Abstract
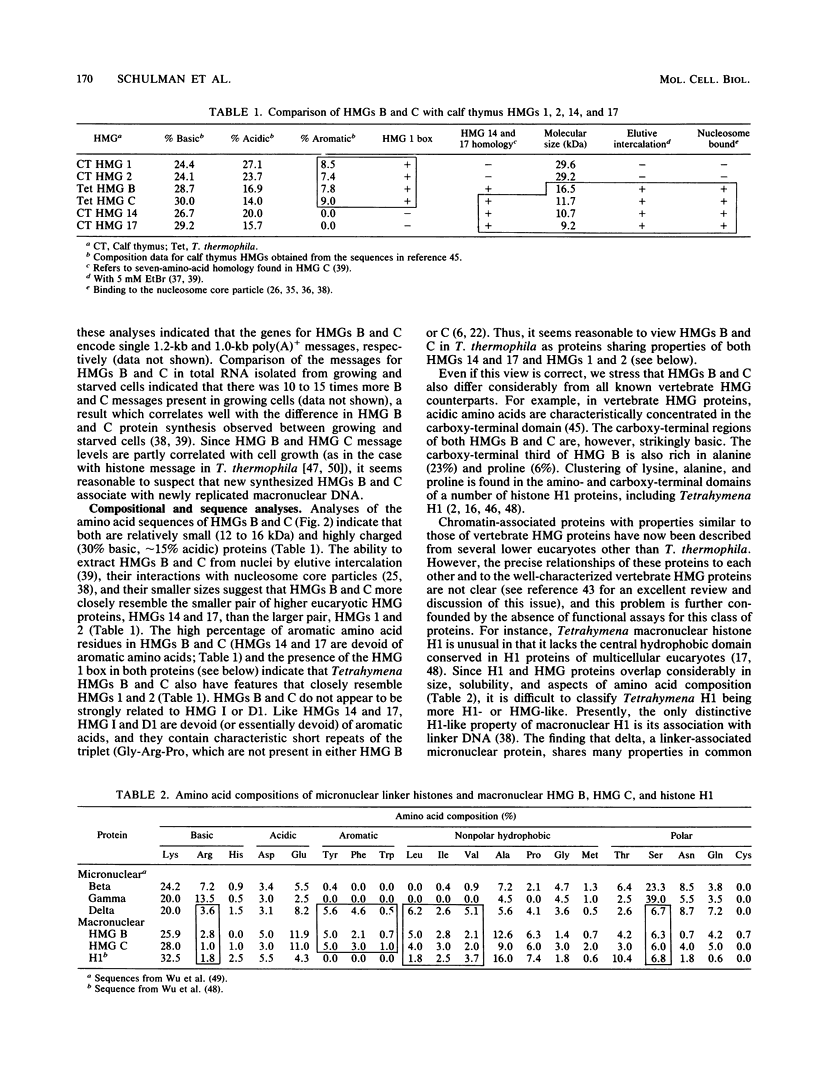
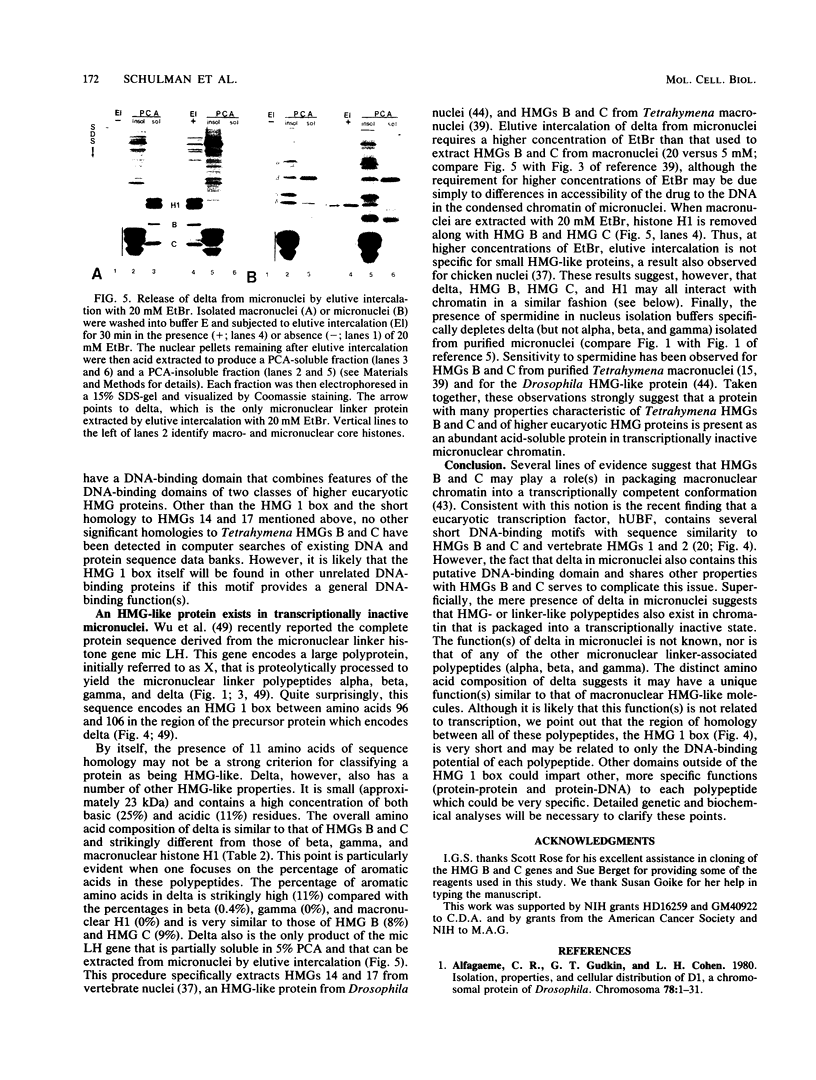
HMG (high-mobility-group protein) B and HMG C are abundant nonhistone chromosomal proteins isolated from Tetrahymena thermophila macronuclei with solubilities, molecular weights, and amino acid compositions like those of vertebrate HMG proteins. Genomic clones encoding each of these proteins have been sequenced. Both are single-copy genes that encode single polyadenylated messages whose amounts are 10 to 15 times greater in growing cells than in starved, nongrowing cells. The derived amino acid sequences of HMG B and HMG C contain a highly conserved sequence, the HMG 1 box, found in vertebrate HMGs 1 and 2, and we speculate that this sequence may represent a novel, previously unrecognized DNA-binding motif in this class of chromosomal proteins. Like HMGs 1 and 2, HMGs B and C contain a high percentage of aromatic amino acids. However, the Tetrahymena HMGs are small, are associated with nucleosome core particles, and can be specifically extracted from macronuclei by elutive intercalation, properties associated with vertebrate HMGs 14 and 17, not HMGs 1 and 2. Thus, it appears that these Tetrahymena proteins have features in common with both of the major subgroups of higher eucaryotic HMG proteins. Surprisingly, a linker histone found exclusively in transcriptionally inactive micronuclei also has several HMG-like characteristics, including the ability to be specifically extracted from nuclei by elutive intercalation and the presence of the HMG 1 box. This finding suggests that at least in T. thermophila, proteins with HMG-like properties are not restricted to regions of transcriptionally active chromatin.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Mitchell T., Harborne N., Bohm L., Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986 Feb 20;187(4):591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Allen R. L., Wiggins J. C., Chicoine L. G., Richman R. Proteolytic processing of h1-like histones in chromatin: a physiologically and developmentally regulated event in Tetrahymena micronuclei. J Cell Biol. 1984 Nov;99(5):1669–1677. doi: 10.1083/jcb.99.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Glover C. V., Gorovsky M. A. Micronuclei of Tetrahymena contain two types of histone H3. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4857–4861. doi: 10.1073/pnas.76.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley C. T., Pendleton C. G., Jennings W. W., Saxena A., Glover C. V. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J Biol Chem. 1989 May 15;264(14):8394–8401. [PubMed] [Google Scholar]
- Blackburn E. H., Karrer K. M. Genomic reorganization in ciliated protozoans. Annu Rev Genet. 1986;20:501–521. doi: 10.1146/annurev.ge.20.120186.002441. [DOI] [PubMed] [Google Scholar]
- Butler A. P., Mardian J. K., Olins D. E. Nonhistone chromosomal protein HMG 1 interactions with DNA. Fluorescence and thermal denaturation studies. J Biol Chem. 1985 Sep 5;260(19):10613–10620. [PubMed] [Google Scholar]
- Elton T. S., Nissen M. S., Reeves R. Specific A . T DNA sequence binding of RP-HPLC purified HMG-I. Biochem Biophys Res Commun. 1987 Feb 27;143(1):260–265. doi: 10.1016/0006-291x(87)90659-0. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Piatak M., Lebowitz P., Weissman S. M. Determination of RNA sequences by primer directed synthesis and sequencing of their cDNA transcripts. Methods Enzymol. 1980;65(1):580–595. doi: 10.1016/s0076-6879(80)65061-7. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Haggren W., Kolodrubetz D. The Saccharomyces cerevisiae ACP2 gene encodes an essential HMG1-like protein. Mol Cell Biol. 1988 Mar;8(3):1282–1289. doi: 10.1128/mcb.8.3.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamana K., Iwai K. High mobility group nonhistone chromosomal proteins also exist in Tetrahymena. J Biochem. 1979 Sep;86(3):789–794. doi: 10.1093/oxfordjournals.jbchem.a132586. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hayashi H., Iwai K. Tetrahymena HMG nonhistone chromosomal protein. Isolation and amino acid sequence lacking the N- and C-terminal domains of vertebrate HMG 1. J Biochem. 1989 Apr;105(4):577–581. doi: 10.1093/oxfordjournals.jbchem.a122707. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hayashi H., Iwai K. Tetrahymena histone H1. Isolation and amino acid sequence lacking the central hydrophobic domain conserved in other H1 histones. J Biochem. 1987 Aug;102(2):369–376. doi: 10.1093/oxfordjournals.jbchem.a122063. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Bowen J. K., Bannon G. A., Gorovsky M. A. Unusual features of transcribed and translated regions of the histone H4 gene family of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jan 12;15(1):141–160. doi: 10.1093/nar/15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isackson P. J., Fishback J. L., Bidney D. L., Reeck G. R. Preferential affinity of high molecular weight high mobility group non-histone chromatin proteins for single-stranded DNA. J Biol Chem. 1979 Jul 10;254(13):5569–5572. [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P., Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Johnson K. R., Lehn D. A., Elton T. S., Barr P. J., Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y). J Biol Chem. 1988 Dec 5;263(34):18338–18342. [PubMed] [Google Scholar]
- Kolodrubetz D., Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990 Feb 25;265(6):3234–3239. [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B., Denker M. S., Ito E. Isolation, characterization, and postsynthetic modifications of tetrahymena high mobility group proteins. Biochemistry. 1983 Mar 29;22(7):1715–1721. doi: 10.1021/bi00276a030. [DOI] [PubMed] [Google Scholar]
- Mardian J. K., Paton A. E., Bunick G. J., Olins D. E. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980 Sep 26;209(4464):1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- Martindale D. W. Codon usage in Tetrahymena and other ciliates. J Protozool. 1989 Jan-Feb;36(1):29–34. doi: 10.1111/j.1550-7408.1989.tb02679.x. [DOI] [PubMed] [Google Scholar]
- McMurray C. T., van Holde K. E. Binding of ethidium bromide causes dissociation of the nucleosome core particle. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8472–8476. doi: 10.1073/pnas.83.22.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentecost B. T., Wright J. M., Dixon G. H. Isolation and sequence of cDNA clones coding for a member of the family of high mobility group proteins (HMG-T) in trout and analysis of HMG-T-mRNA's in trout tissues. Nucleic Acids Res. 1985 Jul 11;13(13):4871–4888. doi: 10.1093/nar/13.13.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Elton T. S., Nissen M. S., Lehn D., Johnson K. R. Posttranscriptional gene regulation and specific binding of the nonhistone protein HMG-I by the 3' untranslated region of bovine interleukin 2 cDNA. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6531–6535. doi: 10.1073/pnas.84.18.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez Alfageme C., Rudkin G. T., Cohen L. H. Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma. 1980;78(1):1–31. doi: 10.1007/BF00291907. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Schulman I. G., Cook R. G., Allis C. D. The complete amino acid sequence of an HMG-like protein isolated from the macronucleus of Tetrahymena. Nucleic Acids Res. 1987 Oct 12;15(19):8112–8112. doi: 10.1093/nar/15.19.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R. H., Candido E. P., Astell C. R. Interaction of the mouse chromosomal protein HMG-I with the 3' ends of genes in vitro. J Biol Chem. 1988 May 5;263(13):6392–6399. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter H., Bode J. The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur J Biochem. 1982 Oct;127(2):429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- Schröter H., Maier G., Ponstingl H., Nordheim A. DNA intercalators induce specific release of HMG 14, HMG 17 and other DNA-binding proteins from chicken erythrocyte chromatin. EMBO J. 1985 Dec 30;4(13B):3867–3872. doi: 10.1002/j.1460-2075.1985.tb04159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman I. G., Cook R. G., Richman R., Allis C. D. Tetrahymena contain two distinct and unusual high mobility group (HMG)-like proteins. J Cell Biol. 1987 Jun;104(6):1485–1494. doi: 10.1083/jcb.104.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Dixon G. H. High mobility group proteins 1 and 2 function as general class II transcription factors. Biochemistry. 1990 Jul 3;29(26):6295–6302. doi: 10.1021/bi00478a026. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Kikuchi M., Mori K., Waga S., Yoshida M. Primary structure of non-histone protein HMG1 revealed by the nucleotide sequence. Biochemistry. 1988 Aug 9;27(16):6159–6163. doi: 10.1021/bi00416a050. [DOI] [PubMed] [Google Scholar]
- Tsunasawa S., Sakiyama F. Amino-terminal acetylation of proteins: an overview. Methods Enzymol. 1984;106:165–170. doi: 10.1016/0076-6879(84)06016-x. [DOI] [PubMed] [Google Scholar]
- Wells D. E. Compilation analysis of histones and histone genes. Nucleic Acids Res. 1986;14 (Suppl):r119–r149. doi: 10.1093/nar/14.suppl.r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Allis C. D., Richman R., Cook R. G., Gorovsky M. A. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8674–8678. doi: 10.1073/pnas.83.22.8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. M., Gorovsky M. A. In situ dot blots: quantitation of mRNA in intact cells. Nucleic Acids Res. 1986 Oct 10;14(19):7597–7615. doi: 10.1093/nar/14.19.7597. [DOI] [PMC free article] [PubMed] [Google Scholar]