fold activation is the ratio between luciferase values obtained in the presence (black bar) or absence (gray bar) of Tat for each siRNA. The graph represents mean and standard error obtained from three independent experiments (see also Figure S4).
(B) AF9 is required for Tat-mediated transcriptional elongation from the HIV-1 promoter. HeLa LTR-luciferase cells were transfected with siRNA against AF9 or control siRNA (SCR) and transduced as in (A). LTR-luciferase transcripts were analyzed by qRT-PCR using proximal (+1/+59) and distal (+1586/+1834) primers; primer 1 and 4 are depicted in (C), respectively. Proximal and distal transcript levels were normalized to those of GAPDH. The graphs represent fold increase relative to mock-treated cells.
(C) AF9, ELL, and PAF-1 associate with the HIV-1 promoter. HeLa-LTR-luciferase cells were transduced as in (A). Fourteen hours after transduction, cells were harvested and analyzed by ChIP using the indicated antibodies. The positions of the primers used in ChIP assays are represented on the scheme (upper left panel). The graph represents the percent of input DNA. Graphs in (B) and (C) are representative of three independent experiments; error bars represent SD.
(D) AF9 and ENL are present at active HIV-1 transcription sites in vivo. U2OS cells containing an integrated HIV-1 reporter (Molle et al., 2007) were transfected with MS2-RFP expression vector to visualize the HIV-1 site of transcription (panel 1) and subsequently transduced with eTat. Endogenous Ser2-phosphorylated RNAPII (RNAPIIS2), AF9, ENL, and eTat were visualized using specific antibodies (panel 2). eTat, RNAPIIS2, AF9, and ENL colocalized with HIV-1 transcription sites (panel 4) with a high frequency (90% of randomly selected cells; n = 50).
We next analyzed the recruitment of Tatcom1 to the integrated HIV-1 promoter. Chromatin IP (chIP) was performed to detect the association of AF9, ELL, and PAF1 with the HIV-1 promoter and throughout the coding region in mock- or eTat-transduced cells (Figure 6C). Consistent with their involvement in basal LTR activity, CDK9, AF9, ELL, and PAF-1 were found associated with both promoters and throughout the transcribed region in absence of Tat (gray bars). Tat treatment (black bars) led to enhanced recruitment of CDK9, AF9, ELL, and PAF-1 and induction of RNAPII Ser2 phosphorylation at the promoter and throughout the transcribed unit. As previously shown (du Chéné et al., 2007), Tat recruitment to the viral promoter results in a reduction of HP1g associated with the reporter. This shows that Tat, CDK9, AF9, ELL, and PAF1 are recruited to the promoter region containing TAR RNA and travel with elongating RNAPII during the elongation step. Additionally, we used the previously described U2OS cells with an integrated HIV-1 vector containing binding sites for the coat protein of phage MS2. This allows the visualization of viral mRNAs at the transcription site upon expression of MS2 protein (Molle et al., 2007). When the HIV-1 vector was activated by Tat, Ser2-phosphorylated RNA-PII, AF9, and ENL accumulated at the HIV-1 transcription site as visualized by MS2 protein (Figure 6D). This experiment shows that Tatcom1 subunits are associated with actively transcribing HIV-1 promoter.
DISCUSSION
Activation of transcription elongation by RNAPII is a highly regulated process involving different classes of elongation factors, including P-TEFb, FACT, TFIIF, elongin, the ELL protein family, ENL, and PAF1, all of which have been shown to employ different mechanisms to enhance RNAPII elongation (Sims et al., 2004). The most important finding from the present work is that HIV-1 Tat assembles a multifunctional transcription elongation complex, Tatcom1, containing P-TEFb, AF9, AFF1, AFF4, ELL, ENL, and PAF1 for optimal activation of the HIV-1 promoter (Figure 7). Our data demonstrate coordinated control of RNAPII elongation by different classes of transcription elongation factors (P-TEFb, ELL, ENL, and PAF1) acting at the same promoter. What might be the benefit of recruiting AFFs/PAF/ELL and ENL to the Tat-P-TEFb complex? First, increased CTD-kinase activity was observed for Tatcom1 as compared to Tat-CDK9/CycT1 complex (Figure 5E). Second, the multifunctional aspect of Tatcom1 may enhance the efficiency of RNAPII elongation and mediate P-TEFb-dependent cotranscriptional histone modifications at the HIV-1 promoter. Indeed, it has been shown that P-TEFb and ELL enhance transcription elongation by different mechanisms (Conaway and Conaway, 1999). While P-TEFb regulates RNAPII processivity through phosphorylation of CTD at Ser2, ELL stimulates RNAPII activity by suppressing transient pausing and preventing backtracking. Functional interaction between P-TEFb and ELL has been reported. Knockdown of CDK9 reduced ELL association with chromatin in Drosophila (Eissenberg et al., 2007). Conversely, knockdown of ELL reduced RNAPII CTD Ser2 phosphorylation (Smith et al., 2008). We show a physical association between endogenous P-TEFb and ELL, which is enhanced by Tat (Figures 4A and 4B), providing evidence for coordinated regulation of transcription elongation by P-TEFb and ELL at the HIV-1 promoter (Figure 6).
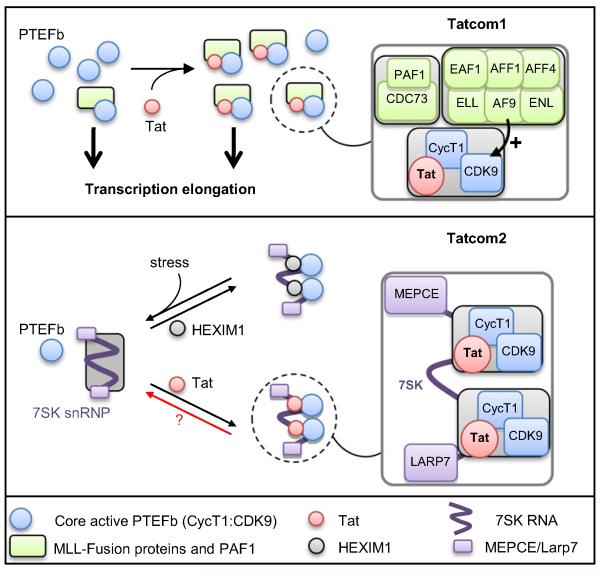
Figure 7. Model for HIV-1 Tatcom1 and Tatcom2 Assembly.
HIV-Tat forms two biochemically distinct complexes. Tatcom1 formation results from the induced binding of AF9/ENL/AFF1/AFF4/ELL and PAF1/CDC73 to P-TEFb, resulting in a kinase-active complex involved in transcription elongation. Certain protein partners, such as AF9, increase CDK9 kinase activity. Expression of Tat in cells leads to remodeling of 7SK snRNP through interaction with CycT1 and the 7SK RNA. The ensuing 7SK snRNP or Tatcom2 is stress resistant and devoid of HEXIM1 and SART3.
Besides regulating RNAPII activity following transcriptional initiation, P-TEFb has been directly involved in cotranscriptional histone modifications. Indeed, knockdown of CDK9 results in loss of global H2BUb, H3K4me3, and H3K36me3 (Pirngruber et al., 2009). PAF1-bound BRE1/RNF20 is responsible for H2B monoubiquitylation at Lys120 (Kim et al., 2009). Additionally, hSet1, responsible for H3K4 methylation, is a member of the PAF1 complex (Krogan et al., 2003). Knockdown of CDK9 resulted in loss of PAF1 recruitment at the HIST1H2BD promoter (Pirngruber et al., 2009). Our data showing physical interaction between P-TEFb and PAF1 may explain how CDK9 directs PAF1 recruitment to transcribing genes and subsequent H2B ubiquitylation. Interestingly, knockdown of hBRE1/RNF20 reduced H2B ubiquitylation within the HIV-1 sequence and diminished HIV-1 promoter activity (Brès et al., 2009). Furthermore, P-TEFb inhibition by flavopiridol reduced H3K4me3 at the HIV-1 promoter (Zhou et al., 2004). Dot1 is responsible for H3K79 methylation (Okada et al., 2005). Interestingly, physical and functional interaction between ENL and Dot1 has been reported (Mueller et al., 2007). Thus, the presence of ENL within Tatcom1 may be responsible for Dot1 recruitment to the viral LTR and subsequent H3K79 methylation.
AF9, AFF1, and AFF4, members of the AF4 family of transcription factors (Slany, 2009), are believed to act as transcription co-activators, despite their mechanism of action being unraveled. Interaction between ENL and AFF1 as well as AFF4 has been demonstrated (Mueller et al., 2007). AFF4/MCEF was first described as a P-TEFb-associated factor (Estable et al., 2002). Recently, using the most common MLL fusions, a super elongation complex containing members of both AF4 and ENL families, together with P-TEFb, was identified and shown to play a role in the pathogenesis of leukemia (Lin et al., 2010; Yokoyama et al., 2010). Knockdown of AF9 reduced CDK9 kinase activity associated with Tat without affecting the amount of Tat-associated P-TEFb (Figure 5F). AF9 also appears to play a role in ELL recruitment to Tatcom1 (Figure 5F). This suggests that AF9, either directly or indirectly, through the recruitment of ELL, regulates CDK9 kinase activity. Further studies are required to understand the role of AF9, AFF1, and AFF4 in Tatcom1 function.
Association of P-TEFb with AF9, AFF1, AFF4, ENL, ELL, and PAF1 occurs in the absence of Tat (Figure 5), and they are likely to assemble in a complex that is physically different from the BRD4/P-TEFb complex (Jang et al., 2005; Yang et al., 2005) (Figure 4E). An important question is whether these two P-TEFb-containing complexes are functionally different. In support of this hypothesis, it has been shown that while BRD4 recruits P-TEFb, it inhibits CDK9 kinase activity by inducing autophosphorylation at Thr29 (Zhou et al., 2009). In contrast, Tat-associated P-TEFb is kinase active (Figure 5E). Another intriguing feature of the BRD4/P-TEFb complex is that it is only recruited to the promoter-proximal region, and unlike P-TEFb, BRD4 does not travel with elongating RNAPII, suggesting that BRD4 does not participate in P-TEFb-mediated transcription elongation (Chen et al., 2008; Zhou et al., 2009). Interestingly, a recent report shows BRD4 accumulation at the 5′ end of the c-fos gene upon mitogen stimulation, while P-TEFb and ELL were highly enriched at the 3′ end (Byun et al., 2009). Thus, it is reasonable to propose that AF9/AFF1/AFF4/ENL/ELL/PAF1/P-TEFb could be the P-TEFb complex involved in transcription elongation per se.
An unexpected finding from our study is that Tat forms a stable complex with 7SK snRNP lacking HEXIM1 and SART3. Formation of Tat 7SK snRNP is likely mediated through direct interaction between Tat and 7SK RNA. Indeed, a TatK50Q mutant unable to bind TAR RNA failed to interact with 7SK RNA and form Tatcom2 (Figure 3B). Additionally, it has been shown that Tat and HEXIM1 compete for 7SKRNA binding in vitro (Sedore et al., 2007). Replacement of HEXIM1 RNA-binding motif by a Tat counterpart is sufficient to restore the 7SK RNA-binding ability of a HEXIM1 RNA-binding-defective mutant (Yik et al., 2004). Formation of Tatcom2 may involve displacement of HEXIM1 from a pre-existing 7SK snRNP by direct competition for 7SK RNA binding. Alternatively, Tat may be recruited to 7SK RNA during the biogenesis of 7SK snRNP particles (Figure 7). Interestingly, we found that, unlike 7SK snRNP, Tatcom2 is resistant to UV, actD, and HMBA (Figures 3C, S3E, and S3F). 7SK snRNP disruption after UV and HMBA treatment is likely due to concerted action of HEXIM1 phosphorylation by AKT (Contreras et al., 2007) and CDK9T186 dephosphorylation by the phosphatase PP1α (Chen et al., 2008). Thus, Tatcom2 resistance to these forms of stress may result from the absence of HEXIM1. What is the biological significance of Tatcom2? Formation of a stable and stress-resistant Tat 7SK snRNP may help maintain the P-TEFb equilibrium and ensure cell survival and/or proper splicing of HIV-1 mRNA during the course of virus production (Haaland et al., 2005; Barboric et al., 2009). Further work will help decipher the role of Tatcom2 in HIV-1 replication.
EXPERIMENTAL PROCEDURES
Purification of Tat-Associated Complexes
Tat complexes were purified from Dignam nuclear extracts derived from 2 × 108 S3 cells stably expressing eTat by two-step affinity chromatography following the standard method (Nakatani and Ogryzko, 2003).
RNase Protection Assay
RNA was extracted from Flag-HA double-purified material with acid phenol:-chloroform (pH 4.5) and precipitated overnight with ammonium-acetate (Ambion; Courtaboeuf, France) and ethanol. The 7SK probe was prepared with the MAXIscript Kit (Ambion) according to the manufacturer’s instructions. RNase protection was performed with the RPA III kit (Ambion) and products were run on a 6% TBE-Urea gel prior to autoradiography.
WCE Preparation and IP
WCEs were prepared by sequential extraction in TETNG-100 buffer (20 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 0.1% Triton X-100, 100 mM NaCl, 10% Glycerol, 2 mM MgCl2, 10 mM β-mercaptoethanol, and 0.5 mM PMSF) and TETNG-400 (same as TETNG-100 but with 400 mM NaCl), supplemented with 1/10 pellet volume 2 M NaCl. WCEs and antibodies were incubated for 3 hr prior to addition of protein A/G beads (Sigma; Saint-Quentin Fallavier, France) for 30 min. Flag IPs were performed using anti-Flag-M2-agarose (Sigma). After three washes in TETNG-100, the beads were incubated with two volumes of Flag peptide (Sigma) diluted in TETNG-100 at 0.2 mg/ml for 1 hr.
In Vitro Kinase Assay
Kinase reactions containing Flag-purified eTat or individual glycerol gradient fractions, 50 ng GST-CTD or 2 μg of YSPTSPS tetrarepeat, 50 mM Tris (pH 7.5), 5 mM MnCl2, 50 mM NaCl, 2 mM DTT, and 2 μCi [γ-32P]ATP were incubated at room temperature for 45 min.
ChIP
HeLa-LTR-Luc cells were mock treated or transduced with an eTat-expressing retrovirus for 14 hr, corresponding to the onset of eTat-induced transcriptional activation of the HIV-1 promoter as measured by qRT-PCR (data not shown). ChIP analysis was performed essentially as described in du Chéné et al., 2007.
Immunofluorescence
Experiments involving the U20S reporter system were performed according to Molle et al. (2007).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of Laboratoire de Virologie Moléculaire and Laboratoire Régulation de l’Expression des Gènes for helpful discussions. We thank U. Fischer, R.K. Slany, J.H. Kersey, and B. Coulombe for the generous gift of antibodies, O. Bensaude for the CTD tetrapeptide, and F. Kashanchi for the BRD4 expression plasmid. Y. Bennasser and D. Latreille kindly provided critical advice and reagents. B.S. was supported by a FRM fellowship. N.L. and M.N. are recipients of ANRS fellowships. Work in M.B.’s laboratory was supported by ERC (250333), ANRS, SIDACTION, ANR, and FRM “Equipe labéllisée FRM” to M.B. and ANRS and SIDACTION to R.K.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, Supplemental References, four figures, and one table and can be found with this article online at doi:10.1016/j.molcel.2010.04.012.
REFERENCES
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Brès V, Kiernan R, Emiliani S, Benkirane M. Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J. Biol. Chem. 2002;277:22215–22221. doi: 10.1074/jbc.M201895200. [DOI] [PubMed] [Google Scholar]
- Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Yoshida T, Pickle L, Jones KA. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol. Cell. 2009;36:75–87. doi: 10.1016/j.molcel.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Transcription. Gene expression—where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun JS, Wong MM, Cui W, Idelman G, Li Q, De Siervi A, Bilke S, Haggerty CM, Player A, Wang YH, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc. Natl. Acad. Sci. USA. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, Zhou Q. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway JW, Conaway RC. Transcription elongation and human disease. Annu. Rev. Biochem. 1999;68:301–319. doi: 10.1146/annurev.biochem.68.1.301. [DOI] [PubMed] [Google Scholar]
- Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol. Genet. Genomics. 2007;277:101–114. doi: 10.1007/s00438-006-0164-2. [DOI] [PubMed] [Google Scholar]
- Estable MC, Naghavi MH, Kato H, Xiao H, Qin J, Vahlne A, Roeder RG. MCEF, the newest member of the AF4 family of transcription factors involved in leukemia, is a positive transcription elongation factor-b-associated protein. J. Biomed. Sci. 2002;9:234–245. doi: 10.1007/BF02256070. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaland RE, Herrmann CH, Rice AP. siRNA depletion of 7SK snRNA induces apoptosis but does not affect expression of the HIV-1 LTR or P-TEFb-dependent cellular genes. J. Cell. Physiol. 2005;205:463–470. doi: 10.1002/jcp.20528. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, Marcello A, Basyuk E. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology. 2007;4:36. doi: 10.1186/1742-4690-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc. Natl. Acad. Sci. USA. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréand C, du Chéné I, Brès V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J. Virol. 2004;78:13522–13533. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Huang K, Jung KJ, Cho WK, Klase Z, Kashanchi F, Pise-Masison CA, Brady JN. Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J. Virol. 2009;83:1036–1044. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.