Abstract
Sulfotransferase isoform 1A1 (SULT1A1) plays a key role in the metabolism of a variety of endo- and xenobiotics and it’s activity could influence response to drugs. Our previous studies have focused on the impact of genetic variants of SULT1A1 on enzymatic activity in Caucasians and African-Americans. However, the contribution of genetic variants to SULT1A1 activity in Asians has not been explored. In this study, we investigated the collective effects of both SULT1A1 copy number variants (CNVs) and single nucleotide polymorphisms (SNPs) in the promoter region, coding region, and 3′ untranslated region on SULT1A1 activity in Japanese subjects. SNPs in the SULT1A1 promoter and 3′ untranslated region were not associated with SULT1A1 activity (P > 0.05). SULT1A1*1/2 (Arg213His) was marginally associated with SULT1A1 activity (P = 0.037). However, SULT1A1 CNVs were strongly associated with SULT1A1 activity (trend test P = 0.008) and accounted for 10% of the observed variability in activity for Japanese subjects. In conclusion, SULT1A1 CNVs play a pivotal role in determination of SULT1A1 activity in Japanese subjects, highlighting the influence of ethnic differences in SULT1A1 genetic variants on drug metabolism and therapeutic efficacy.
Keywords: CNV, genotype, phenotype, SULT1A1, single nucleotide polymorphisms
Introduction
Sulfotransferase isoform 1A1 (SULT1A1) belongs to a family of phase II detoxification enzymes that catalyze the transfer of the sulfonyl group from 3′-phosphoadenosine 5′-phosphosulfate to a variety of endogenous molecules (hormones, neurotransmit-ters, etc) and xenobiotics.1 Sulfation generally enhances the solubility and subsequent excretion of substrates, but it can also catalyze the bio-activation of various carcinogens and mutagens.2,3 The growing field of pharmacogenomics seeks to predict both efficacy and toxicity of therapeutic agents, many of which are substrates for SULT1A1. SULT1A1 metabolizes many drugs, including tamoxifen,4 fulvestrant,5 and toremefine,6 which are used for adjuvant hormonal therapy in breast cancer. Given the central role that SULT1A1 plays in drug metabolism and carcinogenesis, elucidation of the genetic determinants of SULT1A1 activity is essential in assessing the therapeutic efficacy of drugs and estimating cancer risk.
Abnormal expression and/or enzymatic function of SULT1A1 resulting from naturally occurring genetic changes, such as single nucleotide polymorphisms (SNPs) and copy number variants (CNVs), may influence drug metabolism. A common SNP in the coding region of SULT1A1 (638 G > A, SULT1A1*1/SULT1A1*2) is associated with decreased platelet enzymatic activity and thermostability.7,8 This SNP has been investigated in relation to risk of cancer in various organs and tissues in different ethnic groups, with conflicting results in some instances.9 The 638 G > A SNP has been associated with risk of lung cancer,10 colorectal cancer,11,12 urothelial cancer,13 prostate cancer,14 and breast cancer15,16 in relation to smoking status and intake of meat that is cooked well done. SULT1A1 638 G > A has also been reported to influence overall survival in tamoxifen-treated women.17,18 Ning et al19 described four common (−624 G > C, −396 A > G, −341 C > G, and −294 T > C) and one rare SNPs (−358 A > C) in the proximal promoter region and found that allele frequencies varied between Caucasians, African-Americans, and Chinese subjects. These SNPs were associated with platelet SULT1A1 enzymatic activity in Caucasians and African-Americans, but platelets for activity analyses were not available for the Chinese population. Thus, the association of the promoter SNPs with SULT1A1 activity in individuals of Asian descent was not explored. We have recently reported that 902 A > G and 973 C > T in the 3′ untranslated region (3′-UTR) and 1307 G > A in the 3′ flanking region play an important role in determination of SULT1A1 activity in Caucasians and African-Americans. These SNPs, in combination with SULT1A1 CNVs, account for 21% of variability of activity observed in Caucasians.20
In this study, DNA from 97 Japanese subjects was screened for SULT1A1 638 G > A, promoter SNPs, 3′-UTR, and CNVs. Platelets were also obtained and enzymatic activity determined, and genotype–phenotype relationships were examined. There was a significant ethnic difference in the influence of genetic variants on SULT1A1 activity, with copy number variation exhibiting the strongest influence. Hence, pharmacogenomic studies of SULT1A1 substrates should include SULT1A1 CNVs, particularly in Japanese populations.
Materials, subjects, and methods
Materials
Histopaque®-1119 and -1077, 4-nitrophenyl sulfate, and 2-naphthol were obtained from Sigma-Aldrich (St Louis, MO, USA). The 3′-phosphoadenosine 5′-phosphosulfate was obtained from the University of Dayton Chemistry Department (Dayton, OH, USA). Sequencing and polymerase chain reaction (PCR) primers were purchased from Invitrogen (Grant Island, NY, USA). All other chemicals used were of reagent grade and obtained from Fisher Scientific (Houston, TX, USA).
Study subjects
Whole blood specimens (10 mL) were obtained from 101 healthy Japanese subjects recruited at the Chiba Institute of Science. The specimens were drawn using Vacutainer™ tubes (Becton Dickinson, Franklin Lakes, NJ, USA and Fisher Scientific) containing ascorbate, citrate, and dextrose to prevent platelet aggregation. Of the 101 specimens obtained, 97 were evaluable for SULT1A1 genotype–phenotype analysis. There were 55 female and 42 male subjects (age range 22–70 years old, mean 36.4 years). The Institutional Review Board at Chiba Institute of Science approved these study protocols (Ethics Committee approval No 22-8).
Preparation of platelet cytosols and sulfotransferase activity assay
Immediately after collection, the whole blood samples were layered on a discontinuous gradient of Histopaque-1077 and Histopaque-1119, using a modification of the manufacturer’s protocol,8,21 then individual components were separated by differential centrifugation. After separation, platelets were suspended in buffer, membranes were disrupted by sonication, and the cell homogenate was subjected to ultracentrifugation at 100,000 g for 1 hour. Sulfotransferase activity was determined using a simple colorimetric procedure as described by Mulder et al22 with the modifications made by Frame et al.21 Activity was reported as nmol/min/mg protein.
DNA extraction and genotyping
DNA was extracted from lymphocytes isolated from the blood sample provided by the study participants using Qiagen extraction kits according to the manufacturer’s instructions (Valencia, CA, USA). Genotyping for SULT1A1*1/2, promoter SNPs, and 3′-UTR SNPs was performed as previously described.8,19
Copy number variation assay
SULT1A1 copy number was determined by real-time PCR in an ABI PRISM Sequence Detection System 7900 Instrument (Applied Biosystems, Foster City, CA, USA) using TaqMan Gene Expression Absolute Quantification Assay (Applied Biosystems, Foster City, CA, USA). The method has been described by Yu et al.20
Distal promoter SNP identification and genotyping
The human SULT1A1 genomic sequence (GenBank® accession no U52852) was used to design three pairs of primers (P1, P2, and P3) to produce overlapping amplicons spanning from −165 to −2513 bp for distal promoter mutation screening. After mutation screening, two common SNPs, −1975 G > C and −1135 G > A, were identified and a new pair of primers (P4) was designed for sequencing of −1975 G > C. The sequencing primer of P1 was used to identify −1135 G > A. Primer sequences are shown in Table 1. PCR was performed using JumpStart RED Taq-Reaction Mix (Sigma-Aldrich) 5 μL in a total volume of 10 μL containing 3 ng genomic DNA and 0.5 μM each of primers. PCR was amplified under the following thermal cycling conditions: after initial denaturation at 95°C for 4 minutes, the samples were subjected to 35 cycles of 94°C for 50 seconds, 64°C for 50 seconds, and 72°C for 1 minute, followed by a final extension step of 10 minutes at 72°C. Genotype was determined by direct sequencing using a CEQ™ DTCS-Quick Start Sequencing Kit and analyzed on a CEQ 8800 Genetic Analysis System (both Beckman Coulter, Brea, CA, USA).
Table 1.
Primers for genotyping single nucleotide polymorphisms in the distal promoter of sulfotransferase isoform 1A1
| Amplification primer | Sequence (5′-3′) | Orientation |
|---|---|---|
| P1 | cagtcgtggctttggagatc | Forward* |
| gtcgggctctaatgcggtg | Reverse | |
| P2 | atgttgtgtctggttggtcg | Forward* |
| gtgtgtgggcagagtgaag | Reverse | |
| P3 | ctgtggagcctccttcaaac | Forward* |
| acctgagctcttgggaacct | Reverse | |
| P4 | tgggtccgacaggttgttac | Forward* |
| ggaggctccacagacaagag | Reverse |
Note:
Forward primers were used for sequencing.
Statistical analysis
Before data analysis, SNP data were examined for possible genotyping errors by assessing deviation from the Hardy–Weinberg equilibrium. Association of SULT1A1 activity with demographic (age and sex) and genetic (SNPs and copy number) variables was examined using analysis of variance with SAS software (v 9.2; SAS institute, Cary, NC, USA). The independent effects of functional SNPs and copy number were further assessed in analysis of variance and regression models adjusting for age and sex. Individual and collective effects of SNPs and copy number on SULT1A1 activity were assessed from partial sum square, R2, β-coefficient, and P value. In the regression model, the 638 G > A homozygous variant genotype was combined with heterozygous variant genotype due to small sample size and the combined group was treated as a referent. In addition, copy numbers ≥ 4 were combined in one category in the analysis. Statistical significance was set at P < 0.05 (two-sided) and all the statistical analyses were performed using SAS software.
Results
Mutation screening and allele frequency
We screened SNPs in the distal promoter of SULT1A1 in 101 Japanese subjects and identified two common SNPs, −1975 G > C and −1135 G > A (the numbering of bases was designated relative to translation start site). We further characterized four common SNPs, −624 G > C, −396 A > G, −341 C > G, and −294 T > C, in the proximal promoter. Allele frequencies for −341 C > G were 0.5%, so we excluded this rare SNP from further analysis. We then genotyped for 902 A > G and 973 C > T in the 3′-UTR, 1307 G > A in the 3′ flanking region, and SULT1A1*1/2 in the coding region. The variant allele frequencies are shown in Table 2.
Table 2.
Univariate analysis of demographic and genetic factors in relation to platelet sulfotransferase isoform 1A1 activity (nmol/min/mg protein) among Japanese subjects
| Variable | rs number | Allele frequency | P |
|---|---|---|---|
| Age | NA | NA | 0.22 |
| Sex | NA | NA | 0.54 |
| −1975 G > C | rs9922110 | 0.366 | 0.22 |
| −1135 G > A | rs2077412 | 0.381 | 0.87 |
| −624 G > C | rs3760091 | 0.376 | 0.20 |
| −396 A > G | rs750155 | 0.366 | 0.71 |
| −294 T > C | rs4149382 | 0.490 | 0.56 |
| 902 A > G | rs6839 | 0.114 | 0.06 |
| 973 C > T | rs1042157 | 0.173 | 0.35 |
| 1307 G > A | rs4788068 | 0.198 | 0.75 |
| 638 G > A | rs9282861 | 0.109 | 0.037 |
| Copy number | NA | NA | 0.0009 |
Note:P values in bold were significantly different compared to Caucasians.
Abbreviation: NA, not applicable.
Association of SULT1A1 SNPs in the promoter, 3′-UTR, and SULT1A1*1/2 with platelet SULT1A1 enzymatic activity
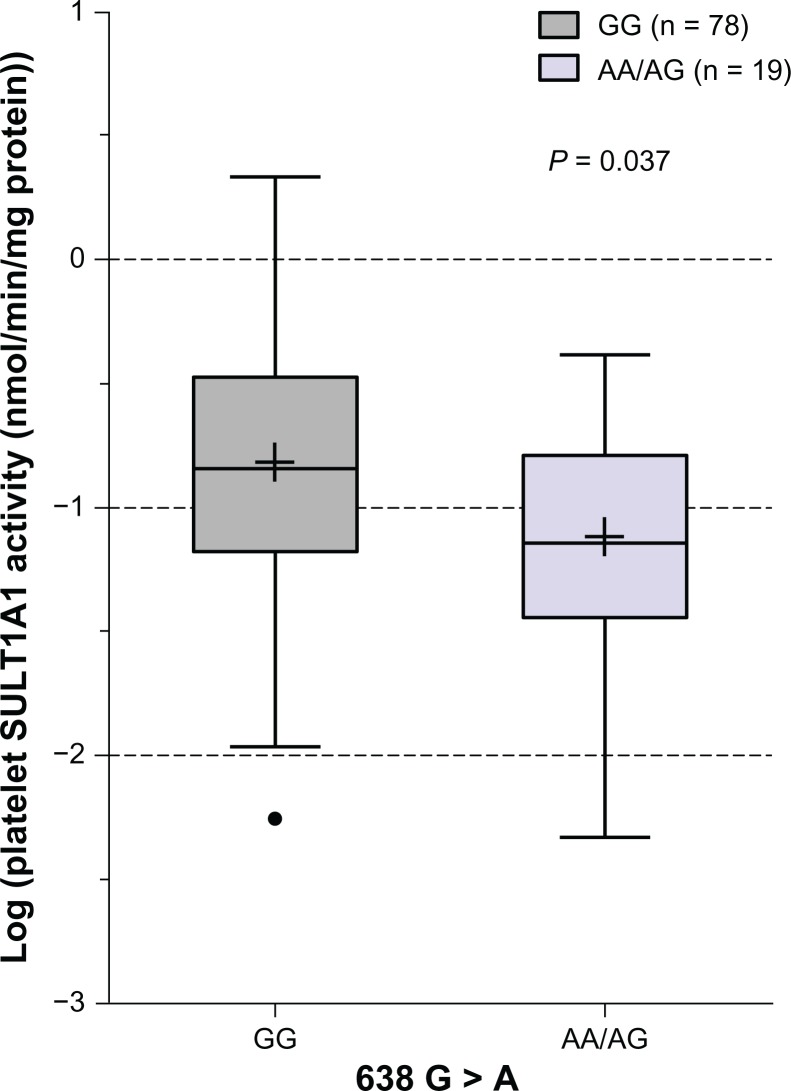
To investigate whether SULT1A13′-UTR SNPs can influence platelet SULT1A1 enzymatic activity in Japanese subjects, we examined the association of individual SNPs with SULT1A1 activity. In this population, 902 A > G, 973 C > T, and 1307 G > A were not associated with SULT1A1 activity (P > 0.05, Table 2). Similarly, neither the SNPs in the proximal promoter (−624 G > C, −396 A > G, and −294 T > C) nor SNPs in the distal promoter (−1975 G > C and −1135 G > A) were associated with SULT1A1 activity (P > 0.05, Table 2). However, SULT1A1*1/2 was marginally associated with SULT1A1 activity in Japanese subjects (P = 0.037). The SULT1A1 activity in the AA/AG group was lower than that in the GG group (Figure 1). In addition, there was no significant difference in platelet SULT1A1 enzymatic activity by sex or age in this population.
Figure 1.
Box plot analysis of the association of SULT1A1*1/2 (G638A) with platelet SULT1A1 activity in Japanese subjects.
Notes: The plus sign and line inside each box indicate mean and median and the upper and lower limits of the box are the seventy-fifth and twenty-fifth percentiles, respectively. The vertical bars above and below show the maximum and minimum values, respectively. The solid circles outside the box are outlier values. Activity is presented in log scale. P values are shown for the genotypes whose differences in mean level of activity were statistically significant.
Abbreviation: SULT1A1, sulfotransferase isoform 1A1.
Copy number variation markedly influences SULT1A1 activity in Japanese subjects
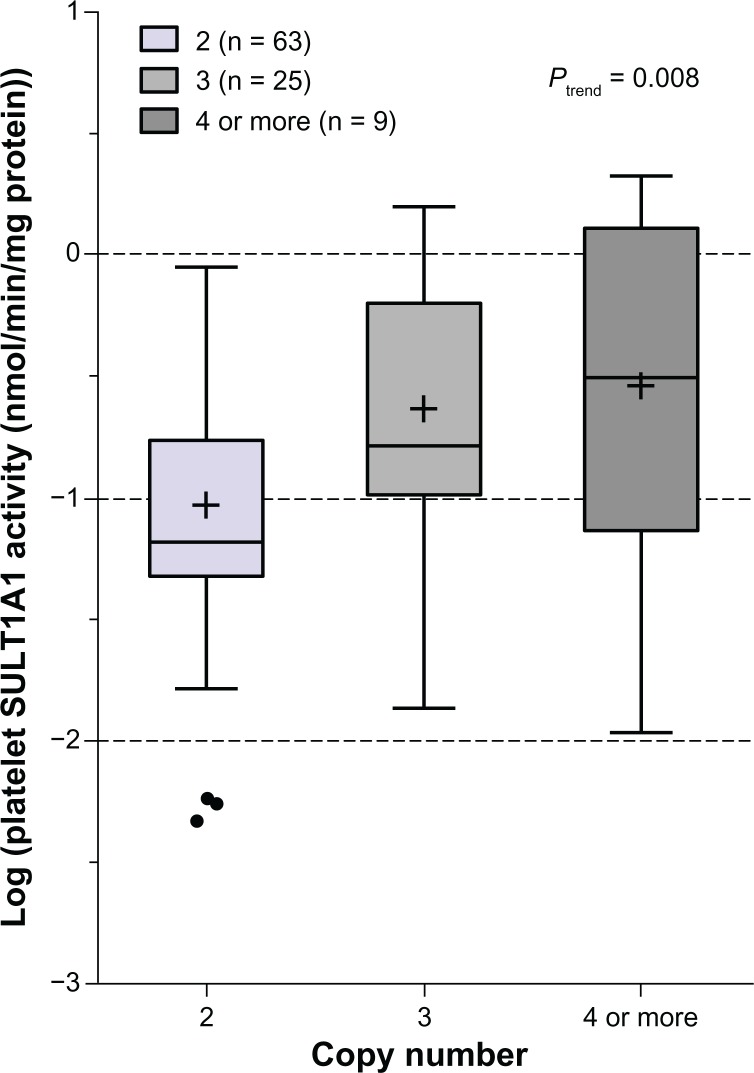
In the 97 Japanese subjects with genotype and phenotype data available, the frequency distribution of SULT1A1 CNVs 2, 3, and ≥4 was 65.0%, 25.8%, and 9.2%, respectively. Notably, SULT1A1 CNVs were significantly associated with platelet SULT1A1 activity (trend test P = 0.008, Figure 2). Univariate analysis indicated that CNVs and the SULT1A1*1 SNP were the only genetic variants tested that were significantly associated with activity in the Japanese population (P = 0.0009 and 0.037, respectively, Table 2). Pair-wise comparison indicated that SULT1A1 activity is higher in subjects carrying higher copy numbers (3 or ≥4) than those with two copies of SULT1A1 (P = 0.005 and P = 0.025, respectively). Copy number variation accounted for 10% of the observed variation in SULT1A1 activity in Japanese subjects (Table 3).
Figure 2.
Box plot analysis of the association of SULT1A1 copy number variant with platelet SULT1A1 activity in Japanese subjects (n = 97).
Notes: Platelet SULT1A1 activity was determined by colorimetrical methods described in the “Materials, subjects, and methods” section and is shown in log scale. The Ptrend test was performed for the three groups with different copies of the SULT1A1 gene. The plus sign and line inside each box indicate mean and median and the upper and lower limits of the box are the seventy-fifth and twenty-fifth percentiles, respectively. The vertical bars above and below show the maximum and minimum values, respectively. The solid circles outside the box are outlier values. Activity is presented in log scale. P values are shown for the genotypes whose differences in mean level of activity were statistically significant.
Abbreviation: SULT1A1, sulfotransferase isoform 1A1.
Table 3.
Results of multivariate analysis of variance and linear regression of platelet sulfotransferase isoform 1A1 (SULT1A1) activity (nmol/min/mg protein) with genetic predictors among Japanese subjectsa
| Variable | n (%) | Partial sum of square | Partial R2 | β coefficientb | P |
|---|---|---|---|---|---|
| 638 G > A (rs9282861) | |||||
| GG | 78 (80.4) | 1.21 | 0.17 | ||
| AA/AG | 19 (19.6) | 1.19 | 0.04 | 1.00c | |
| Copy number | |||||
| 2 | 63 (65) | 0.64 | 0.02 | ||
| 3 | 25 (25.8) | 0.91 | 0.65 | ||
| ≥4 | 9 (9.2) | 3.07 | 0.10 | 1.00c | |
| Total model SS | 29.76 | ||||
Notes:
Models with natural log of SULT1A1 activity as dependent variable and adjusted for demographics;
coefficient transformed back to original scale;
reference category. Bold text indicates the significance of the association with phenotype.
Abbreviation: SS, sum of square.
Discussion
Genetic variation in SULT1A1 is associated with functional effects on enzymatic activity, thermal stability, cellular phenotype, and protein degradation.7,23,24 Associations of genotype with phenotype have only been reported for Caucasian and African-American populations, with the magnitude of effect varying by ethnicity.8 For this reason, we sought to determine the impact of these genetic variants on enzymatic activity in an Asian population. We genotyped two SNPs in the SULT1A1 distal promoter, four SNPs in the proximal promoter, two SNPs in the 3′-UTR, one SNP in the 3′ flanking region, and SULT1A1*1/2 in the coding region. The correlation of SNPs, as well as copy number variation, with variation of SULT1A1 platelet enzymatic activity in Japanese subjects was investigated.
SULT1A1*1/2 has been reported to have frequencies of 0.332, 0.294, and 0.080 in Caucasian, African-American, and Han Chinese subjects, respectively.25 In this study, the allele frequency for SULT1A1*1/2 in Japanese subjects was 0.109, which is consistent with previous reports.13,26,27 Further, genotype–phenotype analysis indicated that SULT1A1*1/2 was only marginally associated with SULT1A1 activity in Japanese subjects, accounting for only 4% of the observed inter-individual variability.
While promoter SNPs have been demonstrated to be significantly associated with enzymatic activity in Caucasians and African-Americans,19 we found no significant associations in Japanese subjects. We further identified two common SNPs in the distal promoter; similarly, these SNPs were not associated with platelet SULT1A1 activity.
We have reported that 3′-UTR SNPs play a central role in the regulation of SULT1A1 activity in both Caucasians and African-Americans and, combined with CNV, they account for the largest percentage of variability in enzymatic activity.20 In this study, the allele frequencies of SNPs in the 3′-UTR were substantially lower than the allele frequencies in Caucasians and African-Americans and no influence on enzymatic activity was evident. Since the allele frequencies were low in Japanese subjects, it is possible that a larger study population could identify significant associations.
SULT1A1 CNVs also display ethnic differences, with 5% of Caucasian subjects possessing a single copy of the gene, 61% with two copies, and 26% with three or more copies, while 63% of African-American subjects had three or more copies.28 This study further documented that the variability in the level of the SULT1A1 enzyme in platelet and liver samples was best explained by gene copy-number differences. In the present study, 65% of the Japanese subjects had two copies of SULT1A1, which was similar to the distribution in Caucasians. Of all the genetic variants examined in the study, copy number variation has the greatest impact on SULT1A1 enzymatic activity in Japanese people, accounting for 10% of the observed inter-individual variability. Although the effects of copy number variation are statistically significant, the overall impact is small, leading to the speculation that environmental influences could be the greatest determinant of variability in SULT1A1 activity. Indeed, some dietary chemicals and environmental phenolic contaminants have been shown to be potent inhibitors of SULT1A1.29,30 Thus, studies of gene–environment interactions in determining SULT1A1 activity warrant further study in all ethnicities.
Conclusion
We found that SULT1A1 CNVs and, to a lesser extent, SULT1A1*1/2, were significantly associated with the SULT1A1 phenotype, while other genetic variants were not. The small magnitude of the contribution of these variants to inter-individual differences in phenotype in Japanese people indicates that results of pharmacogenomic and molecular epidemiological studies involving SULT1A1 genetic variants should be interpreted with caution. Studies of other genetic, epigenetic, and environmental influences on SULT1A1 activity are required to fully understand inter-individual variability in this important enzyme.
Acknowledgments
This work was supported by the National Cancer Institute (grant number R01CA128897) and Susan G Komen for the Cure (grant number BCTR0707584). TK was supported by a Grant-in-Aid for Scientific Research (C:20590156) from the Japan Society for the Promotion of Science.
Footnotes
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Jakoby WB, Ziegler DM. The enzymes of detoxication. J Biol Chem. 1990;265(34):20715–20718. [PubMed] [Google Scholar]
- 2.DeBaun JR, Miller EC, Miller JA. N-hydroxy-2-acetylaminofluorene sulfotransferase: its probable role in carcinogenesis and in protein-(methion-S-yl) binding in rat liver. Cancer Res. 1970;30(3):577–595. [PubMed] [Google Scholar]
- 3.Falany CN. Sulfation and sulfotransferases. Introduction: changing view of sulfation and the cytosolic sulfotransferases. FASEB J. 1997;11(1):1–2. doi: 10.1096/fasebj.11.1.9034159. [DOI] [PubMed] [Google Scholar]
- 4.Nishiyama T, Ogura K, Nakano H, et al. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem Pharmacol. 2002;63(10):1817–1830. doi: 10.1016/s0006-2952(02)00994-2. [DOI] [PubMed] [Google Scholar]
- 5.Edavana VK, Yu X, Dhakal IB, et al. Sulfation of fulvestrant by human liver cytosols and recombinant SULT1A1 and SULT1E1. Pharmacogenomics Pers Med. 2011;4:137–145. doi: 10.2147/PGPM.S25418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edavana VK, Dhakal IB, Yu X, Williams S, Kadlubar S. Sulfation of 4-hydroxy toremifene: individual variability, isoform specificity, and contribution to toremifene pharmacogenomics. Drug Metab Dispos. 2012;40(6):1210–1215. doi: 10.1124/dmd.111.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun. 1997;239(1):298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 8.Nowell S, Ambrosone CB, Ozawa S, et al. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000 Dec;10(9):789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Kotnis A, Kannan S, Sarin R, Mulherkar R. Case-control study and meta-analysis of SULT1A1 Arg213His polymorphism for gene, ethnicity and environment interaction for cancer risk. Br J Cancer. 2008;99(8):1340–1347. doi: 10.1038/sj.bjc.6604683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang G, Miao X, Zhou Y, Tan W, Lin D. A functional polymorphism in the SULT1A1 gene (G638A) is associated with risk of lung cancer in relation to tobacco smoking. Carcinogenesis. 2004;25(5):773–778. doi: 10.1093/carcin/bgh053. [DOI] [PubMed] [Google Scholar]
- 11.Bamber DE, Fryer AA, Strange RC, et al. Phenol sulphotransferase SULT1A1*1 genotype is associated with reduced risk of colorectal cancer. Pharmacogenetics. 2001;11(8):679–685. doi: 10.1097/00008571-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Tiemersma EW, Bunschoten A, Kok FJ, Glatt H, de Boer SY, Kampman E. Effect of SULT1A1 and NAT2 genetic polymorphism on the association between cigarette smoking and colorectal adenomas. Int J Cancer. 2004;108(1):97–103. doi: 10.1002/ijc.11533. [DOI] [PubMed] [Google Scholar]
- 13.Ozawa S, Katoh T, Inatomi H, et al. Association of genotypes of carcinogen-activating enzymes, phenol sulfotransferase SULT1A1 (ST1A3) and arylamine N-acetyltransferase NAT2, with urothelial cancer in a Japanese population. Int J Cancer. 2002;102(4):418–421. doi: 10.1002/ijc.10728. [DOI] [PubMed] [Google Scholar]
- 14.Nowell S, Ratnasinghe DL, Ambrosone CB, et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2004;13(2):270–276. doi: 10.1158/1055-9965.epi-03-0047. [DOI] [PubMed] [Google Scholar]
- 15.Tang D, Rundle A, Mooney L, et al. Sulfotransferase 1A1 (SULT1A1) polymorphism, PAH-DNA adduct levels in breast tissue and breast cancer risk in a case-control study. Breast Cancer Res Treat. 2003;78(2):217–222. doi: 10.1023/a:1022968303118. [DOI] [PubMed] [Google Scholar]
- 16.Zheng W, Xie D, Cerhan JR, Sellers TA, Wen W, Folsom AR. Sulfotransferase 1A1 polymorphism, endogenous estrogen exposure, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10(2):89–94. [PubMed] [Google Scholar]
- 17.Nowell S, Sweeney C, Winters M, et al. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94(21):1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 18.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91(3):249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 19.Ning B, Nowell S, Sweeney C, et al. Common genetic polymorphisms in the 5′-flanking region of the SULT1A1 gene: haplotypes and their association with platelet enzymatic activity. Pharmacogenet Genomics. 2005;15(7):465–473. doi: 10.1097/01.fpc.0000166823.74378.79. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Dhakal IB, Beggs M, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118(2):391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frame LT, Ozawa S, Nowell SA, et al. A simple colorimetric assay for phenotyping the major human thermostable phenol sulfotransferase (SULT1A1) using platelet cytosols. Drug Metab Dispos. 2000;28(9):1063–1068. [PubMed] [Google Scholar]
- 22.Mulder GJ, Hinson JA, Gillette JR. Generation of reactive metabolites of N-hydroxy-phenacetin by glucoronidation and sulfation. Biochem Pharmacol. 1977;26(3):189–196. doi: 10.1016/0006-2952(77)90301-x. [DOI] [PubMed] [Google Scholar]
- 23.Jones AL, Hagen M, Coughtrie MW, Roberts RC, Glatt H. Human platelet phenolsulfotransferases: cDNA cloning, stable expression in V79 cells and identification of a novel allelic variant of the phenol-sulfating form. Biochem Biophys Res Commun. 1995;208(2):855–862. doi: 10.1006/bbrc.1995.1414. [DOI] [PubMed] [Google Scholar]
- 24.Nagar S, Walther S, Blanchard RL. Sulfotransferase (SULT) 1A1 polymorphic variants *1, *2, and *3 are associated with altered enzymatic activity, cellular phenotype, and protein degradation. Mol Pharmacol. 2006;69(6):2084–2092. doi: 10.1124/mol.105.019240. [DOI] [PubMed] [Google Scholar]
- 25.Carlini EJ, Raftogianis RB, Wood TC, et al. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics. 2001;11(1):57–68. doi: 10.1097/00008571-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Ohtake E, Kakihara F, Matsumoto N, et al. Frequency distribution of phenol sulfotransferase 1A1 activity in platelet cells from healthy Japanese subjects. Eur J Pharm Sci. 2006;28(4):272–277. doi: 10.1016/j.ejps.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Ozawa S, Shimizu M, Katoh T, et al. Sulfating-activity and stability of cDNA-expressed allozymes of human phenol sulfotransferase, ST1A3*1 ((213)Arg) and ST1A3*2 ((213)His), both of which exist in Japanese as well as Caucasians. J Biochem. 1999;126(2):271–277. doi: 10.1093/oxfordjournals.jbchem.a022445. [DOI] [PubMed] [Google Scholar]
- 28.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16(5):463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- 29.Coughtrie MW, Johnston LE. Interactions between dietary chemicals and human sulfotransferases-molecular mechanisms and clinical significance. Drug Metab Dispos. 2001;29(4 Pt 2):522–528. [PubMed] [Google Scholar]
- 30.Waring RH, Ayers S, Gescher AJ, et al. Phytoestrogens and xenoestrogens: the contribution of diet and environment to endocrine disruption. J Steroid Biochem Mol Biol. 2008;108(3–5):213–220. doi: 10.1016/j.jsbmb.2007.09.007. [DOI] [PubMed] [Google Scholar]