Summary
Background:
Susceptibility weighted imaging (SWI) is a novel MRI sequence which demonstrates the susceptibility differences between adjacent tissues and it is promising to be a sequence useful in the assessment of brain tumors vascularity. The aim of our study was to demonstrate usefulness of SWI in evaluation of intratumoral vessels in comparison to CET1 sequence in a standardized, objective manner.
Material/Methods:
10 patients with supratentorial brain tumors were included in the study. All of them underwent conventional MRI examination with a 1,5 T scanner. SWI sequence was additionally performed using the following parameters: TR 49 ms,TE 40 ms. We used authors’ personal computer software – Vessels View, to assess the vessels number.
Results:
Comparison of SWI and CET1 sequences was performed using our program. Analysis of all 26 ROIs demonstrated predominance of SWI in the amount of white pixels (vessel cross-sectional) and a similar number of elongated structures (blood vessels).
Conclusions:
To conclude, the results of this study are encouraging; they confirm the added value of SWI as an appropriate and useful sequence in the process of evaluation of intratumoral vascularity. Using our program significantly improved visualization of blood vessels in cerebral tumors. The Vessel View application assists radiologists in demonstrating the vessels and facilitates distinguishing them from adjacent tissues in the image.
Keywords: susceptibility weighted imaging, SWI, brain tumors, MRI
Background
Three-dimensional susceptibility-weighted imaging (HR-SWI) with high spatial resolution is a novel, interesting MRI sequence, which combines filtered phase and magnitude images to demonstrate differences in susceptibility of adjacent tissues [1–6]. Therefore, this technique can be used in non-invasive and non-contrast-enhanced visualization of vascular structures, hemorrhages and iron depositions that are not well visible in other MRI sequences. Usefulness of SWI was confirmed in the diagnosis of wide variety of intracranial pathologies such as vascular malformations, cerebrovascular disease, trauma and neurode-generative disorders [5–8]. Furthermore, SWI is a promising sequence for tumor imaging as it could be used to characterize lesions by discriminating blood products and venous vasculature and visualizing its internal architecture. What is even more important, this sequence can be helpful in grading of brain tumors – information fundamental for determining therapy and survival, as internal vascularization correlates with tumor grade [8,9]. To date, a contrast-enhanced T1 (CET1) sequence was commonly considered to be of the highest value in assessing this feature of central nervous system (CNS) neoplasms. However, it is well known that enhancement of the brain lesion in CET1 is related not only to neoangiogenesis inside the tumor but also to blood-brain barrier disruption. That is why precise evaluation of the number of vessels in CET1 sequence can be difficult and there is interest in using the SWI sequence for differentiation between these two.
Evaluation of the number of vessels in SWI sequence may be difficult to apply in the clinical situation, as it is performed subjectively. Due to that, there are no reports available in the literature establishing one universal method of SWI interpretation. Susceptibility weighted images used to be assessed by individual radiologists in different ways and therefore, the results depended on their methods of interpretation [8–10].
The aim of our study was to demonstrate usefulness of SWI for evaluation of brain tumor vascularization in comparison to CET1 sequence in a standardized, objective way by using authors’ own computer software.
Material and Methods
The ethics committee at the Medical University of Lodz approved the procedure for this study (No RNN/22/12/KE) and it was conducted in accordance with the principles set by the Helsinki Convention. Informed consent was obtained from all participants.
A total of 10 patients (9 men and 1 woman; age: 50–81 y.o.; average: 61.8; median: 58.5) with supratentorial brain tumors were included in the study.
MRI examination
All MR examinations were performed using a 12-channel phased array head coil in a 1.5 T clinical scanner (Avanto, Siemens, Erlangen, Germany) before surgical intervention or x-ray-guided biopsy. MR imaging protocol consisted of standard T2, Flair, T1 and contrast-enhanced T1 sequences; SWI sequence was additionally performed before contrast medium administration.
Imaging parameters of SWI were as follows: TR 49ms, TE 40ms, slice thickness – 2mm, 56 slices in a single slab. Acquisition time was 3:29. Gadobutrol contrast agent (Gadovist 1,0; Bayer) was used at a standard dose of 0.1 ml/kg of body weight as a bolus. Total imaging time of all sequences was approximately 35 min.
The scanner used by the authors was a left-handed system [4]. It means that diamagnetic substances in phase images were presented as dark voxels and paramagnetic substances as bright ones. These phase images or CT scans – if available, allowed us to identify calcifications and hemorrhages. Intratumoral calcifications and hemorrhages that could cause susceptibility effects were excluded from the analysis.
Image analysis
Dedicated computer software – VesselsView, was written in order to perform quantitative analysis of tumor vessels with the use of Python – an interpreted programming language. Enthought distribution that provides a comprehensive, cross-platform environment for scientific computing was used [11].
Our software can be run on the most common operating systems including Windows (XP, Vista, 7), Linux and Mac OS. At the moment, it is a preliminary algorithm of blood vessel 2D image segmentation.
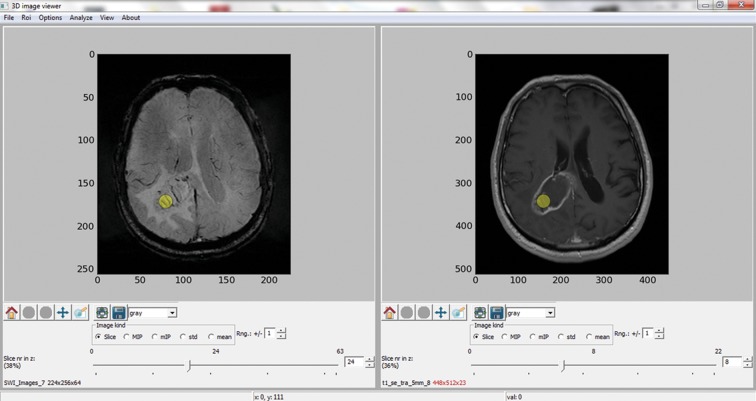
Two different data sets from SWI and CET1 sequences were loaded for each patient in order to perform the analysis. They were displayed as corresponding 2D image slices. The following steps were performed: pressing an icon FILE, choosing OPEN DATA 1 and loading the SWI data of the analyzed study; choosing OPEN DATA 2 and loading the CET1 data of the same patient. It was possible to adjust appropriate 2D image slices with the use of a slider. The next step was to manually draw a circular Region of Interest (ROI) within the tumor on selected sections. The ROI, highlighted with a yellow circle, was automatically set for all loaded images. At least one ROI (from 1 to 6) was selected within each lesion (Figure 1). Choosing an appropriate algorithm from Analyze menu: Frangi or Sato [12,13] was necessary to perform blood vessel segmentation within selected ROI., Both filtration methods were implemented in our application but the Sato segmentation was out of the scope of this paper. Vessels were segmented from surrounding parenchyma using signal intensity differences in the vessel’s neighborhood by calculating second-order derivatives at each voxel (Hessian matrix). A cross-sectional area was chosen for 2D images as a quantitative descriptor, while blood vessel volume was to be used in case of 3D data.
Figure 1.
The VesselsView software. The main panel with slelcted circular region of interest for images of different modalities – SWI sequence on the left side, CET1 on the right.
A “vessleness” function was proposed in the Frangi algorithm that maps some image parameters into probability-like estimates of vesselness according to different criteria [12].
| (1) |
where:
λ1, λ2, λ3 – eigenvalues of the hessian matrix;
α, β, c, – constants responsible for filtration sensibility;
SF – Frobenius matix norm.
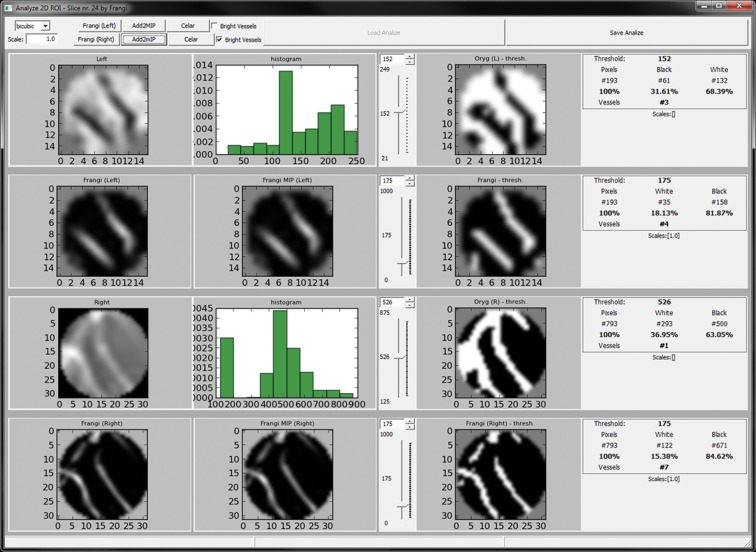
Results of these calculations were presented within seconds in a new, separate window, where the filtration outcome for the first data set (in our examination it was usually SWI) was seen in two top rows. Two bottom rows contained segmented vessels from the CET1 sequence (Figure 2). In the panel at the top of the window, the user had an option of setting some filtration parameters e.g. whether blood vessels are to appear as white on dark tissue like in SWI, or on the contrary, the tissue was to be white while blood vessel would be dark like in CET1.
Figure 2.
The VesselsView software. Analysis of the results window. Top row – original 2D image (ROI), histogram and image after segmentation. Bottom row – highlighted vessels after segmentation and statistical data – number of vessels within ROI, vessel to background ratio.
There was also a possibility to set a scale parameter to segment blood vessels of different sizes. The final image was a superposition of all images for the applied scales.
The segmentation algorithm was dedicated to highlighting elongated, tubular structures and suppressing background. It should be also mentioned that it was very sensitive to brightness variations.
An algorithm of global thresholding was applied with the use of a slider in order to estimate the quantity of blood vessels within the ROI. In the performed analysis, threshold value was set at the level of 20% of maximal brightness, which in authors’ opinion gave optimal vessel visualization.
The analysis was conducted for both, SWI and CET1 sequences in each of 26 chosen ROIs. The outcome of analysis made by the authors’ computer program, which was expressed as the ratio of white pixels to black ones, was compared for both sequences. White pixels stood for the vessels, black ones for the background. A collation between white pixels representing the cross-sectional area of vessels and the number of elongated structures demonstrating the number of blood vessels in CET1 and SWI sequences was performed afterward.
Statistical analysis
Statistical comparison between SWI and CET1 sequences on the visibility and quantity of blood vessels was made with the use of the Wilcoxon test for two samples.
Results
Results obtained after evaluation of chosen ROI’s at SWI and CET1 images with our dedicated software are presented in Table 1.
Table 1.
Results of selected ROI’s.
| Patient no. | ROI nr | SWI | CET1 | ||
|---|---|---|---|---|---|
| White px. (%) | Elongated str.- Vessels (#) | White px. (%) | Elongated str.- Vessels (#) | ||
| I | 1 | 17.10 | 4 | 7.82 | 7 |
| I | 2 | 21.14 | 10 | 15.41 | 13 |
| II | 3 | 22.80 | 8 | 14.06 | 8 |
| II | 4 | 15.54 | 5 | 11.35 | 4 |
| II | 5 | 25.39 | 5 | 15.75 | 13 |
| II | 6 | 25.39 | 6 | 8.32 | 8 |
| III | 7 | 21.24 | 8 | 20.47 | 7 |
| III | 8 | 13.47 | 8 | 12.61 | 6 |
| IV | 9 | 18.13 | 4 | 15.38 | 7 |
| IV | 10 | 17.62 | 4 | 8.21 | 4 |
| IV | 11 | 15.03 | 6 | 10.80 | 8 |
| IV | 12 | 15.03 | 3 | 1.13 | 1 |
| IV | 13 | 15.54 | 2 | 7.99 | 3 |
| IV | 14 | 18.65 | 7 | 14.88 | 4 |
| V | 15 | 10.36 | 2 | 1.13 | 2 |
| VI | 16 | 26.42 | 8 | 16.65 | 12 |
| VI | 17 | 31.09 | 6 | 22.83 | 6 |
| VII | 18 | 16.06 | 9 | 9.08 | 7 |
| VII | 19 | 15.54 | 9 | 13.49 | 12 |
| VII | 20 | 18.13 | 10 | 5.85 | 8 |
| VII | 21 | 22.80 | 10 | 5.17 | 7 |
| VII | 22 | 26.42 | 13 | 11.70 | 6 |
| VIII | 23 | 22.80 | 8 | 7.57 | 6 |
| VIII | 24 | 27.46 | 10 | 6.81 | 3 |
| IX | 25 | 27.46 | 7 | 8.55 | 10 |
| X | 26 | 23.83 | 10 | 16.31 | 13 |
White px. –white pixels; elongated str. – elongated structures.
Comparison of the ratio of white to black pixels obtained in both sequences revealed a statistically significant difference in this aspect (p<0.001). Average proportion of white pixels for SWI and CET1 were 25.44±8.23% and 14.14±7.4% respectively. The ratio was higher in SWI sequence (Table 2).
Table 2.
Comparison of white-to-black pixel ratios (percentages) obtained in SWI and CET1 sequences.
| Sequence | Calculated ratio of white to black pixels (%) | |||||
|---|---|---|---|---|---|---|
| Min. | Max. | x | Me | SD | v(%) | |
| SWI | 11.56 | 44.03 | 25.44 | 24.95 | 8.23 | 32.3 |
| CET1 | 1.14 | 26.68 | 14.14 | 13.90 | 7.40 | 52.4 |
| Comparison | z=4.178; p<0.001 | |||||
Min. – minimum; Max. – maximum; x – arithmetic mean; Me – median; SD – standard deviation (measures the scatter of results); v – variation coefficient (expressed in%); z – value of the test; p – probability of error.
The collation of percentage of white pixels (cross-sectional vessel area) acquired in SWI and CET1 sequences revealed an advantage of SWI sequence (average: 19,96±5,14) over CET1 (average: 12.03±5.72) (p<0.001) (Table 3).
Table 3.
Comparison of the proportion of white pixels obtained in SWI and CET1.
| Sequence | Calculated proportion of white pixels (%) | |||||
|---|---|---|---|---|---|---|
| Min. | Max. | x | Me | SD | v(%) | |
| SWI | 10.36 | 30.57 | 19.96 | 19.95 | 5.14 | 25.8 |
| CET1 | 1.13 | 21.06 | 12.03 | 12.21 | 5.72 | 47.5 |
| Comparison | z=4.153; p<0.001 | |||||
Min. – minimum; Max. – maximum; x – arithmetic mean; Me – median; SD – standard deviation (measures the scatter of results); v – variation coefficient (expressed in%); z – value of the test; p – probability of error.
The number of elongated structures compared for SWI and CET1 indicates the advantage of SWI. However, correlation between both sequences was lesser, without statistical significance (p>0.05). The results for elongated structures in SWI and CET1 were 7.23±2.67 and 6.62±3.2 respectively (Table 4).
Table 4.
Comparison of the number of elongated structures (blood vessels) in SWI and CET1.
| Sequence | Calculated number of elongated structures (%) | |||||
|---|---|---|---|---|---|---|
| Min. | Max. | x | Me | SD | v(%) | |
| SWI | 2 | 13 | 7.23 | 8.0 | 2.67 | 37.0 |
| CET1 | 1 | 15 | 6.62 | 6.0 | 3.20 | 48.4 |
| Comparison | z=0.882; p>0.05 | |||||
Min. – minimum; Max. – maximum; x – arithmetic mean; Me – median; SD – standard deviation (measures the scatter of results); v – variation coefficient (expressed in%); z – value of the test; p – probability of error.
The variation coefficient was higher for the CET1 sequence in all statistical analyses.
Discussion
SWI is a useful neuroimaging technique, which was introduced in 1997 and originally called “high-resolution blood oxygen level – dependent venography” [14]. It was renamed into “susceptibility weighted imaging” in 2004 as it gives much more information about susceptibility effects and goes beyond venography [15]. SWI uses paramagnetic deoxyhemoblobin as intrinsic contrast agent that allows for clear visualization of vein characteristics. In comparison to other routinely used sequences, SWI provides more information in the assessment of brain tumor microvasculature. Thus, it helps to establish the precise diagnosis of brain neoplasms [14,16,17].
Available literature concerning evaluation of brain tumors using SWI sequence was analyzed [9,10,17–20] and no reports were found where the authors would propose any kind of objective method for evaluating brain tumor vascularization through SWI application. The results were based only on visual assessment by neuroradiologists. Moreover, various methods of qualification of intratumoral susceptibility signals were provided in different papers.
The method of assessment used by C. Li et al. involved calculation of hemorrhages – irregular low-signal regions and the number of small vessels – cylindrical, low signal structures that could be followed on contiguous slices [9].
M. Hori et al. performed image evaluation using three methods in order to make qualitative analysis [10]. The first one was a conventional susceptibility grade – from grade 0 defined as no focus of hypointensity in the tumor, through grade 1 – a focus of hypointensity less than 0.5 cm in the axial, largest dimension and grade 2 – 0.5–1 cm in the largest dimension to grade 3 described as focus of hypointensity grater then 1 cm. The second evaluation method was a proportion of hypointense area in the tumor; grade 0 – no focus of hypointensity, grade 1 – total area of hypointensity less than half of the tumor and grade 2 – hypointensity greater than half of the tumor in at least one SW image. The third and the last technique involved discerning a dominant structure of hypointensity; grade 0 was defined as no focus of hypointensity in the tumor, grade 1 as a focus of hypointensity indicating bleeding or hematoma (a dot-like or conglomerated dot-shaped structures), grade 2 as a focus of hypointensity indicating bleeding and vascular structure (linear or tortuous) almost equally presented in the tumor on SWI, and grade 3 as a focus of hypointensity indicating vascular structure (hemorrhage or vascular structure).
Park et al. defined intratumoral susceptibility signal intensity (ITSS) as low signal intensity areas seen within the tumor on HR-SWI and classified them into 3 categories: conglomerated dots, conglomerated fine linear structures and dots mixed with fine linear structures. Afterwards they estimated the incidence of ITSS in chosen tumors [20].
Pinker et al. also made a qualitative evaluation of presence and frequency of intralesional susceptibility effects (SusE) in HR-SWI images on the basis of visual assessment. SusE were described as either tubular or non-tubular foci of low signal intensity in SWI images [19].
Sehgal et al. made a wider analysis of brain tumors performed by three radiologists comparing SWI with other standard sequences. The authors showed that the information received from SWI can be similar or even better than the one obtained from CET1 in terms of detecting boundaries, internal architecture, identification of blood products and veins. Such assessment was also made using individual criteria [16].
Literature review suggests that there is a necessity for creating a method of objective and precise evaluation of vessel number inside brain tumor, which could indicate the degree and intensity of neoangiogenesis closely related to neoplasm aggressiveness. It would allow for a reliable determination of response to treatment: radiotherapy, chemotherapy or immunotherapy in follow-up examinations.
A special, dedicated computer program was created for such a quantitative analysis. Vessel View provides an objective calculation of white pixels, which stand for vessels in selected, analyzed images. Line patterns they formed were chosen as a second condition improving accuracy of the results.
Huge diversity of methods of segmentation of images representing blood vessels are described in the literature and can be divided into three groups. The first group contains image processing-based approaches such as Multiscale filtering, mathematical morphology filtering or evaluation of vascular tree [12,21,22]. Another group represents model-based techniques where a predefined hypersurface tries to fit to the shape of an analyzed vascular structure; among these methods one can distinguish active contour or level sets approaches [23–26]. The last group utilizes some a priori information about properties of vascular system by forming anatomical atlases [27]. The authors of this paper decided to use the method proposed by Frangi that belongs to the first group [12].
Authors referred to CET1 sequence as a method commonly considered to be of the highest value in assessing intratumoral vessels in routine clinical studies in order to determine the value of SWI in this work. Vessel View allows for comparison of CET1 and SWI images in parallel. Regarding our results, the number of white pixels and the number of elongated structures calculated for SWI exceeded those obtained for CET1. The differences of statistical significance between these two contrasts are most likely due to the relatively small size of our study group. The outcome of our research indicates the added value of SWI as sequence applicable in the process of evaluating intratumoral vascularity. SWI sequence allowed to visualize vessels more clearly than CET1 and additionally it facilitated estimation of their number. It corresponds to reports by other authors [9,10,17,19,20].
Lesser value of CET1 observed in our work is undoubtedly due to different ways of visualization of vascular structures in those two MR sequences. Current protocols include use of intravenous contrast agent to fill the vascular bed of tumors for their good visualization in T1-weighted imaging. Obtained images often manifest diffuse signal enhancement lacking proper differentiation of internal tumor structure. In CET1, enhancement might involve the whole lesion due to brain-blood barrier damage, which can impede vessels identification. The great advantage of SWI is such that disruption of brain-blood barrier is not visible, and therefore one can easily distinguish structures that represent vessels. Performed research revealed that visibility of internal tumor architecture varies between SWI and CET1. The difference is much more significant in cases were the blood-brain barrier disruption is present. Moreover, SWI would be particularly important in patients, in whom use of gadolinium contrast is contraindicated as it allows for non-contrast visualization of CNS tumor vessels.
Authors’ attempt to create an objective assessment method for blood vessel calculations was a preliminary step in CNS tumor diagnostics using SWI sequences and gave promising results. However, it should be mentioned that research revealed a downside of the Vessels View program. As it helps to analyze two-dimensional images, there is a possibility that it could calculate a single vessel more than once. Works on creating an improved version of Vessel View to analyze three-dimensional images, in which calculation of blood vessel volume would be used as a quantitative descriptor, are ongoing. The first step of such a quantitative description on the basis of 3D image analysis of simulated vascular tree and in rat brain was already provided [28].
It is also worth mentioning that SWI sequence has a limited value in diagnosis of brain tumors in patients whose intracranial lesions are localized near the skull base and air spaces, as air/tissue boundaries cause severe susceptibility artifacts. In addition, SWI is sensible to motion artifacts. Due to a possible poor general condition of patients with brain tumors, in some cases the evaluation of SWI images can be hindered.
Conclusions
To conclude, the results of this paper are encouraging; they confirm the added value of SWI as a sequence appropriate and useful in the process of evaluation of intratumoral vascularity. Including SWI as a routine sequence when examining the patients with brain tumors as a complement to standard MRI sequences should be considered.
Finally, use of our program significantly improved visualization of blood vessels in cerebral tumors. The Vessel View application serves as an assistance for radiologists, exposing vessels and facilitating their isolation from adjacent tissues in the image.
References:
- 1.Haacke EM, Mittal S, Wu Z, et al. Susceptibility-weighted imaging: technical aspects and clinical applications, Part 1. AJNR Am J Neuroradiol. 2009;30:19–30. doi: 10.3174/ajnr.A1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mittal S, Wu Z, Neelavalli J, et al. Susceptibility-weighted imaging: technical aspects and clinical applications, Part 2. Am J Neuroradiol. 2009;30:232–52. doi: 10.3174/ajnr.A1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Haacke EM. The role of voxel aspect ratio in determining vascular phase behavior in susceptibility weighted imaging. Magn Reson Imaging. 2006;24(2):155–60. doi: 10.1016/j.mri.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 4.Robinson RJ, Bhuta S. Susceptibility-weighted imaging of the brain: current utility and potential applications. J Neuroimaging. 2011;21(4):189–204. doi: 10.1111/j.1552-6569.2010.00516.x. [DOI] [PubMed] [Google Scholar]
- 5.Mittal S, Thomas B, Wu Z, et al. Novel approaches to imaging brain tumors. In: Haacke EM, Reichenbach JR, editors. Susceptibility weighted imaging in MRI Basic concepts and clinical applications. Wiley-Blackwell; 2011. pp. 151–70. [Google Scholar]
- 6.Thomas B, Somasundaram S, Thamburaj K, et al. Clinical applications of susceptibility weighted MR imaging of the brain – a pictorial review. Neuroradiol. 2008;50:105–16. doi: 10.1007/s00234-007-0316-z. [DOI] [PubMed] [Google Scholar]
- 7.Sehgal V, Delproposto Z, Haacke EM, et al. Clinical applications of neuroimaging with susceptibility-weighted imaging. J Magn Reson Imaging. 2005;22(4):439–50. doi: 10.1002/jmri.20404. [DOI] [PubMed] [Google Scholar]
- 8.Ong BC, Stuckey SL. Susceptibility weighted imaging: a pictorial review. J Med Imaging Radiat Oncol. 2010;54(5):435–49. doi: 10.1111/j.1754-9485.2010.02208.x. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Ai B, Li Y, et al. Susceptibility-weighted imaging in grading brain astrocytomas. Eur J Radiol. 2010;75(1):81–85. doi: 10.1016/j.ejrad.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Hori M, Mori H, Aoki S, et al. Three-dimensional susceptibility-weighted imaging at 3 T using various image analysis methods in the estimation of grading intracranial gliomas. Magn Reson Imaging. 2010;28(4):594–98. doi: 10.1016/j.mri.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 11.web site: www.enthought.com (accessed 28.11.2012)
- 12.Frangi AF, Niessen WJ, Vincken KL, et al. Muliscale vessel enhancement filtering. In: Wells WM III, Colchester ACF, Delp SL, editors. MICCAI, vol 1496 serie Lecture Notes in Computer Science. 1998. pp. 130–37. [Google Scholar]
- 13.Sato Y, Nakajima S, Atsumi H, et al. Three-dimensional multiscale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal. 1998;2(2):143–68. doi: 10.1016/s1361-8415(98)80009-1. [DOI] [PubMed] [Google Scholar]
- 14.Reichenbach JR, Venkatesan R, Schillinger DJ, et al. Small vessels in te human brain MR venography with deoxyhemoglobin as an intrinsic contrast agent. Radiology. 1997;204(1):272–77. doi: 10.1148/radiology.204.1.9205259. [DOI] [PubMed] [Google Scholar]
- 15.Haacke EM, Xu Y, Cheng YC, Reichenbach JR. Susceptibility weighted imaging (SWI) Magn Reson Med. 2004;52(3):612–18. doi: 10.1002/mrm.20198. [DOI] [PubMed] [Google Scholar]
- 16.Sehgal V, Delproposto Z, Haddar D, et al. Susceptibility-weighted imaging to visualize blood products and improve tumor contrast in the study of brain masses. J Magn Reson Imaging. 2006;24(1):41–51. doi: 10.1002/jmri.20598. [DOI] [PubMed] [Google Scholar]
- 17.Hori M, Ishigame K, Kabasawa H, Kumagai H, et al. Precontrast and postcontrast susceptibility-weighted imaging in the assessment of intracranial brain neoplasms at 1.5 T. Jpn J Radiol. 2010;28(4):299–304. doi: 10.1007/s11604-010-0427-z. [DOI] [PubMed] [Google Scholar]
- 18.Pinker K, Noebauer-Huhmann IM, Stavrou I, et al. High-field, high-resolution, susceptibility-weighted magnetic resonance imaging: improved image quality by addition of contrast agent and higher field strength in patients with brain tumors. Neuroradiology. 2008;50(1):9–16. doi: 10.1007/s00234-007-0298-x. [DOI] [PubMed] [Google Scholar]
- 19.Pinker K, Noebauer-Huhmann IM, Stavrou I, et al. High-resolution contrast-enhanced, susceptibility-weighted MR imaging at 3T in patients with brain tumors: correlation with positron-emission tomography and histopathologic findings. AJNR Am J Neuroradiol. 2007;28(7):1280–86. doi: 10.3174/ajnr.A0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park MJ, Kim HS, Jahng G-H, Ryu C-W, et al. Semiquantitative assessment of intratumoral susceptibility signs using non-contrast-enhanced high-field high-resolution susceptibility-weighted imaging in patients with gliomas: comparison with MR perfusion imaging. AJNR Am J Neuroradiol. 2009;30(7):1402–8. doi: 10.3174/ajnr.A1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passat N, Ronse C, Baruthio J, et al. Automatic parameterization of grey-level hit-or-miss operators for brain vessel segmentation. Proc of ICASSP. 2005;2:737–40. [Google Scholar]
- 22.Sorantin E, Halmai C, Erdohelyi B, et al. Spiral-CT-based assessment of tracheal stenoses using 3- D- skeletonization. IEEE Trans Med Imaging. 2002;21(3):263–73. doi: 10.1109/42.996344. [DOI] [PubMed] [Google Scholar]
- 23.Hu YL, Rogers WJ, Coast DA, et al. Vessel boundary extraction based on a global and local deformable physical model with variable stiffness. Magn Reson Imaging. 1998;16(8):943–51. doi: 10.1016/s0730-725x(98)00105-2. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Amini AA. Quantifying 3-D vascular structures in MRA images using hybrid PDE and geometric deformable models. IEEE Trans Med Imaging. 2004;23(10):1251–62. doi: 10.1109/TMI.2004.834612. [DOI] [PubMed] [Google Scholar]
- 25.Manniesing R, Velthuis BK, van Leeuwen MS, et al. Level set based cerebral vasculature segmentation and diameter quantification in CT angiography. Med Image Anal. 2006;10(2):200–14. doi: 10.1016/j.media.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Lorigo L, Grimson W, Eric L, et al. Codimension – two geodesic active contours for the segmentation of tubular structures. Proc Comput Vision Pattern Recognition (CVPR) 2000:444–51. [Google Scholar]
- 27.Passat N, Ronse C, Baruthio J, et al. Magnetic resonance angiography: from anatomical knowledge modeling to vessel segmentation. Med Image Anal. 2006;10(2):259–74. doi: 10.1016/j.media.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Kociński M, Klepaczko A, Materka A, et al. 3D image texture analysis of simulated and real-world vascular trees. Comput Methods Programs Biomed. 2012;107(2):140–54. doi: 10.1016/j.cmpb.2011.06.004. [DOI] [PubMed] [Google Scholar]