Abstract
Purpose
Endometriosis, a largely benign, chronic inflammatory disease, is an independent risk factor for endometrioid and clear cell epithelial ovarian tumors. We aimed to identify plasma miRNAs that can be used to differentiate endometriosis and ovarian cancer patients from healthy individuals.
Experimental design
We conducted a two-stage exploratory study to investigate the utility of plasma miRNA profiling to differentiate between endometriosis, endometriosis-associated ovarian cancer (EAOC) and healthy individuals. In the first stage, using global profiling of more than 1,000 miRNAs via reverse transcriptase quantitative PCR (RT-qPCR) in a 20-patient initial screening cohort, we identified 23 candidate miRNAs, which are differentially expressed between healthy controls (n=6), endometriosis (n=7), and EAOC (n=7) patients based on the fold changes. In the second stage, the 23 miRNAs were further tested in an expanded cohort (n=88) of healthy individuals (n =20), endometriosis (n = 33), EAOC (n = 14), and serous ovarian cancer cases (SOC, n= 21, included as controls).
Results
We identified three distinct miRNA signatures with reliable differential expression between healthy individuals, endometriosis, and EAOC patients. When profiled against the control SOC category, our results revealed different miRNAs, suggesting that the identified signatures are reflective of disease-specific pathogenic mechanisms. This was further supported by the fact that the majority of miRNAs differentially expressed in human EAOC were mirrored in a double transgenic mouse EAOC model.
Conclusion
Our study reports for the first time that distinct plasma miRNA expression patterns may serve as highly specific and sensitive diagnostic biomarkers to discriminate between healthy, endometriosis, and EAOC cases.
Keywords: Endometriosis, endometriosis-associated ovarian cancer, microRNA, plasma
Introduction
Epithelial ovarian cancer (EOC) is the fifth leading cause of cancer death in women in the United States (1). It is often called a “silent killer” due to advanced stage at presentation when the disease has spread beyond the pelvis (stage III or IV) (2). Women with stage I disease have a five-year survival of approximately 95%, which is in stark contrast to the considerably lower survival of only 33% associated with advanced stage disease (3). Thus, the key to increase the overall survival of women with ovarian cancer lies in early detection and screening, especially in patients with precursor lesions.
Currently, two major screening approaches have been utilized for early detection of EOC: transvaginal sonography (TVS) and serum protein biomarker testing (2). Both approaches have been found to have low sensitivity and specificity for early EOC screening and are therefore not recommended for screening the general population with average risk (2). CA125 is the most widely used serum biomarker in EOC (4). However, less than 50% of early stage EOC patients have augmented CA125 levels, and elevated circulating CA125 levels can result from many other medical/physiological conditions (2). As a result, CA125 has been mostly used for monitoring EOC progression (5).
MicroRNAs (miRNAs) are single-stranded non-coding small RNA molecules that regulate gene expression by inhibiting mRNA translation or by facilitating cleavage of the target mRNA (6). Past studies have demonstrated that miRNAs are frequently dysregulated in human cancers, including EOC (7, 8). Tumor-specific miRNA expression signatures have been reported in numerous malignancies and have been utilized to classify normal and malignant tissues as well as cancer subtypes (9). When used to classify poorly differentiated tumors, miRNA expression profiling outperformed mRNA expression profiling (10), suggesting that the former may be a superior approach in classifying human malignancies. Although tissue miRNA expression signatures have shown great promise as a new class of biomarkers, they cannot be used to address the current challenge of early detection of EOC because tissue samples at or post diagnosis are required.
Recent discoveries demonstrate that miRNAs are exceptionally stable and can be readily and reliably detected in the systematic circulation (11, 12), raising the possibility of using blood-based miRNA assays to develop novel biomarkers for cancer detection, diagnosis, and prognosis (13, 14). In addition, blood-based miRNA expression profiling has several crucial advantages, such as easy accessibility using a minimally invasive method, and the potential of developing a test for population screening.
EOC encompasses four major histotypes: serous, mucinous, clear cell, and endometrioid. Progressively accumulating evidence from epidemiological and molecular studies demonstrates the role of endometriosis as a potential precursor of endometrioid and clear cell adenocarcinoma of the ovary, also known as endometriosis-associated ovarian cancer (EAOC) (15, 16). Endometriosis, a common gynecological disorder affecting up to 10% to 15% of women in the reproductive age group, consists of ectopic endometrial glands surrounded by stroma, found at locations outside uterine cavity (17). While largely benign, the lesions often show characteristics similar to those of malignancy such as cellular atypia, proliferation, invasion, tissue remodeling, and neovascularization (18). Around 60–80% of EAOC occur in the presence of atypical ovarian endometriosis (19–21). Importantly, in about 25% of these cases, pathologists often describe direct continuity of the atypical ovarian endometriosis with ovarian cancer (20), suggesting a transition spectrum of non-atypical to atypical and malignant variants (21). The risk of direct malignant transformation of ovarian endometriosis has been estimated as 0.7–1.6% over an average of eight years (22, 23).
As with early stage EOC, diagnosis of endometriosis is often difficult, with patients having to undergo invasive procedures via laparoscopy or laparotomy (17). Even in diagnosed patients, it remains a challenge to predict who is at risk to develop EAOC later in life, demonstrating a clear need for developing specific and minimally invasive biomarker assays for endometriosis and early stage EAOC.
In this study, we focused on plasma miRNAs as potential biomarkers that can be used to differentiate endometriosis and EAOC patients from healthy individuals. We performed a global profiling of plasma miRNA expression by RT-qPCR, using plasma samples from patients diagnosed with EAOC, patients with documented endometriosis, and healthy individuals. Our results reveal several distinct plasma miRNA expression patterns that may serve as specific and sensitive diagnostic biomarkers to discriminate between these different disease categories.
Material and Methods
Ethics statement
This research study protocol was approved by the institutional review board (IRB) at the University of Pittsburgh, and informed consent was obtained from all study participants prior to blood collection.
Patients
The clinical and demographic characteristics of patients with endometriosis (n=33), EAOC (n=14, of which six were endometrioid, seven were clear cell tumors and one had mixed endometrioid/clear cell histology), and serous ovarian cancer (SOC, n=21) are shown in Supplementary Table 1. All cancer patients were treated at Magee-Womens Hospital of the University of Pittsburgh Medical Center (UPMC) between 2006–2011. The inclusion criteria consisted of patients with primary ovarian tumors and confirmed histology of EAOC or SOC. In the EAOC group, five patients had endometriosis on the final pathology. Of the five, two had endometrioid, two had clear cell tumors, and one had mixed histology of clear cell and endometrioid type.
The endometriosis patients were treated at Magee-Womens Hospital of UPMC between 2006–2011. Samples from patients with confirmed histology of endometriosis were included in this study. The cases where endometriosis could not be histologically confirmed on surgically removed tissues were excluded from this study.
Plasma samples from healthy women (controls, n=20) were purchased from Innovative Research Labs (Seattle, WA). Women without any current clinical conditions and without family history of diseases such as cancer, HIV, diabetes, and autoimmune diseases, were qualified as healthy individuals by Innovative Research Labs.
Specimen characteristics
Peripheral blood was drawn in heparinized tubes (BD Biosciences, San Jose, CA) and processed at Magee-Womens Research Institute within eight hours from collection. The tubes were centrifuged at 2,300 rpm for 20 minutes at room temperature. Plasma was collected in a sterile biohazard cabinet, aliquoted, and cryopreserved at −80°C until ready to use. Processing of blood by Innovative Research Labs was similarly performed.
Study design
Of the total 88 retrospectively collected plasma samples, 20 were randomly selected for the initial discovery phase (healthy controls, n=6; endometriosis, n=7; EAOC, n=7, of which four were endometrioid and three were clear cell tumors). We used all of the 88 samples for our validation analyses. Study design is illustrated in Supplementary Figure 1. In the discovery phase, miRNAs extracted from 20 plasma samples were used to quantify a total of 1113 miRNAs by RT-qPCR. Expression of 23 candidate miRNAs that are differentially expressed in these three categories of samples and expression of an endogenous control miRNA, miR-132, were confirmed by an independent RT-qPCR in these 20 samples. Finally, expression of the 24 miRNAs was further studied in the complete cohort of 88 plasma samples, by RT-qPCR.
miRNA isolation and RT-qPCR assay
RNA was isolated from 88 plasma samples using the mirVana miRNA Isolation Kit (Life Technologies, Carlsbad, CA). Concentrations of RNA were measured by a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). Sixty nanograms of purified RNA were used for RT using the QuantiMir Kit (System Biosciences, Mountain View, CA). One microliter of cDNA was then diluted 1:160 and 1.1 μl of diluted cDNA was used in each qPCR reaction for a genome-wide expression profiling of 1113 miRNAs (Sanger miRBase Version 15) using the Human miRNome Profiler kit (System Biosciences). qPCR was performed on an ABI7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA) using the RT2 SYBR Green ROX qPCR master mix (Qiagen, Valencia, CA) under the following conditions:50°C for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds followed by 60°C for 10 seconds, and a standard dissociation stage. The RT-qPCR data were analyzed according to the comparative CT method (24).
NanoString nCounter miRNA assay
The miRNeasy FFPE Kit (Qiagen) was used to isolate miRNAs from formalin fixed paraffin embedded (FFPE) tissues for global miRNA profiling using matching plasma-tissue of endometriosis or EAOC samples. The nCounter Human miRNA Panel v2 that evaluates 800 miRNAs was used (NanoString, Seattle, WA). miRNAs extracted from plasma and tissue samples were subjected to nCounter miRNA sample preparation according to the manufacturer’s instructions. This was followed by ligation of 100 ng of miRNA and hybridization to probes at 65°C for 18 hours following the manufacturer’s protocol. Next day, the hybridized probes were purified and counted on nCounter Prep Station and Digital Analyzer. The data obtained from Analyzer contained counts of individual fluorescent barcodes and thus, a count of miRNAs present in the sample. The nCounter results were analyzed by the nSolver software according to the manufacturer’s instructions.
Plasma miRNA measurements in LSL-KrasG12D/+/Ptenloxp/loxp mice with endometrioid ovarian cancer
All animal experiments were performed according to a protocol approved by the University of Pittsburgh International Animal Care and Use Committee (IACUC). The mice were originally provided by Dr. Dinulescu (25), and the colony was maintained at the Magee-Womens Research Institute. Genotyping for the identification of LSL-KrasG12D/+/Ptenloxp/loxp mice was performed as previously described (25).
Survival surgery procedure and administration of recombinant adenovirus encoding for Cre recombinase [Ad5CMVCre (AdCre)] (University of Iowa Gene Transfer Vector Core) was performed in synchronized animals as previously described by us and others (25, 26). Briefly, seven to nine weeks old KrasG12D/+/Ptenloxp/loxp virgin females were injected i.p. with 5 U pregnant mare’s serum gonadotropin (PMSG) followed by 5 U human chorionic gonadotropin (hCG) 48 hours later. Thirty-six hours later, mice received 5 μl of 2.5×107 plaque-forming units (p.f.u.) Ad5CMVCre delivered to the ovary surface epithelium (OSE) of the left ovary only, via intrabursal injection. The contra-lateral ovary served as a control.
Mice were sacrificed when disease was clinically evident (tumor mass on the injected side and/or ascites accumulation) or when mice were moribund (hunched appearance, ruffled fur, unable to reach for food or water). Blood was collected by cardiac puncture at necropsy and serum cryopreserved until ready to use. Expression of mouse miRNAs, mmu-miR-15b, 16, 21, 191, and 195 were measured by RT-qPCR as described above.
Statistical analysis and sample size justification
The candidate demographic variables analyzed were: age, race, history of alcohol and tobacco consumption. We also compared stage of disease in EAOC versus SOC. For the baseline characteristics, we conducted univariate comparisons using Chi-square tests, ANOVA test, or their nonparametric equivalents, as appropriate. We determined the medians and interquartile range as measures of central tendency for variables with highly skewed distribution (not normally distributed such as gravidity or parity). All analyses were performed using SAS 9.3 (SAS Institute, Cary, NC) assuming statistical significance at p<0.05.
All qPCR CT values were normalized to miR-132. The ΔCT of sample i marker j is defined as ΔCTij = CTij–CT imir132. Here CTij and CTimir132 are CT values of marker j and miR-132 for sample i,, respectively. The fold change (FC) between two specific groups for a specific marker was calculated as . To test the reliability of the assay, the 23 miRNAs of interest were tested twice on 20 randomly selected samples. The mixed effect model based method was used to calculate the coefficient of variation (CV) and intra-class correlation (ICC) on the ΔCT values of each marker (27). Only samples with detected values were used in the calculation. An average CV of 2.44% (range: 0.016%–12.6%) was observed across the markers, indicating good reproducibility of the assay. The ICC ranges from 8% to 99.9%, with a median value of 83%. ICC measures the percentage of variation contributing to the variation among individuals. This value is high when the variance component associated with individual samples greatly exceeds the variation of the assay. ICCs of 15 of the markers exceed 70% in our data.
For the discovery cohort we used n=20 samples. We estimated that the mean for the coefficients of variation (CV) of the assay to be 2.44%. The calculation is based on the method described by Gail et al (27). By the delta method, we could approximate the variance of the ln(ΔCT) by the CV of the measurement. The effect size (ES) is defined as , thus, the fold change can be calculated as .
For the validation cohort, we used a total of n=88 cases. We applied the Bonferroni adjustment, with a p-value cut off of 0.05/23=0.002 to control the overall family wise error rate (FWER) at 0.05. A total of 23 markers passed the initial screening and were formally tested using all 88 samples. With 20 samples from the healthy individuals and at least 14 samples in the disease group, there is at least 90% power to detect an effect size of 1.72 at the log level with a two-sided Wilcoxon rank-sum test (α=0.05/23=0.002). This translates into a FC of 1.3.
Hierarchical clustering analysis was applied to the ΔCT values. Markers that were consistently detected across all groups in the expansion cohort (missing data rate < 30%) were used in the clustering analysis.
Wilcoxon rank-sum tests were used to search for differentially expressed miRNAs between two groups. Only markers that were consistently detected in both groups (total number of samples with non-missing values > 5) were used in the differential gene analysis. The Benjamini and Hochberg’s method was used to control the false discovery rate (FDR) at 20% (28). Principle Component Analysis (PCA) was applied to the differentially expressed miRNAs to reduce the dimension and to visualize the clusters. Linear Discriminant Analysis (LDA) was used to generate a three-marker model to classify samples of different groups (29, 30). Area Under the Receiver Operating Characteristic (AUROC) curve, sensitivity and specificity were calculated for each model. We used leaved-one-out cross validation (LOOCV) to avoid overfitting of the data.
Results
Reproducibility and reliability of our approach
Our overall experimental design for miRNA profiling is outlined in Supplementary Figure 1. First, we confirmed that our extracted plasma miRNAs accurately reflect the original plasma miRNA population by adding spike-ins of serially diluted, synthetic miR-210, into aliquots of a randomly selected plasma sample from a healthy subject. Following miRNA extraction, we consistently detected miR-210 in a linear fashion by RT-qPCR, validating our miRNA extraction method (Supplementary Fig. 2A). Next, we examined the reproducibility of our RT-qPCR protocol by two approaches. First, expression of three randomly selected miRNAs, miR-132, 362-5p, and 1974 was measured in three independent RT-qPCR assays using miRNAs extracted from six plasma samples. The CT values of all three runs in each plasma sample for each of the three miRNAs were highly consistent among measurements (Supplementary Fig. 2B), demonstrating that extracted plasma miRNAs can be reproducibly detected by our RT-qPCR approach. Second, we examined miR-132 expression in three consecutive plasma samples collected one month apart from each of three preoperative EAOC patients. Remarkably, consistent expression of miR-132 is detected in all three samples from each of these patients (Supplementary Fig. 2C).
Because the peripheral blood used in this study was drawn in heparinized tubes and heparin is a known inhibitor of RT-PCR reaction, the robustness of our RT-qPCR was tested by comparing the expression of miR-16, 21, and 195 in three plasma samples collected in heparinized tubes and in matched serum samples collected in tubes without anticoagulant from EAOC patients. Comparable expression levels of the three miRNAs between plasma and serum samples suggest that the influence of heparin on our RT-qPCR is minimal (Supplementary Fig. 3A). Despite similar blood collection protocols, our case and control plasma samples are from different institutions. Thus, we examined whether different sources of plasma samples may affect miRNA measurements in our study. Clearly, no difference was detected when miR-132 expression was examined by RT-qPCR between purchased healthy controls and endometriosis and EAOC samples collected at our institution (Supplementary Fig. 3B).
Global miRNA profiling in patient plasma
The clinical demographics of all cases used in this study are summarized in Supplementary Table 1. The majority of patients in the EAOC, endometriosis, and SOC categories were Caucasian (100%, 88%, and 100%, respectively) whereas the control group contained individuals more evenly distributed among the Caucasian, African-American, and Hispanic categories (45%, 35%, and 20%, respectively, p<0.0001). As expected, the endometriosis patients were younger than those with cancer and had lower gravidity and parity scores. Sixty-four percent of the EAOC cases were stage I or II whereas only 14 % of the SOC cases were early stages, consistent with the more aggressive phenotype and late diagnosis seen in patients with SOC (31). There were no significant associations with body mass index, tobacco or alcohol use. Of the EAOC cases (n=14), 43% (n=6) were clear cell and 50% (n=7) were endometrioid tumors, while 7% (n=1) was with mixed clear cell/endometrioid histology. Presence of concurrent endometriosis at the time of cancer diagnosis was confirmed by pathology in 36% EAOC cases (n=5), of which two were endometrioid, two were clear cell, and one was mixed endometrioid/clear cell tumor.
Since the genome-wide circulating miRNA expression profile in endometriosis and EAOC has not been established to date, we performed global plasma miRNA expression profiling by RT-qPCR in the discovery phase of our study. Because the purpose of our study is to identify plasma miRNAs that can serve as biomarkers to distinguish endometriosis and EAOC samples from healthy controls, expression of 1113 human miRNAs was measured in 20 plasma samples from healthy individuals (n=6), endometriosis patients (n=7), and EAOC patients (n = 7) to pick candidate miRNAs that are expressed in these sample categories and have potential to differentiate different groups of tissues, and to evaluate the reproducibility of our assay. Of the 1113 miRNAs, miRNAs that were not expressed in any of the samples were eliminated first. Of the remaining 286 miRNAs, miRNAs that were not expressed in at least four samples in any sample category were further removed. Finally, 23 miRNAs with a minimal differential average CT value of two (ΔCT ≥ 2) among EAOC, endometriosis, and healthy controls, and miR-132, a potential endogenous control miRNA, were selected as candidate biomarkers for further study (Supplementary Table 2). Expression of these 24 miRNAs was confirmed in independent RT-qPCR runs using the 20 samples in the discovery cohort.
Unique plasma miRNA signatures can differentiate between patient categories
The 24 candidate miRNAs were next validated by RT-qPCR in a total of 67 samples (healthy, n = 20; endometriosis, n= 33; EAOC, n = 14), including the 20 samples from the discovery phase. The serous ovarian cancer (SOC) cases (n=21) were added as non-endometriosis-associated controls (15). Currently, there is no consensus on housekeeping miRNAs used for plasma miRNA RT-qPCR data normalization. In our study, we found that miR-132 is the most consistently expressed miRNA across all samples from all categories (Supplementary Fig. 4). Thus, miR-132 was subsequently employed as an endogenous control for plasma miRNA RT-qPCR data normalization.
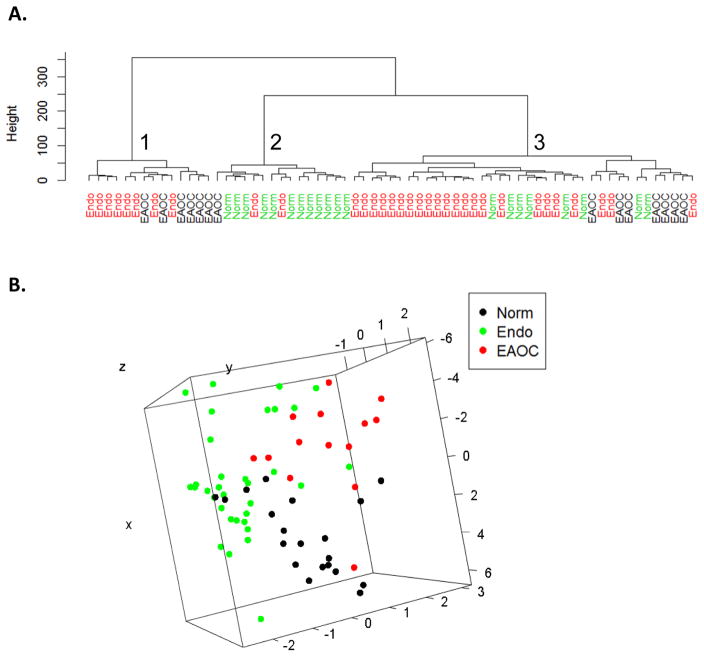
The unsupervised clustering analysis using the expression of the candidate plasma miRNAs in healthy controls (n=21), endometriosis (n=33), and EAOC (n=14) shows three distinct clusters: cluster 1 is enriched with endometriosis (n=8) and EAOC (n=6) samples; cluster 2 is enriched with normal samples with two endometriosis cases and one EAOC case misclassified in this cluster; cluster 3 has two sub-clusters. One is enriched with endometriosis samples with six healthy controls misclassified in this sub-cluster; the other is enriched with EAOC cases (n=7) but with misclassified endometriosis (n=3) and healthy controls (n=2) (Fig. 1A). We also used principle component analysis (PCA) to aid the visualization of the data at lower dimension. Consistent with our clustering analysis, the plot of the first three principle components of the data also shows that the three groups form three distinct but not completely separated clouds (Fig. 1B). These results reveal the potential of using plasma miRNA expression patterns for classification of these diseases. When the unsupervised clustering analysis was performed in all cases (n=88, including SOC), the 21 SOC cases were found to be interspersed into the other three categories and cannot be separated from other categories of diseases (Supplementary Fig. 5A). Similarly, an unsupervised clustering in pair-wise comparison of endometriosis/EAOC or endometriosis/SOC (Supplementary Fig. 5B & C) showed that while the majority of endometriosis and EAOC samples can be classified into relatively distinct clusters, the SOC samples are mixed with endometriosis samples. However, when EAOC and SOC samples were compared, the 23-miRNA signature correctly classified the majority of them into two major clusters (93% and 81%, respectively, Supplementary Fig. 5D), and when unsupervised clustering was performed between healthy controls and SOC or healthy controls and EAOC, the 23-miRNA signature can also correctly classify cancer samples from controls (Supplementary Fig. 5E & F). Overall, these results suggest that although the 23-miRNA signature is reflective of its originating clinical entity (endometriosis and EAOC from the discovery cohort), it also contains commonly dysregulated plasma miRNAs across different EOC histotypes. Thus, optimization of this list of candidate miRNAs may provide novel plasma biomarkers for detection and classification of endometriosis and EAOC.
Figure 1.
Plasma miRNA expression profiles can distinguish different disease categories. A) Unsupervised hierarchical clustering was applied to miRNAs with < 30% missing values in healthy controls, endometriosis, and EAOC samples (n=20, 33, and 14, respectively). Different distance measure and link were explored. Samples are classified into three clusters based on the expression signature of 23 plasma miRNAs. B) Principal component analysis was applied to markers with adjusted p value < 0.2 in either one of the three groups’ pair-wised comparisons. First three components were used for the three-dimensional plot. Norm, healthy controls; Endo, endometriosis.
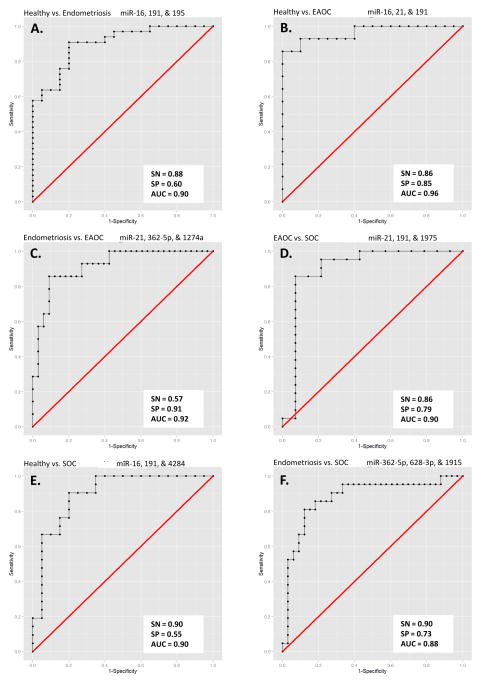
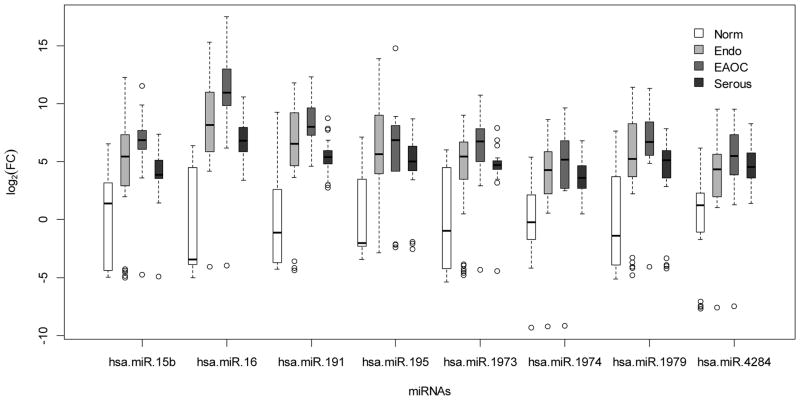
We then compared the expression of the 23 miRNAs between all categories (healthy controls, n=20; endometriosis, n=33; EAOC, n=14; and SOC, n=21) in the validation cohort to identify the top 10 differentially expressed miRNAs between any two categories by Wilcoxon Rank Sum test (Table 1 and Supplementary Table 3). The top three candidate miRNAs for each of the pair-wise comparisons are shown in Supplementary Figure 6. While the majority of the top differentially expressed plasma miRNAs in both EAOC and SOC samples are overexpressed compared to healthy controls, the level of expression in EAOC samples is generally much higher than in SOC samples (Supplementary Fig. 6F). Next, we examined which combination of miRNAs among the candidate miRNAs could differentiate between the sample groups with the highest predictive power by linear discriminant analysis (LDA). Leave-one-out cross validation (LOOCV) was used to avoid overfitting of the data. Application of LDA revealed three plasma miRNAs, miR-16, 191, and 195, all upregulated in endometriosis, that could differentiate between healthy and endometriosis cases with 88% sensitivity (SN), 60% specificity (SP) (Fig. 2A). A combination of miR-16, 21, and 191 can differentiate between healthy and EAOC with 86% SN and 85% SP (Fig. 2B), while miR-21, 362-5p, and 1274a can differentiate between endometriosis and EAOC with 57% SN and 91% SP (Fig. 2C). miR-21, 191, and 1975 together could distinguish between EAOC and SOC with 86% SN and 79% SP (Fig. 2D). Expression signature of miR-16, 191, and 4284 could be used for discerning healthy individuals from SOC patients with 90% SN and 55% SP (Fig. 2E), while miR-362-5p, 628-3p, and 1915 can differentiate endometriosis and SOC with 90% SN and 73% SP (Fig. 2F). Interestingly, we also noticed a general trend of elevated plasma miRNA expression from healthy controls to endometriosis to EAOC but not in SOC samples (Fig. 3), suggesting that these miRNAs may serve as novel biomarkers that reflect the pathological progression from benign to precursor lesion to fully developed EAOC. Altogether, we have identified different panels of plasma miRNAs that may serve as novel biomarkers to discriminate between healthy, endometriosis, EAOC, and SOC patients.
Table 1.
The 10 most differentially expressed miRNAs in pair-wise comparisons among healthy controls, endometriosis, and EAOC samples.
| Endometriosis (n=33) vs. Healthy Controls (n=20) | W.pvalue1 | a.W.pvalue2 | FC3 |
|---|---|---|---|
| hsa-miR-16 | 0.00000 | 0.00001 | 396.62 |
| hsa-miR-195 | 0.00001 | 0.00011 | 62.02 |
| hsa-miR-191 | 0.00004 | 0.00028 | 79.05 |
| hsa-miR-1974 | 0.00007 | 0.00034 | 15.43 |
| hsa-miR-4284 | 0.00025 | 0.00101 | 16.79 |
| hsa-miR-15b | 0.00041 | 0.00124 | 26.27 |
| hsa-miR-1978 | 0.00041 | 0.00124 | 51.29 |
| hsa.miR-1979 | 0.00051 | 0.00135 | 30.54 |
| hsa-miR-362-5p | 0.00062 | 0.00149 | 3.41 |
| hsa-miR-1973 | 0.00113 | 0.00259 | 16.69 |
| EAOC (n =14) vs. Healthy Controls (n=20) | W.pvalue1 | a.W.pvalue2 | FC3 |
|---|---|---|---|
| hsa-miR-21 | 0.00001 | 0.00014 | 147.40 |
| hsa-miR-191 | 0.00001 | 0.00014 | 380.01 |
| hsa-miR-16 | 0.00002 | 0.00015 | 1323.18 |
| hsa-miR-15b | 0.00005 | 0.00032 | 80.50 |
| hsa-miR-1977 | 0.00029 | 0.00117 | 11.48 |
| hsa-miR-1979 | 0.00029 | 0.00117 | 99.11 |
| hsa-miR-1973 | 0.00038 | 0.00131 | 58.96 |
| hsa-miR-1974 | 0.00057 | 0.00170 | 20.75 |
| hsa-miR-4284 | 0.00073 | 0.00196 | 26.58 |
| hsa-miR-195 | 0.00137 | 0.00273 | 48.02 |
| EAOC (n=14) vs. Endometriosis (n =33) | W.pvalue1 | a.W.pvalue2 | FC3 |
|---|---|---|---|
| hsa-miR-362-5p | 0.00014 | 0.00343 | 0.14 |
| hsa-miR-1274a | 0.01323 | 0.09634 | 0.13 |
| hsa-miR-21 | 0.01606 | 0.09634 | 13.84 |
| hsa-miR-766 | 0.02195 | 0.10534 | 15.11 |
| hsa-miR-1975 | 0.03142 | 0.11204 | 7.75 |
| hsa-miR-1308 | 0.03527 | 0.11204 | 0.13 |
| hsa-miR-191 | 0.03735 | 0.11204 | 4.81 |
| hsa-miR-744 | 0.05209 | 0.13192 | 4.89 |
| hsa-miR-376a | 0.05497 | 0.13192 | 6.38 |
| hsa-miR-1246 | 0.06112 | 0.14058 | 1.36 |
Wilcoxon p value,
adjusted Wilcoxon p value,
Fold change
Figure 2.
The leave-one-out cross validation receiver operating characteristic (ROC) curves of Logistic regression model for four groups’ pair-wised comparisons are plotted based on the top three markers. Area under curve (AUC) is also provided. SN, sensitivity; SP, specificity.
Figure 3.
Increased expression of plasma miRNAs along with the progression of diseases from endometriosis to EAOC, but not in SOC samples. The eight miRNAs are derived from shared top 10 most differentially expressed miRNAs in both endometriosis and EAOC samples compared to healthy controls. y-axis, log2 of folder changes of plasma miRNA expression (log2(FC)). Norm, healthy controls; Serous, SOC; Endo, endometriosis.
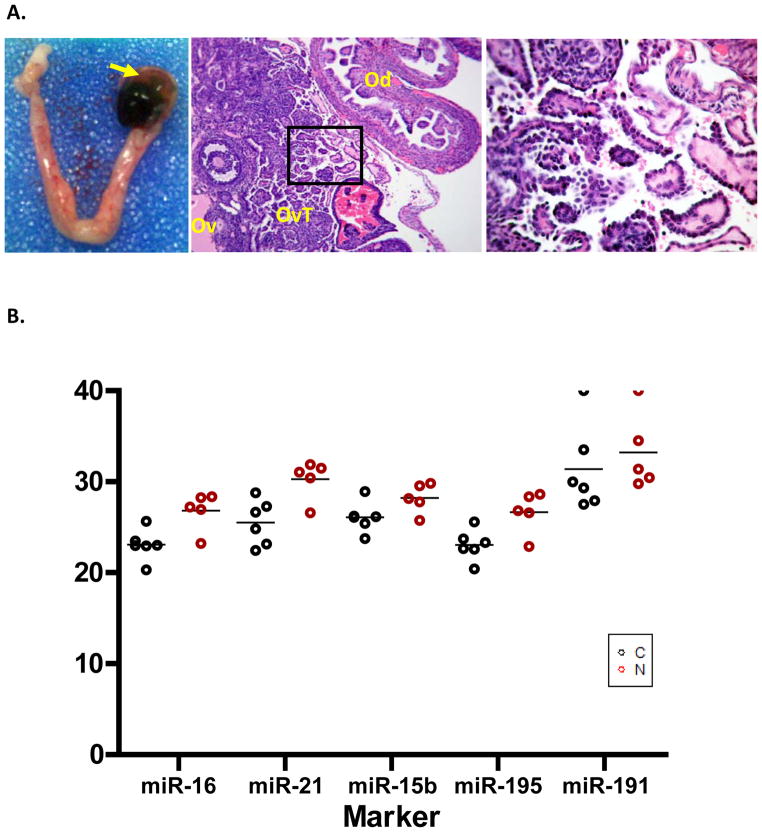
Validation of human miRNA signature in a preclinical mouse model
Mouse models of human diseases are powerful tools to study the mechanism of disease and test the efficacy of pre-clinical therapeutics. We next tested whether some of the most differentially expressed miRNAs between healthy controls and EAOC patients (Table 1) are also differentially expressed between healthy mice and mice with endometrioid ovarian tumors. To achieve this, we employed a previously described, conditional mouse model for endometriosis-associated endometrioid ovarian cancer (25). The LSL-KrasG12D/+/Ptenloxp/loxp conditional mice develop orthotopic tumors 12 weeks post AdCre injection under the ovarian bursa (Fig. 4A) (25). We induced tumors in six female mice, sacrificed the mice when moribund, and collected serum at necropsy. Five healthy (non-injected), age-matched female mice were sacrificed as controls. Serum miRNAs were extracted and subjected to RT-qPCR analysis to measure expression of miR-15b, 16, 21, 191, and 195. Among the top 10 differentially expressed human miRNAs between healthy controls and EAOC patients (Table 1), these five are the only miRNAs that have mouse orthologs. Our results demonstrate that four of the five miRNAs can also delineate the EAOC mice from healthy controls (Fig. 4B), suggesting a potential EAOC-specific pathogenesis leading to the dysregulation of miR-15b, 16, 21, and 195, and further validating the biological relevance of the plasma miRNA expression signature we have identified as biomarkers of human EAOC.
Figure 4.
The plasma miRNA expression signature that differentiates healthy controls from EAOC patients can be detected in a mouse endometrioid ovarian cancer model. A) Left panel shows ovarian tumor at the Ad-Cre injected site (arrow), but no tumor formation seen on non-injected left ovary. Middle panel is 10X HE staining of a cross section of mouse ovarian tumor with ovary (Ov), oviduct (Od), and ovarian tumor (OvT). Right panel shows magnified image of ovarian tumor with endometrioid histology (40X). B) Expression of orthologous miRNAs in healthy mice (n=5) and mice with EAOC (n=6). Four out of the five miRNAs are significantly upregulated in most of the mice with EAOC as compared to normal (p = 0.00009, 0.000433, 0.000209, and 0.00014 for miR-16, 21, 15b, and 195, respectively; student’s t-test), similar to the profiles in human EAOC samples. miR-191 did not reach statistical significance, although there is also a trend that expression of plasma miR-191 is elevated in EAOC mice compared to that of in normal mice. Equal amount of RNA extracted from mouse serum was used for RT and qPCR. The raw CT values were plotted because currently there is no consensus on endogenous plasma miRNA in mouse that can be used for normalization. C, mice with EAOC tumors; N, normal control mice.
Ovarian tumor tissue and corresponding plasma have distinct miRNA expression profiles
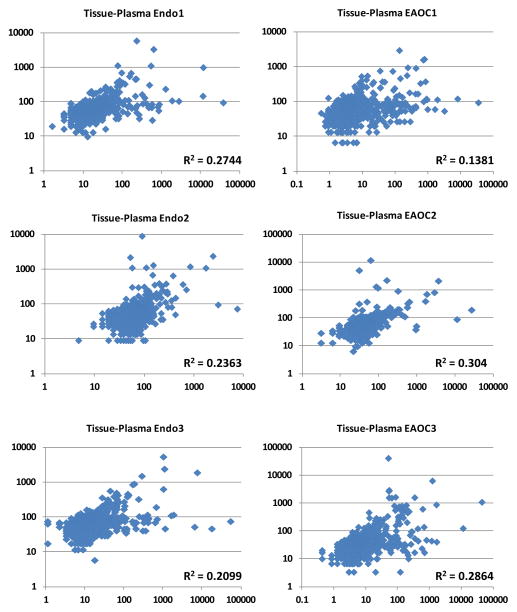
Numerous miRNAs have been reported to be dysregulated in ovarian tumors (8, 32). Despite the great potential circulating miRNAs hold as novel biomarkers for classification and early diagnosis of EOC, it remains unclear whether miRNAs in patient plasma reflect miRNA expression occurring in corresponding diseased tissues. To address this question, we profiled miRNA expression in six pairs of endometriosis tissue or EAOC primary tumors and corresponding plasma samples using the NanoString technology (33), which provides digital counting of miRNA copy numbers without the need for miRNA amplification. While we detected a very modest correlation of overall miRNA expression in paired tissue and plasma samples, we also observed distinct miRNA expression profiles, especially among the highly expressed miRNAs (Fig. 5). Specifically, while miR-16, 21, and 132 were consistently ranked as the top three most highly expressed plasma miRNAs, only miR-21 was consistently ranked among the top five most highly expressed tissue miRNAs. To further support our conclusion, we examined the expression of 10 miRNAs with mouse orthologs (miR-15b, 16, 21, 132, 191, 195, 362-5p, 652, 744, and 1274a) from the 24 miRNAs identified from the discovery cohort (Supplementary Table 2) in five paired tumor and plasma samples from tumor-bearing LSL-KrasG12D/+/Ptenloxp/loxp mice. No significant correlation was detected in the paired samples from all five mice (Supplementary Figure 7), mirroring the findings in humans. Our data suggest that plasma and tissue samples have distinct miRNA expression profiles. Thus, differentially expressed miRNAs identified through comparing normal and tumor tissues as reported in numerous studies cannot be simply applied to study plasma/serum samples.
Figure 5.
NanoString analysis reveals very low correlation of miRNA expression between matching tissue and plasma of EAOC and endometriosis patients. Left panel, comparison of three matched endometriosis tissue and plasma samples; Right panel, comparison of three matched EAOC tissue and plasma samples. The NanoString data were normalized using the nSolver software. x-axis, copy number of miRNAs in tissue samples; y-axis, copy number of miRNAs in plasma samples. Endo, endometriosis.
Discussion
In this study, we performed, to our knowledge, the first global profiling of circulating miRNAs in endometriosis and EAOC samples. We have identified unique plasma miRNA expression signatures that distinguish endometriosis and EAOC patients from healthy controls, suggesting that circulating miRNAs may serve as promising biomarkers with high sensitivity and specificity for early detection and diagnosis of endometriosis and EAOC. Remarkably, four out of five miRNAs (miR-15b, 16, 21, and 195) we found differentially expressed in human EAOC from healthy controls show a similar expression pattern in a preclinical mouse model for EAOC, providing a strong support for the validity of our results and also suggesting that changes in these miRNA levels are likely due to disease-specific pathogenesis. This is also the first time that plasma miRNA biomarkers are validated in an ovarian cancer mouse model.
In addition to causing pain and infertility, endometriosis is also considered a precursor of EAOC, as supported by a growing number of epidemiological and molecular studies (15, 34). Frequent mutations of a tumor suppressor gene, ARID1A, have not only been identified in clear cell and endometrioid ovarian tumors, but have also been found in concurrent endometriosis and atypical endometriosis lesions (34), suggesting loss of ARID1A function to be an early step in the transformation of endometriosis to EAOC (34). Although the complete pathway remains unclear, these notable findings strongly support the molecular links between endometriosis and EAOC (35), and pave the way for developing new, reliable biomarkers that can not only aid in the diagnosis of endometriosis and EAOC, but also identify endometriosis patients at risk for developing EAOC. The plasma miRNA signatures reported in our study, focused on distinguishing endometriosis or EAOC from healthy controls, is particularly valuable since these biomarkers may potentially be utilized in a well-defined high-risk population, such as patients with prolonged history of suspected or confirmed endometriosis and with other ovarian cancer predisposing risk factors like age, reproductive history, or family history of ovarian cancer.
It is estimated that a screening test for ovarian cancer would require a sensitivity of at least 75% and a specificity of more than 99.6% to achieve a positive predictive value (PPV) of 10%, the minimum PPV required for a screening test. Despite extensive efforts to develop new protein biomarkers for early detection of ovarian cancer, CA125 still stands to be the most dependable of all biomarkers examined to date (36). However, even CA125 falls short of the requirement for sensitivity and specificity to be useful as a biomarker for ovarian cancer screening (36, 37). Ovarian cancer is a highly heterogeneous disease and the four main histological subtypes of ovarian cancer are now considered different diseases, which may develop differently, respond differently to chemotherapy, and express different sets of biomarkers (38). However, in the majority of past biomarker development studies, ovarian cancer has been largely regarded as a single entity. This may at least partially account for the failed effort to develop biomarkers for early detection of ovarian cancer. Our results further support this concept by demonstrating that EAOC and SOC are different clinical entities and can be distinguished based on plasma miRNA expression profiles (Supplementary Fig. 5D). Although the sensitivity and specificity values of the plasma miRNA signatures reported here are lower than required to be applied in clinical practice yet, our study nevertheless serves as a foundation for future follow-up studies with larger sample sizes. In addition, combining histotype-specific plasma miRNA expression signatures with CA125 may be a promising strategy to improve the sensitivity and specificity of ovarian cancer early detection.
The panel of plasma miRNAs we identified clearly demonstrates that circulating miRNAs are promising novel biomarkers for early detection of EAOC. Surprisingly, when we compared the global miRNA expression in primary EAOC tumors or endometriosis tissues to corresponding plasma samples, only a very modest correlation was observed (Fig. 5). Interestingly, neovascularization is important for pathogenesis of endometriosis and EAOC and the lesions of endometriosis and EAOC are highly vascular (39, 40). Despite no consensus on the cellular origin of circulating miRNAs at present (41), the lack of correlation between paired tissue-plasma miRNA expression profiles in both endometriosis and EAOC patients and in EAOC mouse model (Fig. 5 and Supplementary Fig. 7) strongly suggests that disease tissue or malignant tumor cells are not the sole source of circulating miRNAs. Since loco-regional inflammation plays an important role in endometriosis and EAOC pathogenesis (42, 43), it is more likely that the miRNA signatures we detected in patient plasma actually reflect the output of a systematic response of host microenvironment to the disease. Currently, the majority of circulating miRNA biomarker studies is based on primary tumor miRNA expression profiles. In light of our results, by not performing independent global miRNA profiling in plasma/serum samples, these studies may have missed many relevant biomarker candidates.
Numerous miRNAs have been reported to be dysregulated in EOC (8, 44), among which miR-21 and members of the miR-200 family are the most consistently upregulated compared to normal controls. Because SOC accounts for a majority of EOC cases, few reports have focused on identifying miRNA expression signatures in other EOC histotypes. Upregulation of miR-21, miR-203, and miR-205 were found to be specific to the endometrioid histotype and miR-222 was downregulated in EAOC samples (8). By using next generation sequencing, a different set of miRNAs that are specifically upregulated in endometrioid and clear cell histotypes have also been reported recently (44), of which miR-9, 96, 182, 183, 196a, 196b, 205, and 375 are specifically upregulated in endometrioid histotype, and miR-30a, 30a*, and 486-5p are upregulated in clear cell histotype. Among the top 10 plasma miRNAs that are most differentially expressed between EAOC and healthy controls (Table 1), miR-21 is the only miRNA that overlaps with the EAOC-specific miRNA signature derived from tumor tissues (8). This discrepancy further supports our conclusion that tumor cells are not a major source of circulating miRNAs. Interestingly, miR-21 has been reported as one of the most consistently overexpressed oncomiRs in almost all tumor types (45), raising the possibility that the highly elevated miR-21 expression in the plasma of EAOC patients may reflect activation of a common oncogenic pathway that contributes to EAOC pathogenesis, despite the source of circulating miR-21 remains unknown.
Circulating miRNAs hold great promise as biomarkers on cancer early detection, diagnosis, and prognosis. Plasma/serum miRNA signatures have been reported in almost all tumor types, such as in lung (13, 46), gastric (47), breast (48), pancreatic (14), and ovarian cancers (49, 50). Among the three miRNAs that comprise the signature that distinguishes EAOC from healthy controls (Fig. 2B), overexpression of plasma/serum miR-16 and miR-21 has been reported in many tumor types (14, 46, 47), including in ovarian cancer (49, 50). However, dysregulated circulating miR-191 expression has not been implicated in any cancers to date. Thus, the combination of miR-16, 21, and 191 may represent a unique signature to EAOC.
Despite our rigorous statistical methods for signature identification, we acknowledge the limited sample size of our studies and the need for further validation of findings in larger cohorts. In addition, inclusion of cases with atypical endometriosis, concurrent endometriosis-EAOC cases, and of early stage EAOC is warranted in future studies to further validate our miRNA signatures. The plasma miRNAs identified in this study are promising biomarkers of endometriosis-to-EAOC progression and ultimately, will be extremely useful to improve the clinical outcome of ovarian cancer patients.
Supplementary Material
Translational Relevance.
Patients with epithelial ovarian cancer (EOC) typically have a poor prognosis because most cases are not diagnosed until they have reached advanced stages. The key to increasing the overall survival of women with EOC lies in early detection and screening, especially of patients with precursor lesions. In this study, we demonstrate that plasma miRNA expression signatures can be used as biomarkers to distinguish endometriosis, a precursor of endometrioid and clear cell adenocarcinoma of the ovary, and different histotypes of EOC from healthy controls. Importantly, the plasma miRNA signatures we identified can be mirrored in an EOC mouse model, suggesting that they are reflective of EOC pathogenesis. Taken together, the plasma miRNAs identified in this study are promising biomarkers of endometriosis-to-EOC progression, which may ultimately be used to improve the clinical outcome of ovarian cancer patients.
Acknowledgments
We would like to thank Lindsay Mock, Louise Mazur, Julia Thaller, Joan Brozick, and Sean Soisson for technical support.This work has been supported in part by UPMC grant # 02.93530 (A.M. Vlad, R.P. Edwards, X. Huang), Pennsylvania Department of Health and Scaife Foundation (A.M. Vlad and X. Huang), Department of Defense Ovarian Cancer Academy Award and NIH/NCI 1 R01 CA163462-01 (A. M. Vlad). This project used the UPCI Biostatistics Facility and was supported in part by NCI award P30 CA047904 (HL, YL).
Footnotes
Statement of Conflict of Interest: None
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Clarke-Pearson DL. Screening for Ovarian Cancer. N Engl J Med. 2009;361:170–7. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 3.Engel J, Eckel R, Schubert-Fritschle G, Kerr J, Kuhn W, Diebold J, et al. Moderate progress for ovarian cancer in the last 20 years: prolongation of survival, but no improvement in the cure rate. Eur J Cancer. 2002;38:2435–45. doi: 10.1016/s0959-8049(02)00495-1. [DOI] [PubMed] [Google Scholar]
- 4.Bast RC, Jr, Badgwell D, Lu Z, Marquez R, Rosen D, Liu J, et al. New tumor markers: CA125 and beyond. Int J Gynecol Cancer. 2005;15:274–81. doi: 10.1111/j.1525-1438.2005.00441.x. [DOI] [PubMed] [Google Scholar]
- 5.Gagnon A, Ye B. Discovery and application of protein biomarkers for ovarian cancer. Curr Opin Obstet Gynecol. 2008;20:9–13. doi: 10.1097/GCO.0b013e3282f226a5. [DOI] [PubMed] [Google Scholar]
- 6.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 7.Ventura A, Jacks T. MicroRNAs and Cancer: Short RNAs Go a Long Way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum MicroRNAs Are Promising Novel Biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum MicroRNA Signatures Identified in a Genome-Wide Serum MicroRNA Expression Profiling Predict Survival of Non-Small-Cell Lung Cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in Plasma of Pancreatic Ductal Adenocarcinoma Patients as Novel Blood-Based Biomarkers of Disease. Cancer Prev Res. 2009;2:807–13. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce CL, Templeman C, Rossing MA, Lee A, Near AM, Webb PM, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol. 2012;13:385–94. doi: 10.1016/S1470-2045(11)70404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ness RB. Endometriosis and ovarian cancer: Thoughts on shared pathophysiology. Am J Obstet Gynecol. 2003;189:280–94. doi: 10.1067/mob.2003.408. [DOI] [PubMed] [Google Scholar]
- 17.Giudice LC. Endometriosis. N Engl J Med. 2010;362:2389–98. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garry R. Endometriosis: an invasive disease. Gynaecol Endoscopy. 2001;10:79–82. [Google Scholar]
- 19.Oral E, Ilvan S, Tustas E, Korbeyli B, Bese T, Demirkiran F, et al. Prevalence of endometriosis in malignant epithelial ovary tumours. Eur J Obstet Gynecol Reprod Biol. 2003;109:97–101. doi: 10.1016/s0301-2115(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 20.Fukunaga M, Nomura K, Ishikawa E, Ushigome S. Ovarian atypical endometriosis: its close association with malignant epithelial tumours. Histopathology. 1997;30:249–55. doi: 10.1046/j.1365-2559.1997.d01-592.x. [DOI] [PubMed] [Google Scholar]
- 21.Varma R, Rollason T, Gupta JK, Maher ER. Endometriosis and the neoplastic process. Reproduction. 2004;127:293–304. doi: 10.1530/rep.1.00020. [DOI] [PubMed] [Google Scholar]
- 22.Nishida M, Watanabe K, Sato N, Ichikawa Y. Malignant transformation of ovarian endometriosis. Gynecol Obstet Invest. 2000;50:18–25. doi: 10.1159/000052874. [DOI] [PubMed] [Google Scholar]
- 23.Seidman JD. Prognostic importance of hyperplasia and atypia in endometriosis. Int J Gynecol Pathol. 1996;15:1–9. doi: 10.1097/00004347-199601000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protocols. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 25.Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- 26.Budiu RA, Diaconu I, Chrissluis R, Dricu A, Edwards RP, Vlad AM. A conditional mouse model for human MUC1-positive endometriosis shows the presence of anti-MUC1 antibodies and Foxp3+ regulatory T cells. Dis Model Mech. 2009;2:593–603. doi: 10.1242/dmm.002535. [DOI] [PubMed] [Google Scholar]
- 27.Gail MH, Fears TR, Hoover RN, Chandler DW, Donaldson JL, Hyer MB, et al. Reproducibility studies and interlaboratory concordance for assays of serum hormone levels: estrone, estradiol, estrone sulfate, and progesterone. Cancer Epidemiol Biomarkers Prev. 1996;5:835–44. [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 29.McLachlan GJ. Cluster analysis and related techniques in medical research. Stat Methods Med Res. 1992;1:27–48. doi: 10.1177/096228029200100103. [DOI] [PubMed] [Google Scholar]
- 30.McLachlan GJ. Discriminant analysis and statistical pattern recognition. New York: Wiley; 1992. [Google Scholar]
- 31.Cannistra SA. Cancer of the Ovary. N Engl J Med. 2004;351:2519–29. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 32.Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer. 2010;17:F77–F89. doi: 10.1677/ERC-09-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotech. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dzatic-Smiljkovic O, Vasiljevic M, Djukic M, Vugdelic R, Vugdelic J. Frequency of ovarian endometriosis in epithelial ovarian cancer patients. Clin Exp Obstet Gynecol. 2011;38:394–8. [PubMed] [Google Scholar]
- 36.Cramer DW, Bast RC, Berg CD, Diamandis EP, Godwin AK, Hartge P, et al. Ovarian Cancer Biomarker Performance in Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial Specimens. Cancer Prev Res. 2011;4:365–74. doi: 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss EL, Hollingworth J, Reynolds TM. The role of CA125 in clinical practice. J Clin Pathol. 2005;58:308–12. doi: 10.1136/jcp.2004.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, et al. Ovarian Carcinoma Subtypes Are Different Diseases: Implications for Biomarker Studies. PLoS Med. 2008;5:e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laschke MW, Menger MD. Anti-angiogenic treatment strategies for the therapy of endometriosis. Hum Reprod Update. 2012;18:682–702. doi: 10.1093/humupd/dms026. [DOI] [PubMed] [Google Scholar]
- 40.Kandalaft LE, Motz GT, Busch J, Coukos G. Angiogenesis and the tumor vasculature as antitumor immune modulators: the role of vascular endothelial growth factor and endothelin. Curr Top Microbiol Immunol. 2011;344:129–48. doi: 10.1007/82_2010_95. [DOI] [PubMed] [Google Scholar]
- 41.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37:460–5. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Nezhat F, Datta MS, Hanson V, Pejovic T, Nezhat C, Nezhat C. The relationship of endometriosis and ovarian malignancy: a review. Fertil Steril. 2008;90:1559–70. doi: 10.1016/j.fertnstert.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Wei J-J, William J, Bulun S. Endometriosis and Ovarian Cancer: A Review of Clinical, Pathologic, and Molecular Aspects. Int J Gynecol Pathol. 2011;30:553–68. doi: 10.1097/PGP.0b013e31821f4b85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, et al. Repertoire of microRNAs in Epithelial Ovarian Cancer as Determined by Next Generation Sequencing of Small RNA cDNA Libraries. PLoS ONE. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–26. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 47.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as Novel Potential Biomarkers for Gastric Cancer Detection. PLoS ONE. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schrauder MG, Strick R, Schulz-Wendtland Rd, Strissel PL, Kahmann L, Loehberg CR, et al. Circulating Micro-RNAs as Potential Blood-Based Markers for Early Stage Breast Cancer Detection. PLoS ONE. 2012;7:e29770. doi: 10.1371/journal.pone.0029770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–9. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 50.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.