Abstract
To elucidate the role of the chromatin environment in the regulation of replication origin activation, autonomously replicating sequences were inserted into identical locations in the budding yeast genome and their activation times in S phase determined. Chromatin-dependent origins adopt to the firing time of the surrounding locus. In contrast, the origins containing two binding sites for Forkhead transcription factors are activated early in the S phase regardless of their location in the genome. Our results also show that genuinely late-replicating parts of the genome can be converted into early-replicating loci by insertion of a chromatin-independent early replication origin, ARS607, whereas insertion of two Forkhead-binding sites is not sufficient for conversion.
Keywords: Cdc45, DNA replication, Forkhead transcription factors, Saccharomyces cerevisiae , replication origin timing
Introduction
Eukaryotic DNA replication is initiated from multiple origins that are defined as autonomously replicating sequences (ARSs) in budding yeast. The essential sequence motifs of replication origins are B1, B2, B3, B4 boxes and the ARS consensus sequence (ACS). While ACS, B1 and B2 elements participate in recruitment of factors needed for pre-replicative complex (pre-RC) formation, the B3 and B4 boxes are involved in regulation of origin efficiency [1, 2, 3, 4, 5, 6, 7]. In the G1 phase of cell cycle, the MCM (minichromosome maintenance) helicase complex is recruited to origins in an origin recognition complex (ORC)-dependent manner and pre-RCs are formed [8]. On entrance into S phase, pre-RCs are activated by recruitment of cofactors Cdc45 and the GINS complex to the origins [9, 10]. The following unwinding of DNA and initiation of DNA synthesis is a rapid process, as the time of Cdc45 association with an origin correlates with its firing [11, 12, 13, 14, 15]. The general activation mechanisms for individual replication origins are thought to be common. However, the activation time of replication origins in S phase is different. The mechanisms leading to the selective activation of a subset of origins to fire in early S phase and others in late S are not fully understood. It has been shown that chromatin structure in the origin’s genomic locus can regulate its activation time, and that to some extent the firing of origins can be influenced by their location within chromatin: origins in subtelomeric regions are generally late firing, whereas origins in centromeric regions are typically early firing [16, 17]. Additionally, nucleosome positioning and post-translational modifications of histones can change the activity of origins. Generally, nucleosomes are depleted from regions of replication origins [18] and the activity of origins can be inhibited by unfavourable nucleosome positioning in a Sir2-dependent manner [19]. In budding yeast, the global increase of histone acetylation in rpd3Δ cells shifts origins to fire earlier [13, 20, 21], whereas loading of Cdc45 to origins correlates with enrichment of H3 K36 monomethylation in the locus [14]. A recent study indicates that Forkhead transcription factor-dependent clustering of replication origins is required for early firing of some origins, indicating that the spatial distribution of replication origins in the cell nucleus might also influence their activation [22].
To explore how the chromosomal positioning and the consequent epigenetic environment can regulate the firing of replication origins, we relocated different ARS sequences to a common chromosomal position and determined their activation time in S phase. Reasoning that if the timing of origin activation is determined solely by chromatin context, all origins should fire at the same time in their new position, and if the timing is derived from their intrinsic DNA sequence elements, the origins should maintain their own original firing times regardless of their location in genome. The origins that fired very early in S phase in their native sites retained the early activation pattern also in the ectopic location, whereas the activation time of other origins was changed in a location-dependent manner. Detailed analysis of the early-replicating origins revealed that chromatin-independent firing of origins was mediated by two consensus binding sites for Fkh1/2 proteins, confirming the role of Forkhead factors in regulation of early-firing origins [22]. Our results also show that early-firing origins can override the original replication timing pattern of the locus by converting genuinely late-replicating chromatin into early-replicating regions.
Results and Discussion
Location-dependent origins
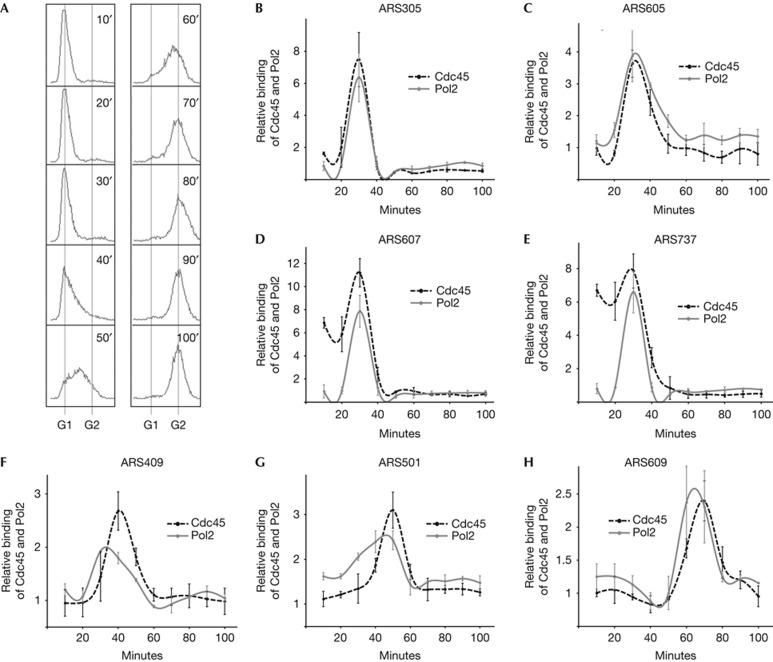
To compare the activation kinetics of replication origins simultaneously in several yeast strains, we measured the recruitment of the Cdc45 protein to the origins as an indicator of origin activation. Cdc45 is required for activation of MCM helicase and several studies have shown that association of Cdc45 with pre-RC is a highly reliable marker for origin firing [11, 12, 13, 14, 15, 20]. To follow the firing of origins, cells were arrested in the late G1 phase with α-factor, released synchronously into S phase (Fig 1A) and Cdc45 binding to the origins was determined by chromatin immunoprecipitation (ChIP) assay. Association of Cdc45 with early-firing origins peaked at 30 min after release from G1 arrest (Fig 1B–E), whereas late-firing origins were bound by Cdc45 at 40–70 min after the release (Fig 1F–H), which is in good agreement with previously reported firing times of these origins [13, 14, 15, 23]. To further validate the idea that recruitment of Cdc45 is tightly coupled to initiation of DNA synthesis, we confirmed that its binding to the origins occurs simultaneously with that of Pol2, the catalytic subunit of replicative DNA polymerase ε (Fig 1B–H). As the ChIP-signals of Cdc45 were stronger and less diffused compared with Pol2 signals on late-firing origins, we preferred to follow Cdc45 recruitment as an indicator of origin firing throughout the rest of the study.
Figure 1.
Recruitment of Cdc45 and Pol2 to replication origins. (A) DNA content analysis of AKY543 strain at indicated time-points after release from G1 arrest. (B–H) Recruitment of Cdc45 and Pol2 proteins to various replication origins throughout the S phase. The relative binding of the factors to the origins at indicated time-points are shown, 1 was defined as the average value of unbound samples. Error bars indicate s.d. of three experiments. ARS, autonomously replicating sequence.
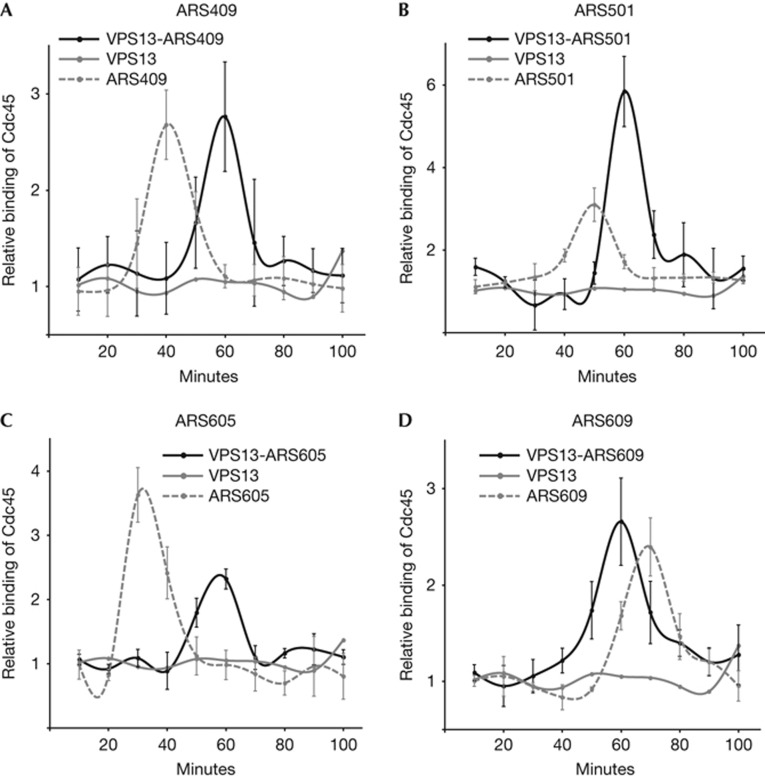
To explore whether the firing time of replication origin is determined by its primary sequence or by its location in the genome, we constructed a panel of yeast strains where seven different ARS elements were inserted into the VPS13 locus in chromosome XII. This locus contains no other DNA replication origins in a 60-kb region and is a genuinely late-replicating part of the chromosome in wild-type cells [21]. We have previously shown that DNA replication origins relocated to the VPS13 locus are efficiently loaded with pre-RC protein complexes ORC and MCM, and can initiate DNA synthesis in S phase [11]. To elucidate the role of chromatin environment in the timing of replication origin activation, the binding profiles of Cdc45 to the replication origins in their genuine locations were compared with the recruitment of Cdc45 to the identical ARS elements in the VPS13 locus. On relocation into the VPS13 locus, the dynamics of Cdc45 recruitment were changed on four replication origins. In their native locations, the peak of Cdc45 binding was observed at 30 min after G1 release on ARS605, at 40 min on ARS409, at 50 min on ARS501 and at 70 min on ARS609. However, when inserted into the VPS13 locus, the activation time of these origins was changed, suggesting that their firing was determined primarily by the genomic location rather than by the intrinsic properties of their sequences (Fig 2). When no replication origin was inserted into VPS13, no recruitment of Cdc45 to the locus was observed, confirming the functionality of inserted origins in the VPS13 locus. Furthermore, the new activation times of ARS sequences were synchronized in VPS13 indicating that regardless of their original timing, different replication origins can adopt similar firing patterns when inserted into identical chromatin environment (Fig 2). On the basis of these experiments, we conclude that the location-derived firing of replication origins in the VPS13 locus occurs approximately at 60 min after release into S phase.
Figure 2.
Synchronization of chromatin-dependent origins in the VPS13 locus. Relative binding of Cdc45 to origins is shown. (A) VPS13-ARS409 and ARS409; (B) VPS13-ARS501 and ARS501; (C) VPS13-ARS605 and ARS605; (D) VPS13-ARS609 and ARS609. The binding of Cdc45 to the origin-free VPS13 locus is shown for reference in all panels. Error bars indicate s.d. of three experiments. ARS, autonomously replicating sequence.
Location-independent origins
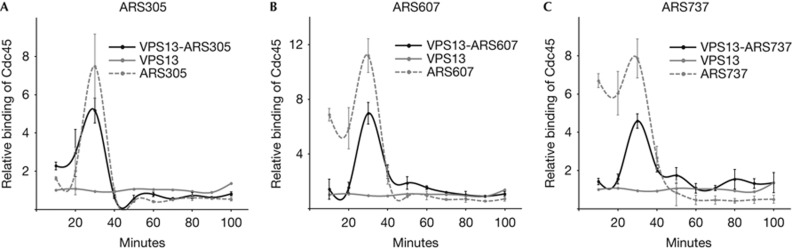
In addition to chromatin-synchronized origins, another set of replication origins emerged from the VPS13-ARS experiments. In contrast to the first set of origins, no shift in Cdc45-binding peak was detected on origins ARS305, ARS607 or ARS737 when inserted into the VPS13 locus. All these origins were early firing, both in their original and in the VPS13 loci, as the binding of Cdc45 to these origins peaked at 30 min after the release from G1 arrest (Fig 3). These results demonstrate that some early-firing origins can override the chromatin-derived control of replication timing and retain their activation pattern in new environment. However, it should be emphasized that not all early-firing origins can execute their timing programme in foreign locations, as the ARS605 shifted from early- to late-firing on relocation into the VPS13 locus (Fig 2C). Therefore, we propose that a set of early-firing ARS sequences should possess specific sequence elements that ensure their early firing even when relocated to different sites in the genome.
Figure 3.
Location-independent activation of early origins in the VPS13 locus. Relative binding of Cdc45 to origins is shown. (A) VPS13-ARS305 and ARS305; (B) VPS13-ARS607 and ARS607; (C) VPS13-ARS737 and ARS737. The binding of Cdc45 to the origin-free VPS13 locus is shown for reference in all panels. Error bars indicate s.d. of three experiments. ARS, autonomously replicating sequence.
Forkhead sites ensure early activation
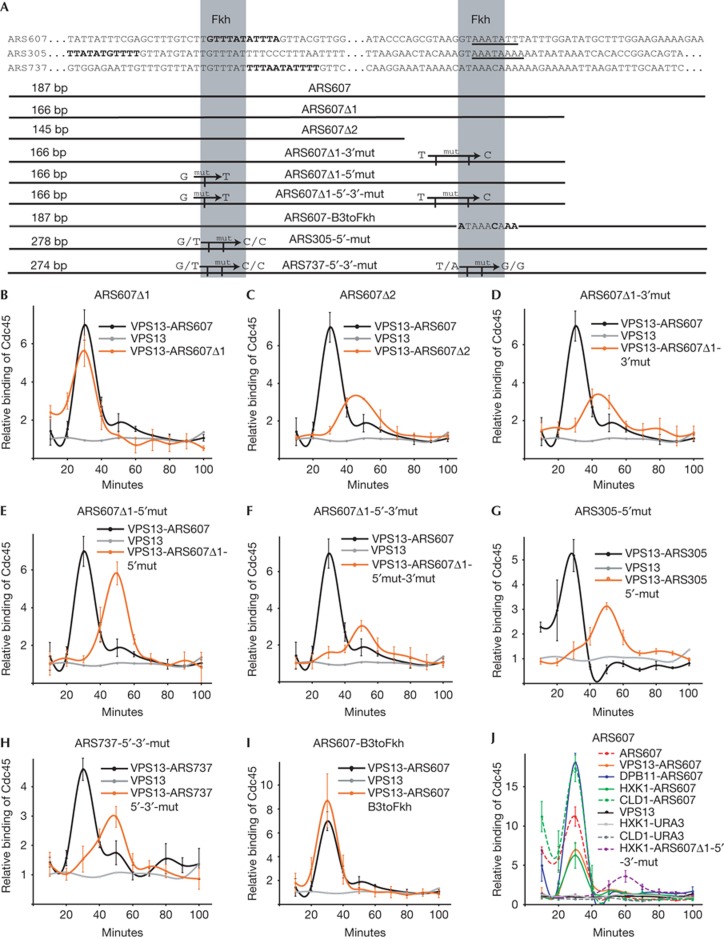
To find out the sequence elements required for chromatin-independent activation of the origin, we made truncated variants of the ARS607 sequence. In the ARS607Δ1 mutant, 21 nucleotides were removed from the 3′-end of the origin. In the second mutant, ARS607Δ2, extra 21 nucleotides were removed from ARS607Δ1 (Fig 4A). Both deletion mutants were inserted into the VPS13 locus and Cdc45 binding to the origins was monitored throughout S phase. Activation of VPS13-ARS607Δ1 was indistinguishable from the full-length ARS607, indicating that the last 21 nucleotides from the origin’s 3′-end were unnecessary for its function and regulation of its timing (Fig 4B). However, ARS607Δ2 was activated 20 min later in the VPS13 locus compared with ARS607, VPS13-ARS607 and VPS13-ARS607Δ1, implying that the deleted region was required for the early firing of ARS607 (Fig 4C). A recent study has shown that deletion of Forkhead transcription factors leads to genome-wide deregulation of origin-firing pattern in budding yeast, resulting in delayed activation of early origins and shifting many late origins to fire earlier [22]. ARS607 contains two putative binding sites for Fkh1/2 proteins, one of which is deleted in the ARS607Δ2 mutant, whereas both sites are present in the ARS607Δ1. To confirm the importance of Forkhead factors for early firing of origins, we introduced point mutations into the Fkh1/2-binding sites in ARS607, ARS305 and ARS737 (Fig 4A). When any one of the Forkhead-binding sites was mutated, the early activation of the origins was lost, suggesting that both Fhk1/2-binding sites were necessary to ensure early firing of these origins (Fig 4D–H).
Figure 4.
Activation of ARS607 in ectopic loci. (A) Schematic representation of ARS305, ARS737, ARS607 and their mutants. Deletions of ARS607 and the sites of point mutations of all origins are shown. Sequences corresponding to the Fkh1/2 consensus sequence RTAAAYA [33] are indicated as grey boxes, ACS is shown in bold, B3 and B4 boxes in ARS607 [7] and ARS305 [3] are underlined. (B–I) Relative binding of Cdc45 to indicated wild-type and mutated ARS sequences in the VPS13 locus. The binding of Cdc45 to the origin-free VPS13 is shown for reference in all panels. (J) Activation of ARS607 in different genomic loci. Relative binding of the Cdc45 protein to the ARS607 sequence inserted into VPS13, DPB11, HXK1 or CLD1 loci was determined. The binding of Cdc45 to genuine ARS607 and origin-free VPS13, HXK1 and CLD1 loci is shown for reference. Error bars indicate s.d. of three experiments. ARS, autonomously replicating sequence; mut, mutation.
Systematic analysis of the sequence elements required for efficient replication of plasmids with ARS607 or ARS305 as sole replication origins has lead to the identification of B3 and B4 boxes in ARS607 and ARS305, respectively [3, 7]. Both elements are largely overlapping with the 3′-binding sites of Fkh1/2 proteins in these origins (Fig 4A). To test whether the Fkh1/2-binding site can functionally substitute for the entire B3 box, we replaced the B3 sequence in ARS607 with a different, but functional Fkh1/2-binding site (‘ARS607-B3toFkh’; Fig 4A). This mutant also retained its early-firing profile in VPS13 locus, indicating that the Fkh1/2-binding site can substitute for the function of the B3 box (Fig 4I).
Translocation of ARS607 to other loci
As ARS607 was activated early in the VPS13 locus (Fig 3B), we tested whether it is able to retain its early firing also in other loci. When ARS607 was inserted into the DPB11 locus on chromosome X, which resides in late-replicating chromatin with no other adjacent replication origins [21], Cdc45 was recruited to the locus 30 min after the release from G1 arrest as it was in VPS13 and in its native locus (Fig 4J). As neither the VPS13 nor the DPB11 locus contain active replication origins in wild-type cells, it is possible that activation of ARS607 is not properly regulated in these loci, because some of the distal regulatory sequences might be missing in genuinely origin-free regions. To explore the activation profile of ARS607 in genomic regions that contain native replication origins, we replaced an early-firing origin ARS728 near the CLD1 locus (chromosome VII) and a late-firing ARS609 in the HXK1 locus (chromosome VI) with the ARS607 sequence. As expected, the CLD1-ARS607 locus was activated early in S phase, as both replaced ARS728 and inserted ARS607 were early-firing origins. Replication was activated early in S phase also in the HXK1 locus on replacement of late-firing ARS609 with early-firing ARS607 (Fig 4J). While the firing time of the genuine ARS609 locus was around 70 min after the release from G1 arrest (Figs 1H, 2D), the recruitment of Cdc45 was shifted 40 min forward in the HXK1-ARS607 locus and peaked at 30 min after the release from G1 (Fig 4J). Conversion of the HXK1 locus into early-replicating region was dependent on functional Fkh1/2 sites in ARS607, as the point-mutated version of ARS607 fired late in this locus (Fig 4J). Notably, the replaced ARS609 itself did not harbour any information for very late firing, as on insertion into the VPS13 locus, its activation shifted earlier and it was synchronized with the timing of other location-dependent origins in the same locus (Fig 2D). Taken together, these experiments confirm that ARS607 is activated early in the S phase regardless of its location in the genome, and that it can convert late-replicating regions into early-replicating ones. In our experimental system, this origin fires 30 min after release from the G1 arrest in all tested loci, which demonstrates that early firing of ARS607 is an intrinsic property of its sequence, enabling early initiation of DNA synthesis independently from its location in the genome.
Early origins have two Forkhead sites
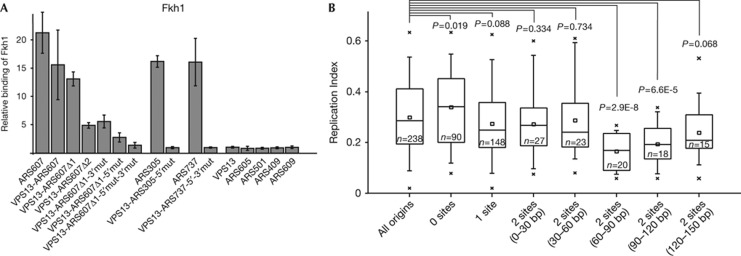
We also followed the binding of the Fkh1 protein to replication origins by ChIP assay. Fkh1 was detected in the early-replicating ARS305, ARS737, ARS607 and VPS13-ARS607 loci, but not in ARS409, ARS501, ARS605, ARS609 or origin-free VPS13 loci (Fig 5A). When either one or both Fhk1/2 sites were disrupted in ARS607, ARS305 or ARS737, the recruitment of Fkh1 to the origins was severely reduced (Fig 5A). Possibly, the binding of Forkhead proteins to replication origins must exceed some threshold level to commit the origins to early activation and two Forkhead-binding sites ensure efficient recruitment of these factors through cooperative binding to the target DNA sequence as shown by in vitro assays [24]. We also confirmed that mutations of Fkh1/2 sites in ARS607 did not abolish the recruitment of the ORC complex to the origin, indicating that Forkhead binding to ARS607 is not prerequisite for its functioning as a DNA replication origin (supplementary Fig S1 online). As the presence of two Forkhead sites was necessary for early firing of chromatin-independent origins, we also tried to convert a late-firing ARS609 to an early-firing origin by introducing two Forkhead-binding sites into its sequence, either in its native or in the VPS13 locus (supplementary Fig S2A online), but this did not change the firing pattern of ARS609 (supplementary Fig S2B,C online). However, we could not detect the binding of Fkh1 protein to these loci (supplementary Fig S2D online), indicating that solely the presence of Fkh1/2-recognition sequences in the locus is not sufficient for efficient binding of Forkhead factors. We propose that the accessibility of Fkh1/2 sites might be hindered by nucleosomes in these loci. This possibility is supported by the genome-wide nucleosome localization data indicating that the ARS609 locus is covered by nucleosomes, whereas ARS305, ARS607 and ARS737 are located in nucleosome-free regions (NFRs) [25]. Therefore, at least one of the Fkh1/2 sites inserted into the ARS609 locus is probably covered by a nucleosome, which in turn might make the site inaccessible for Forkhead factors. Earlier studies have shown that the localization of ARS1 regulatory sequences in the NFR is crucial for the function of the origin [26], and the ORC is a key factor for precise nucleosome positioning at the borders of origins [27, 28]. However, the NFRs are maintained in the ARS loci even in the absence of ORC [28], indicating that the establishment of NFR is directed by the origin itself. Therefore, we propose that the sequences of chromatin-independent origins are unfavourable for nucleosome formation, which ensures the accessibility of Fkh1/2 sites in these loci.
Figure 5.
Binding of Fkh1 to ARS loci. (A) Relative binding of Fkh1 protein to different native replication origins and mutants of ARS607, ARS305 and ARS737 in VPS13 locus was determined by ChIP assay in G1-arrested cells. ChIP signal from the origin-free VPS13 locus was set to 1. Error bars indicate s.d. of three experiments. (B) Box plots of replication indexes of budding yeast ARS sequences containing 0–2 binding sites for Forkhead factors. Origins containing two Fkh1/2 sites at various distances from each other were analysed. T-test P-values for the difference in the means between all origins and each selected group are shown. ARS, autonomously replicating sequence.
Next, we analysed whether the presence of Fkh1/2 sites in an origin sequence might be a general characteristic of early replication origins. Because of high similarity of the presence and spacing of Forkhead-binding sites in chromatin-independent origins ARS305, ARS607 and ARS737 (Fig 4A), we assumed that at least two Fkh1/2 sites are required for the early firing of the origin. We compared the average values of replication indexes [17, 29, 30] of origins containing different numbers of Forkhead-binding sites. The mean activation time of replication origins harbouring zero or one Fkh1/2 site did not differ from the activation time of all origins. Further analysis revealed that the origins containing two Forkhead sites separated from each other by 60–120 bp were almost exclusively early-firing (Fig 5B). However, the differences in origins’ firing times were not statistically significant if the distance between Fkh1/2 sites was shorter than 60 bp or longer than 120 bp (Fig 5B).
Taken together, we propose that immediate-early replication origins contain specific sequence motifs that lead to their activation in the beginning of S phase, and that these origins ensure initiation of DNA synthesis regardless of their location in genome. Central features of these motifs are two Fkh1/2-binding sites, one of which is found in close proximity to the ACS and another is located ∼60–120 bp away. We suggest that activation of other origins is determined mostly by their location in the genome, where they adopt the firing programme that is characteristic of the surrounding locus.
Methods
Yeast strains. All Saccharomyces cerevisiae strains were congenic with W303 and are described in the supplementary information online.
ChIP assay. Cells were grown in yeast extract peptone medium containing 2% glucose as a carbon source for designated periods of time before fixation for the ChIP assay. Cell cycle arrest in G1 was achieved by addition of α-factor-mating pheromone (Zymo Research) with the final concentration of 100 nM to the growth media for 3 h. For efficient release into S phase, cells were washed with water and pronase (Applichem) 50 μg/ml was added to the release media. Cell cycle status during the experiment was confirmed by flow cytometry. ChIP assays were performed as described previously [31]. Shortly, whole-cell extract from 107 cells was used for ChIP assays with 0.5 μg of 1E2 or 5E11 antibodies (Icosagen). Co-precipitated DNA was analysed by quantitative PCR (qPCR) using ABI Prism 7900HT real-time PCR system under standard conditions (40 cycles; 95 °C for 15 s and 60 °C for 1 min). Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) was used. qPCRs were done with primer pairs covering regions of VPS13, DBP11, CLD1 and HXK1 loci. Also native origins ARS605, ARS607, ARS609, ARS409, ARS305, ARS737 and ARS501 were analysed. Signals were normalized with high copy-number telomeric PAU1 gene. Sequences of primers are listed in supplementary Table S2 online.
Flow cytometry. Samples for DNA content analysis were gathered together with all ChIP samples. A volume of 0.5 ml of yeast culture was fixed in 10 ml of ice-cold 70% ethanol for at least 15 min and washed once with 50 mM citric acid. RNA was degraded with RNase A (10 μg/ml) in 50 mM citric acid overnight at 37 °C. DNA was stained with 10 × SYBR Green I (Invitrogen) in 50 mM citric acid for 30 min.
Computational analysis of ARS sequences. The ARS sequences were downloaded from the Saccharomyces Genome Database [32]. Fuzznuc program from EMBOSS suite was used for motif search. Forkhead-binding site was defined as RTAAAYA and the search was carried out on both strands. When searching for two binding sites in ARS sequences, following distances between the two sites were defined: 0–30 bp, 30–60 bp, 60–90 bp, 90–120 bp or 120–150 bp. Next the average values of replication indexes (RIs) [17, 29, 30] were assigned to origins and the RI values of each group were plotted. T-test was used to compare the difference in the means of each selected group with all origins.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank J. Sedaman and N. Avvakumov for critical reading of the manuscript. This work was supported by the Wellcome Trust International Senior Research Fellowship grant no. 081756, the European Molecular Biology Organization Installation grant no. 1454, Estonian Science Foundation grant no. 9188 and the Estonian Ministry of Education and Research grant SF0180028s12.
Author contributions: M.L. designed and performed experiments, analysed data and wrote the manuscript; K.K. and S.V. performed the experiments; A.K. designed experiments, analysed data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823 [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B (1995) The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA 92: 2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RY, Kowalski D (1996) Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucl Acids Res 24: 816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Kowalski D (1997) Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol 17: 5473–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Stillman B (2007) Cdc6 ATPase activity regulates ORC x Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem 282: 11705–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bell SP (2002) The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc Natl Acad Sci USA 99: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, May CD, Hoggard T, Miller J, Fox CA, Weinreich M (2011) High-resolution analysis of four efficient yeast replication origins reveals new insights into the ORC and putative MCM binding elements. Nucl Acids Res 39: 6523–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR (2010) Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 37: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lõoke M, Reimand J, Sedman T, Sedman J, Järvinen L, Värv S, Peil K, Kristjuhan K, Vilo J, Kristjuhan A (2010) Relicensing of transcriptionally inactivated replication origins in budding yeast. J Biol Chem 285: 40004–40011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Stout AM, Bell SP (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA 96: 9130–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Pryde F, Jain D, Kerr A, Curley R, Mariotti FR, Vogelauer M (2009) H3 K36 methylation helps determine the timing of Cdc45 association with replication origins. PloS One 4: e5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl TJ, Brewer BJ, Raghuraman MK (2012) Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet 8: e1002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E (2008) Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comp Biol 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton A, Chang F, Pappas DL, Frisch RL, Weinreich M (2008) An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol Cell 30: 156–166 [DOI] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM (2004) The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavaré S, Aparicio OM (2009) Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavaré S, Aparicio OM (2012) Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavaré S (2006) Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Pietz G, Fox CA (2001) Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev 15: 2445–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF (2009) A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT (1990) Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature 343: 387–389 [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP (2001) Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7: 21–30 [DOI] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM (2010) Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD (2007) OriDB: a DNA replication origin database. Nucl Acids Res 35: D40–D46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]
- Kristjuhan A, Walker J, Suka N, Grunstein M, Roberts D, Cairns BR, Svejstrup JQ (2002) Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol Cell 10: 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucl Acids Res 40: D700–D705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, West AG, Johnston LH, Sharrocks AD, Morgan BA (2000) The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J 19: 3750–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.