Abstract
Between 2004 and 2008, the NIH molecular libraries and imaging initiative (MLI) pilot phase funded ten high-throughput Screening Centers, resulting in the deposition of 691 assays into PubChem and the nomination of 64 chemical probes. We crowdsourced the MLI output to 11 experts, who expressed medium or high levels of confidence in 48 of these 64 probes.
A new kind of science
The NIH (National Institutes of Health) Molecular Libraries and Imaging (MLI) initiative1 was designed as a cross-institutional effort to stimulate the discovery of novel small molecule tools for chemical biology. Supported by all 27 NIH institutes for a limited (up to 9 years) period, the MLI pilot phase (2004-2008) stimulated the development of ten national high-throughput screening (HTS) centers known as the Molecular Libraries Screening Centers Network (MLSCN). The MLI also encouraged scientists to submit in a peer-reviewed process via MLSCN, assays amenable to medium- and high- throughput, novel substances to be tested, novel experimental data curated for MLSCN, as well as novel experimental results – all these to become freely accessible via the PubChem system2. Key to the open-access, free access system of the MLI is the intention to complement, rather than duplicate drug discovery efforts currently carried out in a pharmaceutical industrial setting. At the heart of this complementary process is the creation and screening of a compound library aimed at the identification of small molecule modulators of biological functions, suitable for basic research yet not necessarily ready to serve as starting points (leads) for drug discovery. It was envisioned that the MLSCN should ‘not [be] interested primarily in drug discovery but in the elucidation of biochemical pathways’1, by discovering small molecule ‘tools’ or ‘probes’ to enable and support research in chemical biology.
Having recognized that chemical biology relies on small molecule tools to ‘probe” biochemical aspects for all gene-encoded proteins, the majority of which the pharmaceutical industry considers yet “undruggable”1, the NIH aimed not only to provide a suitable framework for the development of such “chemical probes”, but also to encourage a cultural shift towards open, cross-disciplinary, communication within multiple scientific and technological disciplines to facilitate probe discovery. These chemical tools are aimed at filling the gap between target identification, often regarded as an early step in drug discovery whereby a certain macromolecule or process is “targeted” for therapeutic manipulation via (often) small molecules, and lead discovery – an ulterior step that is often preceeded by HTS, thus based on “hit” identification and optimization. Hence, the MLI framework set the stage for a new phase in academic science by broadly enabling this intermediate step of “chemical probe” discovery1. The MLI further strengthened the “community resource” aspect of the initiative by mandating rapid dissemination of all MLI-generated data, and ensured freedom to operate by eliminating trade secrets and intellectual property claims.
Pilot tools
Essential to this pilot phase was the creation of a chemical library, the Molecular Libraries Small Molecule Repository, or MLSMR. This library, accessible by querying “MLSMR” in PubChem Substance, was subjected to MLSCN bioactivity screening between 2004 and 2008, for a total of 691 assays that were uploaded to PubChem. These included 242 primary, 402 confirmatory and 8 summary assays, which covered 171 targets and 29 phenotypic screens, respectively. These numbers continue to grow as Pilot Phase projects are completed.
Within this pilot phase, these NIH chemical probes were envisioned as having “adequate potency and adequate solubility to be useful for in vitro (e.g., cell-based experimentation” (Box 1), but requiring further “chemical modifications […] to confer the selectivity, pharmacokinetic, and metabolic properties required for in vivo use”1. The criteria for a chemical probes evolved further, and currently requires a certain potency (up to 100 nM), selectivity, aqueous solubility, and an improvement over existing probes for that target/assay (Box 1),
BOX 1. CHEMICAL PROBE EVOLUTION.
Chemical Probe 1.0 (2005)
…a chemical compound with activity in the primary and any secondary assays with adequate potency and aqueous solubility to be useful for in vitro (i.e., cellbased) experimentation. The specifications for a probe are likely to vary depending on the target and may need to be set jointly by the assay provider and the MLSCN Steering Committee. An example of the specifications for a chemical probe that would define an endpoint for MLSCN activities would be: expected potency of 1 μM and solubility in 1% DMSO.
Source: http://mli.nih.gov/mli/wp-content/uploads/data-sharing-and-ip.pdf
Probe Definition 2.0 (MLSCN 2007)
An MLSCN probe is a compound which represents an improvement over the existing art. Supporting information is required showing currently available probes, their properties and how the new probe is clearly an improvement [first or best in class]
Potency
Criteria for potency is context dependent, varying with the assay and target type, and current state of the art. In general, probes identified via biochemical assays are expected to exhibit potencies of < 500nM and, more ideally in the 100 nM range. Those identified in cell based assays are expected to exhibit potencies at a level of < 1 μM, however in certain instances potencies in the 10 μM may be acceptable. Criteria for whole organism screening is less stringent.
Solubility
Sufficient solubility in relevant solvents
Availability
The probe molecule should be accessible in amounts to allow advanced studies (15-20mg), and protocols for its preparation or isolation should be made available
SAR, mode of action
(e.g. evidence of binding to target, characterization of mechanism of action) and awareness of selectivity against relevant and/ or related targets is expected Appropriate data on toxicity, permeability, etc. of probes are strongly encouraged.
Chemical Probe 3.0 (2008)
One of the primary goals of the MLPCN (Molecular Libraries Probe Production Centers Network) is the production of new and useful chemical probes for biomedical research. The minimum characteristics that a probe compound will need to have to be a useful research tool has been determined by the MLPCN Steering Committee and the MLP Project Team. According to the definition currently used by the MLSCN, these characteristics include <100 nM affinity, >10-fold selectivity against related targets, and solubility in aqueous solutions (possibly including a low concentration of DMSO). Most importantly, a chemical probe must represent an improvement over existing probes for the designated target (https://mli.nih.gov/resources/MLPCN_U54_FAQ.htm. NIH recognizes that whatever the characteristics of the probes, further modification may be necessary to produce compounds that are useful for in vivo studies. However, such additional efforts are outside of the activities called for in this FOA. It is the desire of NIH that during the production phase, all probes and all related biological and chemical data will be made available to all researchers
Source: http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-08-005.html
The review procedure for the MLPCN Probe Reports [https://mli.nih.gov/mli/mlpcn/documents-definitions/?dl_id=883] details aspects related to scientific merit, information completeness and the approval process. A template for the Probe Reports is available on-line. [https://mli.nih.gov/mli/mlpcn/documents-definitions/?dl_id=841]
Given the interdependence between the library being screened and a particular assay, not all assays can yield novel chemical probes. Yet MLSCN scientists were tasked with nominating suitable chemical probes, based on the emergent results. Beyond these general guidelines for chemical probes, the Centers were given considerable autonomy in determining the quality and druggability necessary for a compound to be nominated as a probe.
As output, this US$ 385 million initiative produced “a compound repository, a database and technology” as well as 64 chemical probes so far, with additional probes pending approval, which subjected it to critical analysisError! Bookmark not defined.3. The initiative is also intended to nucleate discovery science throughout the research community by providing for individual investigators as well as centers to produce targets, probes, chemistry, novel hardware and novel software tools. In this Commentary, we seek two goals: First, we evaluate the quality of the NIH chemical probes, as perceived by a network of experts (‘wisdom of the crowd’ or ‘crowdsourcing’). Second, as the MLI enters a second phase, we discuss strategies for increasing the quality and usefulness of nominated chemical probes including some of those implemented already by the MLPCN.
Molecular Confidence or Dubiosity
Molecular confidence or dubiosity (Box 2) is a subjective or objective property that can be attributed to small molecules in certain cycles of preclinical drug discovery. Its subjective aspect relates to what skilled medicinal chemists would describe as “gut reaction”, an intuitive or emotional response they experience when deciding which HTS hits, or confirmed actives, should be progressed further from a particular HTS assay. In its more subjective form, the scientific opinion of a medicinal chemist regarding the potential of a compound to be optimized into a clinical candidate drug3 is based on what Polanyi described4 as ‘tacit’ knowledge based on learning from experience. In its more objective aspect, molecular confidence or dubiosity is rooted in empirical observations, and it is usually derived by filtering HTS hits using a variety of parameters: i) a pre-determined list of substructures to remove reactive species5, frequent hitters6, aggregators7,8 or fluorescent compounds (as observed, e.g., via flow cytometry9); ii) a pre-determined list of toxicity filters10, Lipinski’s rule-of-five11 (Ro5) and Pfizer-like12 rules, CNS penetration13, druglike14,15 or leadlike16,17 properties; iii) criteria for enrichment or removal of chemotypes based on similarity or dissimilarity to desired or unwanted chemicals; iv) additional model-based filters18 for down-stream drug safety issues such as hERG binding (to rule out cardiac toxicity), cytochrome P450 inhibition (to reduce the risk of drug-drug interactions), and other potential antitargets19; v) all of the above. Thus, confidence or doubt in a molecule stems from any number of undesired properties, and the potential to circumvent them within a reasonably small period (typically, less than six months), as judged by drug discovery scientists.
BOX 2. DEFINITIONS.
Dubiosity (plural dubiosities)
(rare) The state or characteristic of being dubious. Synonyms for dubiosity include doubtfulness, incertitude, skepticism, suspicion, and uncertainty. In Ulysses, episode 16, James Joyce (1922) writes: “Possibly perceiving an expression of dubiosity on their faces, the globetrotter went on adhering to his adventures” (James Joyce: http://en.wikipedia.org/wiki/James_Joyce).
Crowdsourcing
First suggested by Jeff Howe25, crowdsourcing refers to an outsourcing ‘open innovation’26 model that utilizes distributed problem-solving27. Its R&D applications are utilized by InnoCentive, YourEncore, and NineSigma, among others. In its simplest form, problems are “crowdsourced” when a large (often unknown) group of solvers, the “crowd”, is tasked with solving a problem or a particular set of problems. Often, the crowd ranks the possible solutions and selects the optimal one(s). If crowdsourcing occurs within a business parameter, the crowdsourcer owns the solution, while the best-ranked solutions are compensated financially or otherwise.28
InnoCentive: http://www.innocentive.com/
YourEncore: http://www.yourencore.com/jsp/index.html
NineSigma: http://www.ninesigma.net/
Widsom of Crowds
This concept, popularized by Surowiecki29, describes the group decision-making based on the aggregation of independent, individual decisions, where the average decision is more accurate than any individual decision. The four elements of a wise crowd are independence, diversity of opinion, decentralization and aggregation. The survey on the NIH chemical probes fulfils the requirements of a wise crowd.
Crowdsourcing
A key challenge in evaluating the quality of the chemical probes resulting from the MLI initiative is a lack of skilled experts who can individually determine, with a reasonable expectation of success, the quality of these probes. In the absence of a completely objective way to evaluate these chemical probes, we used the ‘wisdom of the crowd’ to crowdsource (Box 2) this evaluation to a team of eleven scientists with diverse backgrounds in small molecule discovery, most of them experienced in industrial drug discovery and currently engaged in both academia and industry. Their task? Express their level of confidence in the 64 NIH chemical probes.
The UNM team, which did not vote, conducted exhaustive (as of October 10, 2008) queries for each chemical probe; the results were synthesized in tabular format and provided to the voting group. The “crowdsourced group” (CSG) was then asked to evaluate these probes in the absence of any information related to the principal investigator (PI) or the institution that performed the assay and proposed the chemical probe. As a final step to ensure a blind evaluation, voters were asked not to use PubChem or any other database during the evaluation.
We anticipate that this crowdsourcing approach will accomplish two goals: i) to address the question “what is the quality of the output” given the magnitude of the investment in the MLI; and ii) to provide an individual, analytical, overview of these probes from well-known experts in preclinical drug discovery.
Last but not least, each of the CSG experts was informed that their individual contribution would be made available as Supplementary Information, and could potentially serve as the basis of a retrospective analysis in a decade – a period of time judged by most to be ample enough to confirm (or not) the quality of these chemical probes for the targets for which they were proposed.
Confidence scores
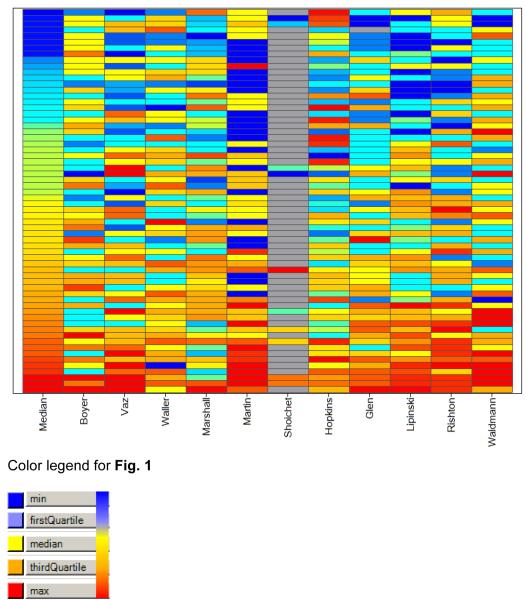
Since molecular confidence is neither exact nor accurate, the NIH chemical probes evaluation was performed on a qualitative ranking of 0 (high confidence, low dubiosity) to 10 (low confidence, high dubiosity), as described in Box 3. Of the 64 probes, 16 (25%) were evaluated as having low confidence or high dubiosity, i.e., their empirical rank scores are between 5 and 6; 16 probes (25%) were assigned “medium”, i.e., their empirical rank scores are 3 or 4; while half the probes (32) were considered “high in confidence or low in dubiosity”, i.e., their score was ranked between 0 and 2. A visual overview of the individual confidence scores is provided as a heatmap in Fig. 1. Exact scores, together with comments inserted by the 11 members of the voting group, are provided as Supplementary Table 1.
BOX 3. THE EVALUATION OF CONFIDENCE.
By definition, low confidence would make us doubt the overall potential of a chemical probe related to, e.g., selectivity, its probability to act as promiscuous binder, as well as its future use as chemical probe and starting structure for lead discovery. Each voting member was asked to assign score, from 0 to 10, to each of the chemical probes according to their own judgment. However, they were under no obligation to score each probe; indeed, 2 voters did not score all chemicals (7.8% missing values). Higher scores imply higher dubiosity for the molecule, whereas low values imply higher confidence. To derive a qualitative evaluation of confidence, we applied the following steps and criteria:
Compute the average score across all voters; see Box 2 Table; note that the average across all averaged scores was 4.54335
Compute the median score across all voters; see Box 2 Table;
Count the number of times a probe was scored above the average of all scores (4.54335), i.e., the “Score above average”; see also Box 2 Table;
Empirically rank confidence by adding 3, if the Score Above Average or the Score Median is above the 75% quartile; 2, if the Score Above Average or the Score Median is between the median and 75% quartile; 1, if the Score Above Average or the Score Median is between the 25% quartile and the median; and adding 0 if the Score Above Average or the Score Median is below the 25% quartile, respectively.
Qualitatively rank probes as follows: If the empirical rank is 5 or 6, “high dubiosity” (16 probes); if it’s 3 or 4, “medium” (16 probes); if it’s 0, 1 or 2, “high confidence” (32 probes). Further discussions are based on this qualitative rank.
Table.
Distribution of Scores
| Distribution Type | Score Average | Score Median | Score Above Average |
|---|---|---|---|
| Minimum | 2.50 | 2.97 | 1 |
| 25% quartile | 3.78 | 3.49 | 3 |
| Median | 4.19 | 4.18 | 5 |
| 75% quartile | 5.13 | 5.25 | 6 |
| Maximum | 7.84 | 8.00 | 11 |
Figure 1.
Heatmap of the confidence scores, by voting member, sorted by the median value. Red and orange indicate high dubiosity, yellow reflects medium values, whereas shades of blue indicate high confidence scores; grey indicates absence of score. The heatmap is sorted by the median score on the vertical axis, and by the research area of the CSG experts on the horizontal axis. The first two columns reflect pharmacokinetics and toxicology, the last four columns reflect a chemical / HTS perspective, whereas the middle five columns are based on cheminformatics tools and experience.
For some probes, most of the CSG members were in agreement (horizontal lines that are predominantly one color in Fig. 1). Such consensus implies that some of these molecules should perhaps not have been nominated as probes, e.g., where predominantly red and orange lines are found). Eight probes ranked lowest in confidence (six), and had a median score of 5 or higher. The CSG experts are in agreement that these structures should not have been nominated as chemical probes since their bioactivity spectrum is quite non-specific, making them useful only under carefully controlled biochemical conditions. Some of them share a polyphenolic, dye character: Alizarin Yellow (CID 5281855) and Myricetin (CID 5281672), and the diketone CID 665013. A known fungicide (Dazomet; CID 10788) contains both an aminal and a thiourea group, while an immunosuppresant drug (azathioprine; CID 2265) is known to hydrolyze in vivo to 6-mercaptopurine, a thiol-reacting group. Idarubicin (CID 151582) is a well characterized cytotoxin. CSG members expressed concern that Ebselen (CID 3194) may be a high-liability compound, as reflected by its Se-N moiety. Compound CID 2980973 may be a false positive due to its aliphatic ester / pyrimidinyl sulfone combination, while CID 16725315 is an acyl-hydrazine. Finally, two probes contain what toxicologists agree are troublesome chemical moieties (an ammonium group, CID 5716367; and an N-oxide-nitrile, CID 1756).
In some cases, e.g., CID 665013, CSG questioned if compounds with such chemical liabilities (polyphenol and diketone) are indeed an improvement over state-of-the-art, since the intended target, HIV-1 Reverse Transcriptase, has already been targeted successfully by known drugs. Of the 16 low-confidence probes, 8 (12.5% of the total) are flagged for potential toxicity alerts and one is a pesticide (Dazomet). Three (4.7%) of these probes are, or have been, marketed drugs (Azathioprine, Idarubicin and Ebselen), while another 4 (6.25%) are perceived as druglike. The low confidence score awarded to these 16 chemical probes is primarily owed to innate chemical structure liabilities as well as to potential or known toxicity issues. However, one cannot a priori exclude the possibility that, under controlled conditions, they could serve as chemical probes. The CSG evaluation was based on the time- and data- limited information that was gathered as of October 10, 2008. Given that additional data have been collected since, some of these votes could change significantly, making these compounds more valuable as probes than they appear. Our goal is to preserve this “time capsule” of the voting process, not only to allow the community at large to discuss this with a ten-year timeframe in mind, but also to document steps in a decision-making process regarding the evaluation of these compounds.
At the other end of the scale, 32 probes ranked high in confidence (low dubiosity): 14 probes were scored zero; 10 probes received a score of one; and 8 probes a score of two. The CSG has higher confidence in the overall qualities of these chemical probes (in particular those ranked 0 and 1), although most of them are likely to encounter downstream liabilities, should they be chosen as starting points for drug discovery. Of the 32 high confidence chemical probes, 14 (21.88%) are flagged for potential toxicity alerts, 17 (26.56%) are perceived as druglike and one (1.56%) is a marketed drug (Benzbromarone). The high-confidence, low-dubiosity perception attributed to half (32) of the chemical probes is primarily rooted in their apparent lack of immediate chemical liabilities. Although 12 (18.75%) of them are aromatic amines, this potential mutagenicity risk was not deemed relevant at the chemical probe stage by the CSG.
Can we bring objectivity to this process?
One of the frustrating aspects of chemical probe evaluation is the lack of an objective function, or metric, by which their quality can be judged. How did the few empirical criteria for chemical probes correlate with the results of the voting group? Potency for the designated target and the estimated aqueous solubility (computed via ALOGPS20) were central to the NIH requirements (Probe 2.0, Box 1). Current NIH guidelines mandate that probes should have affinity below 100 nM (Probe 3.0, Box 1). Yet 45 out of 64 probes (70.3%) have activity above 100 nM, with 3 probes above 10 μM. Among high-confidence, low-dubiosity probes, 11 (17.2% of the total) have potency below 100 nM, and another 13 (20.3%) are between 100 nM and 1 μM. Since 7 of the 17 low-confidence, high-dubiosity probes were reported as having bioactivity of 100 nM or lower, while 8 high confidence and 11 medium confidence probes have activity worse than 1 μM, it appears that target potency is regarded as a tunable parameter, i.e., one that can be further optimized. Although 70% of the probes would not meet the current (Probe 3.0) activity cut-off, this aspect should to be regarded in the context of the pilot phase (MLI), where fewer resources were available to meet this desirable goal. As the production phase proceeds, no doubt more medicinal chemistry resources will be allocated for improving affinity and selectivity. However, potency was deemed as less important in both the MLSCN nomination process, and in our own CSG evaluation. In this context, perhaps the NIH should consider reverting to Probe 2.0 criteria with respect to affinity.
The lack of an apparent relationship between activity vs. computed solubility is shown in Fig. 2. All probes except one low confidence probe have estimated (ALOGPS) solubility above 1 μM, so this criterion was largely met, but does not distinguish high from low ranked probes. Even the Ro5 criteria11, graphically summarized in Fig. 3, fail to discriminate between low and high probes. Sixty chemical probes violate none of the Ro5 criteria, while two violate one rule. Only two probes are outside the Ro5 parameters – indicative perhaps of the profound effect that Ro5 criteria had on the lead and drug discovery community. These two exceptions are peptidomimetic Cathepsin L inhibitors that owe their low confidence score in part to an acyl-hydrazine moiety. By setting the polar surface area (PSA) value above 97 Å2, 8 low confidence probes are filtered, in contrast to 4 high and 4 middle confidence probes, respectively. Twenty-eight high confidence and 11 medium confidence probes, as well as 9 low confidence probes (75% of the total) satisfy the condition PSA ≤ 97 Å2. Based on a simple set of 2D descriptors21, and in the absence of supervised learning tools, we could find no criterion that adequately separates probes based on assessed quality.
Figure 2.
Measured activity (−log10 of the reported target activity, for the intended target) versus estimated aqueous solubility (computed with ALOGPSError! Bookmark not defined.20) for the 64 chemical probes, colored by their qualitative score (red, “high dubiosity”; yellow, “middle”; and blue, “high confidence”, respectively), and labeled by Screening Center.
Figure 3.
Rule-of-fiveError! Bookmark not defined.11 overview by molecular weight (MW), computed octanol/water partition coefficient (ClogP1), and polar surface area (PSA) for the 64 chemical probes, colored by their qualitative score (see Fig. 2).
Lessons learned?
Having completed the pilot phase (MLSCN), the NIH Molecular Libraries Program (MLP) moved into the production phase by launching the Molecular Libraries Probe Production Centers Network (MLPCN) in September 2008. MLPCN aims to explore the interaction between small molecules and biological function at the molecular, cellular and in vivo levels, by encouraging biomedical researchers to combine large-scale screening with medicinal chemistry and informatics, in order to identify chemical probes to study the functions of genes, cells, and biochemical pathways. The MLPCN has four Comprehensive Centers (Broad, Burnham, NCGC, and Scripps) that will provide all 3 types of services (assay, cheminformatics/informatics, and medicinal chemistry); three Specialized Screening Centers (Johns Hopkins, SRI, and UNM), focused on specific assay technologies and informatics; and two Specialized Chemistry Centers (Kansas and Vanderbilt), which will provide medicinal chemistry and cheminformatics support for chemical probe optimization.
Since low confidence was expressed in only 25% of the MLSCN chemical probes, we anticipate that through collective experience, chemical probe optimization will become an effective effort both in the Chemistry Centers, and in the Comprehensive Centers as well. We also envision a dynamic MLSMR library, where protein-reactive electrophiles, warhead-containing agents, known frequent hitters and aggregators will gradually disappear as our collective knowledge of artifact-generating chemicals and their negative impact on chemical biology interactions increases. Indeed, the fact that, despite more than a decade of cumulative experience from the pharmaceutical industry, such chemical structures are even found among the NIH chemical probes suggests that a rigorous evaluation of this problem, and how it influences the MLSMR library, is appropriate.
With few exceptions, each MLSCN center had a share of “high” and “low” confidence chemical probes. Low confidence probe nominations could perhaps be explained by a variety of factors: Confidence in the chemists’ ability to overcome potential liabilities; no known promiscuity at the time of filing the probe; or perhaps knowledge that the perturbation of the intended biological target/process is not interfered with by these liabilities. MLPCN has already instigated a more rigorous review process for vetting both probe development plans and probes themselves before they are accepted.
From our experience, rigorous assessment of probe quality requires multiple communication rounds performed in an integrative manner involving: 1) chemical and drug informatics, including data mining, liabilities, virtual screening; 2) biology, from cellular processes to in vivo observations wherever possible and 3) chemistry, with respect to structure-activity analyses, synthesis planning, solubility and purity information. First and foremost, both the biology underlying the assay and the chemistry on which the probes are derived need to be ascertained and confirmed in independent experiments, according to strict guidelines22. Furthermore, any information regarding frequent hitter behavior, scaffold-specific chemical liabilities as well as potential and known off-target activities for the compounds of interest need to be discussed in the context of structure-activity relationships23. Finally, the biological activity of the putative probe should be confirmed by a third party, using fresh samples of the chemical in question. These types of cycles of probe testing at individual Centers, as well as at the site originating the bioassay, or “target provider”, are now commonplace.
This type of further integration between biological experiments and chemical synthesis, together with intensive data mining and in silico tools, should lead to substantial improvements in chemical probe discovery. This approach is likely to be well-complemented by some of the Challenge (RC1; http://grants.nih.gov/grants/funding/challenge_award/) and Grand Challenge (RC2; http://grants1.nih.gov/grants/guide/rfa-files/RFA-OD-09-004.html) grants, currently launched by the NIH under the American Recovery and Reinvestment Act [http://frwebgate.access.gpo.gov/cgi-bin/getdoc.cgi?dbname=111_cong_bills&docid=f:h1enr.pdf]. The NIGMS has also launched a “Grand Opportunity” (RC2) challenge related to the “Development and application of statistical and computational data analysis methods for DNA sequence, variation, GWAS, genomic function, chemical biology and related genomic data sets”, specifically for the development of “chemist- and biologist-friendly” tools, to support medicinal chemistry optimization by further improving the data analysis and integration components of screening [http://www.genome.gov/27530674#2].
What are the measures of MLI success?
Despite significant increases in R&D funds in the top 50 major pharmaceutical companies over the last decade, the number of new chemical entities is somewhere between 15 and 30 per year. A rigorous analysis24 indicates that the total cost of developing new drugs increased from $350 million per drug in 1991 to over $800 million in 2003 (normalized for US$ in 2000). Though MLI, its pilot phase budget and superficially modest productivity were subject to industrial criticismError! Bookmark not defined.3, it can be noted that small molecule discovery and innovation in both academia and the pharmaceutical sector can substantially benefit from a focus on innovative discovery science. Through this public initiative, all the results can be subjected to peer review through traditional academic mechanisms including publications and meeting presentations.
Although objective functions to assess confidence, for both chemical probes and drug leads, are not available, we offer crowdsourcing as a cross-disciplinary alternative that pools multiple levels of expertise from translational disciplines, to provide a more rigorous chemical probe evaluation process within MLPCN. We anticipate that the progressive removal of artifactual chemicals from the MLSMR, coupled with substantial improvements in integrative communication tools and data-to-knowledge transformative technologies, will result in significant improvements in chemical probe output. The 3 year MLI pilot phase MLSCN was extended by one year, running in parallel with the MLPCN, giving the original ten centers a chance to complete projects that were in process. These additional probe projects are now being vetted according to the more rigorous standards instituted for the MLPCN.
The MLI has provided a blueprint for many (planned) activities in different countries. Several national chemical library initiatives are in progress, and even a European library is currently under consideration via a consortium. As a community experiment, the MLI has not only delivered some interesting chemical probes, but also has altered the chemical biology community by providing an open-access data repository system (PubChem), a wide array of associated chemical and biological data for a large chemical library (MLSMR) and an increasingly larger number of assays. While not all chemical probes may stand the test of time, nonetheless, as the NIH program continues, we expect that academic probe discovery efforts will have an increasingly high impact in the public health sector. In the end, the MLI will be judged by the ultimate crowdsourcing experiment when scientists around the world choose whether or not to use these chemical tools in their own research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grant 1U54MH084690 (UNM). We thank Dr Evan Bolton (NIH/PubChem) for promptly answering questions related to the MLI. Dr. Brian K. Shoichet (UCSF) voted on some of these chemical probes, as reflected in Figure 1 and the Supplementary Material.
Footnotes
COMPETING INTERESTS STATEMENT Tudor Oprea is the Founder and CEO of Sunset Molecular Discovery LLC, and co-founder of Drug4Cast LLC; both companies provide services related to the pharmaceutical sector. He also serves on the Scientific Advisory Board for ChemDiv, Inc. Larry A. Sklar is a founder of IntelliCyt, a company that provides hardware and software for the HyperCyt HTS flow cytometry platform.
References
- 1.Austin CP, Brady LS, Insel TR, Collins FS. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 2. [9/04/09];The PubChem database is available online at the National Center for Biotechnology Information. Retrieved from http://pubchem.ncbi.nlm.nih.gov/
- 3.Hopkins AL, Polinsky A. Knowledge and Intelligence in Drug Design, Ann. Rep. Med. Chem. (41):425–437. [Google Scholar]
- 4.Polanyi M. Personal Knowledge: Towards a Post-Critical Philosophy. University of Chicago Press; Chicago: 1974. [Google Scholar]
- 5.Rishton GM. Drug Discov. Today. 1997;2:382–384. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- 6.Roche O, et al. J. Med. Chem. 2002;45:137–142. doi: 10.1021/jm010934d. [DOI] [PubMed] [Google Scholar]
- 7.McGovern SL, Helfand BT, Feng B, Shoichet BK. J. Med. Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 8.Seidler J, McGovern SL, Doman TN, Shoichet BK. J. Med. Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 9.Young SM, Bologa C, Oprea TI, Prossnitz ER, Sklar LA, Edwards BS. J. Biomol. Screening. 2005;10:374–382. doi: 10.1177/1087057105274532. [DOI] [PubMed] [Google Scholar]
- 10.Kramer JA, Sagartz JE, Morris DL. Nature Rev. Drug Discov. 2007;6:636–649. doi: 10.1038/nrd2378. [DOI] [PubMed] [Google Scholar]
- 11.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug. Deliv. Reviews. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 12.Keserü GM, Makara GM. Nature Rev. Drug Discov. 2009;8:203–212. doi: 10.1038/nrd2796. [DOI] [PubMed] [Google Scholar]
- 13.Norinder U, Haeberlein M. Adv. Drug. Deliv. Rev. 2002;54:291–313. doi: 10.1016/s0169-409x(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 14.Ajay, Walters WP, Murcko MA. J. Med. Chem. 1998;41:3314–3324. doi: 10.1021/jm970666c. [DOI] [PubMed] [Google Scholar]
- 15.Sadowski J, Kubinyi HJ. Med. Chem. 1998;41:3325–3329. doi: 10.1021/jm9706776. [DOI] [PubMed] [Google Scholar]
- 16.Teague SJ, Davis AM, Leeson PD, Oprea TI. Angew. Chem. Int. Ed. 1999;38:3743–3748. doi: 10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Hann MM, Leach AR, Harper G. J. Chem. Inf. Comput. Sci. 2001;41:856–64. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 18.Oprea TI, Matter H. Curr. Opin. Chem. Biol. 2004;8:349–358. doi: 10.1016/j.cbpa.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Vaz RJ, Klabunde T. Antitargets. Wiley-VCH; Weinheim: 2008. [Google Scholar]
- 20.Tetko IV, Tanchuk VY, Villa AE. J. Chem. Inf. Comput. Sci. 2001;41:1407–1421. doi: 10.1021/ci010368v. [DOI] [PubMed] [Google Scholar]
- 21.Oprea TI. J. Braz. Chem. Soc. 2002;13:811–815. [Google Scholar]
- 22.Inglese J, Shamu CE, Guy RK. Nature Chem. Biol. 2007;3:438–441. doi: 10.1038/nchembio0807-438. [DOI] [PubMed] [Google Scholar]
- 23.Rishton GM. Curr. Opin. Chem. Biol. 2008;12:1–12. doi: 10.1016/j.cbpa.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Adams CP, Brantner VV. Health Aff. 2006;25:420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- 25.Howe J. [9/04/09];Wired Magazine 14.06. 2006 Retrieved from http://www.wired.com/wired/archive/14.06/crowds.html.
- 26.Chesbrough HW. Open Innovation: The New Imperative for Creating and Profiting from Technology. Harvard Business School Press; 2003. [Google Scholar]
- 27. [Retrieved 9/04/09];Anonymous. http://en.wikipedia.org/wiki/Crowdsourcing.
- 28.Howe J. Crowdsourcing: Why the Power of the Crowd Is Driving the Future of Business. Crown Business; 2008. [Google Scholar]
- 29.Surowiecki J. The Wisdom of Crowds: Why the Many are Smarter Than the Few and How Collective Wisdom Shapes Business, Economies, Societies and Nations, Little, Brown. 2004 [Google Scholar]
- 30.Leo A. Chem. Rev. 1993;5:1281–1306. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.