Abstract
Background
Candidate gene association studies for peripheral artery disease (PAD), including subclinical disease assessed with the ankle-brachial index (ABI), have been limited by the modest number of genes examined. We conducted a two stage meta-analysis of ~50,000 SNPs across ~2100 candidate genes to identify genetic variants for ABI.
Methods and results
We studied subjects of European ancestry from 8 studies (n = 21,547, 55% women, mean age 44–73 years) and African American ancestry from 5 studies (n = 7267, 60% women, mean age 41–73 years) involved in the candidate gene association resource (CARe) consortium. In each ethnic group, additive genetic models were used (with each additional copy of the minor allele corresponding to the given beta) to test each SNP for association with continuous ABI (excluding ABI > 1.40) and PAD (defined as ABI < 0.90) using linear or logistic regression with adjustment for known PAD risk factors and population stratification. We then conducted a fixed-effects inverse-variance weighted meta-analyses considering a p < 2 × 10−6 to denote statistical significance.
Results
In the European ancestry discovery meta-analyses, rs2171209 in SYTL3 (β = −0.007, p = 6.02 × 10−7) and rs290481 in TCF7L2 (β = −0.008, p = 7.01 × 10−7) were significantly associated with ABI. None of the SNP associations for PAD were significant, though a SNP in CYP2B6 (p = 4.99 × 10−5) was among the strongest associations. These 3 genes are linked to key PAD risk factors (lipoprotein(a), type 2 diabetes, and smoking behavior, respectively). We sought replication in 6 population-based and 3 clinical samples (n = 15,440) for rs290481 and rs2171209. However, in the replication stage (rs2171209, p = 0.75; rs290481, p = 0.19) and in the combined discovery and replication analysis the SNP–ABI associations were no longer significant (rs2171209, p = 1.14 × 10−3; rs290481, p = 8.88 × 10−5). In African Americans, none of the SNP associations for ABI or PAD achieved an experiment-wide level of significance.
Conclusions
Genetic determinants of ABI and PAD remain elusive. Follow-up of these preliminary findings may uncover important biology given the known gene-risk factor associations. New and more powerful approaches to PAD gene discovery are warranted.
Keywords: Ankle brachial index, Peripheral artery disease, Genetics, Candidate gene array, Meta-analysis, Ethnicity
1. Introduction
Peripheral artery disease (PAD) is associated with an increased risk for incident cardiovascular disease events and mortality [1,2]. In the reduction of atherothrombosis for continued health (REACH) registry, almost two-thirds of the individuals with PAD had concomitant clinically evident atherosclerotic disease in the cerebrovascular or coronary artery disease (CAD) territories whereas only one-quarter of the individuals with coronary disease had clinically evident atherosclerotic involvement of other arterial beds [3]. While PAD and CAD share many common risk factors, cigarette smoking and type 2 diabetes are stronger risk factors for PAD than for coronary artery disease [4]. The variable distribution of the burden of atherosclerosis across vascular beds among subjects at risk suggests that other factors exist, including possibly genetic factors, that may contribute to the predilection of atherosclerosis to develop in a given anatomic location. Currently, little is known about the genetic susceptibility to PAD but familial aggregation of PAD and heritability estimates suggest a significant genetic contribution [5–10].
The ankle brachial index (ABI) is an easy and reliable diagnostic test used to detect symptomatic as well as asymptomatic PAD [11]. A genome-wide linkage scan for ABI identified several potential candidate genes under six linkage signals in pathways of inflammation, coagulation, blood pressure regulation, and lipid metabolism [8]. A recent large genome-wide association study (GWAS) meta-analysis of European descent participants found variants in the 9p21 locus significantly associated with ABI [12]. However, there have been few candidate gene association studies for PAD, most of which have been limited by small sample size, modest number of genes examined, and lack of robust independent replication of initial findings [13]. We conducted a two stage large scale candidate gene association study of the ~2100 candidate genes included in a cardiovascular gene-centric 50K single nucleotide polymorphism (SNP) array [14] within the candidate gene association resource (CARe) consortium that included 21,547 individuals of European ancestry (1190 with ABI < 0.9) from eight cohorts and 7267 African American individuals (594 with ABI < 0.90) from six cohorts. In the second stage of the investigation, we sought to replicate our significant associations among individuals of European ancestry in 13,524 individuals from population-based cohort studies and 1916 individuals from clinically based studies. We hypothesized that this approach would lead to the identification of novel genetic variants associated with ABI and PAD (as defined by an ABI < 0.90). Furthermore, we hypothesized that some variants may influence both PAD risk factors and PAD itself, as has been observed in genome-wide studies of lipids and coronary artery disease [15,16].
2. Methods
2.1. Discovery studies: CARe consortium and additional studies
The CARe consortium (http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx) was funded by the National Heart Lung and Blood Institute (NHLBI) in 2006 to explore the association of a custom cardiovascular gene centric SNP array [14] with a broad set of cardiovascular, metabolic, and inflammatory phenotypes collected across nine longitudinal cohort studies [17]. The following CARe consortium studies contributed data to the present analysis (Tables 1A and 1B): atherosclerosis risk in communities study (ARIC, n = 9031 European Americans, n = 2853 African Americans), the Cleveland family study (CFS, n = 275 European Americans, n = 365 African Americans), the cardiovascular health study (CHS, n = 3826 European Americans, n = 722 African Americans), the Framingham heart study (FHS, n = 2701 European Americans), the Jackson heart study (JHS, n = 1734 African Americans), and the multi-ethnic study of atherosclerosis (MESA, n = 2280 European Americans, n = 1593 African Americans). The Amish study (n = 1008), the cooperative research in the region of Augsburg (KORA F3, n = 1807), and the Penn diabetes heart study (PDHS, n = 622) cohort all used the same cardiovascular gene centric SNP array and joined the discovery stage analyses conducted in European Americans.
Table 1A.
Characteristics of discovery samples at the time of ankle brachial index (ABI) measurement. European and European Americans.a
| Characteristic mean (SD) or n (%) | ARIC n = 9031 | CFS n = 275 | CHS n = 3826 | FHSb n = 2701 | MESA n = 2280 | Amish study n = 1008 | KORA F3 n = 1807 | PDHS n = 622 |
|---|---|---|---|---|---|---|---|---|
| Age (years) | 5±6 | 44±19 | 73±6 | 61±9 | 63±10 | 44±14 | 62±11 | 59±9 |
| Women (%) | 4871 (54%) | 145 (53%) | 2152 (56%) | 1453 (54%) | 1196 (52%) | 484 (48%) | 965 (53%) | 429 (69%) |
| ABI (mean) | 1.13±0.13 | 1.08±0.10 | 1.06±0.16 | 1.13±0.12 | 1.11±0.12 | 1.10±0.10 | 1.11±0.13 | 1.12±0.14 |
| PAD (ABI < 0.9) (%) | 334 (4%) | 12 (4%) | 453 (12%) | 100 (4%) | 105 (5%) | 36 (3.6%) | 92 (5%) | 40 (6.5%) |
| Hypertension (%) | 2410 (27%) | 85 (31%) | 2136 (56%) | 629 (23%) | 882 (39%) | 125 (12%) | 1046 (58%) | 361 (58%) |
| Diabetes (%) | 785 (9%) | 34 (12%) | 547 (14%) | 288 (11%) | 141 (6%) | 6 (0.6%) | 168 (9%) | 622 (100%) |
| Body mass index (kg/m2) | 27±5 | 32±9 | 26±4 | 28±5 | 28±5 | 27±5 | 28±5 | 33±6 |
| Ever smoker (%) | 5387 (60%) | 120 (44%) | 2076 (54%) | 1595 (59%) | 1274 (56%) | 239 (24%) | 929 (51%) | 361 (58%) |
| Total cholesterol (mg/dL) | 215±41 | 192±42 | 211±39 | 200±36 | 196±36 | 209±47 | 222±40 | 177±38 |
| LDL cholesterol (mg/dL) | 137±38 | 102±30 | 130±35 | 119±33 | 117±30 | 139±4 | 130±33 | 99±30 |
| HDL cholesterol (mg/dL) | 51±17 | 44±12 | 54±16 | 54±17 | 52±16 | 56±15 | 59±17 | 46±13 |
| Triglyceride (mg/dL) | 137±93 | 139±101 | 144±79 | 137±89 | 134±92 | 69±41 | 171±133 | 155±88 |
| Lipid lowering meds (%) | 297 (3%) | 40 (15%) | 73 (2%) | 559 (21%) | 414 (18%) | 32 (2%) | 247 (14%) | 386 (62%) |
| Claudication (%) | 70 (0.8%) | 3 (1%) | 79 (2%) | 45 (2%) | 9 (0.4%) | N/Ac | 67 (3.7%) | 0 (0%) |
| Prevalent CVD (%) | 0 (0%) | 14 (5%) | 879 (23%) | 110 (4%) | 0 (0%) | 28 (2.9%) | 227 (13%) | 0 (0%) |
ARIC = atherosclerosis risk in communities, CFS = Cleveland family study, CHS = cardiovascular health study, FHS = Framingham heart study, MESA = multi-ethnic study of atherosclerosis, KORA F3 = cooperative research in the region of Augsburg, PDHS = Penn diabetes heart study.
Includes both original cohort and offspring cohort.
N/A = not measured or assessed in the study.
Table 1B.
Characteristics of discovery samples at the time of ankle brachial index (ABI) measurement. African Americans.a
| Characteristic mean (SD) or n (%) | ARIC n = 2853 | CFS n = 365 | CHS n = 722 | JHS n = 1734 | MESA n = 1593 |
|---|---|---|---|---|---|
| Age (years) | 53±6 | 41±19 | 73±6 | 50±12 | 62±10 |
| Women (%) | 1798 (63%) | 208 (57%) | 454 (63%) | 1051 (61%) | 867 (54%) |
| ABI (mean) | 1.1±0.1 | 1.0±0.1 | 1.0±0.2 | 1.1±0.1 | 1.1±0.1 |
| PAD (ABI < 0.9) (%) | 148 (5%) | 45 (12%) | 153 (21%) | 105 (6%) | 143 (9%) |
| Hypertension (%) | 1578 (55%) | 155 (42%) | 520 (72%) | 970 (56%) | 959 (60%) |
| Diabetes (%) | 543 (19%) | 72 (20%) | 170 (24%) | 251 (14%) | 281 (18%) |
| Body mass index (kg/m2) | 30±6 | 33±9 | 28±6 | 32±7 | 30±6 |
| Ever smoker (%) | 1500 (53%) | 175 (48%) | 367 (51%) | 544 (31%) | 860 (54%) |
| Total cholesterol (mg/dL) | 215±45 | 183±43 | 209±39 | 197±39 | 189±36 |
| LDL cholesterol (mg/dL) | 138±43 | 97±34 | 128±36 | 126±36 | 116±33 |
| HDL cholesterol (mg/dL) | 55±18 | 44±13 | 58±16 | 51±14 | 52±15 |
| Triglyceride (mg/dL) | 113±74 | 103±63 | 116±63 | 106±89 | 105±71 |
| Lipid lowering meds (%) | 39 (1%) | 50 (14%) | 32 (4%) | 154 (9%) | 252 (16%) |
| Claudication (%) | 18 (0.6%) | 3 (0.8%) | 11 (2%) | 534 (30%) | 10 (0.6%) |
| Prevalent CVD (%) | 0 (0%) | 11 (3%) | 191 (26%) | 98 (6%) | 0 (0%) |
ARIC = atherosclerosis risk in communities, CFS = Cleveland family study, CHS = cardiovascular heart study, JHS = Jackson heart study, MESA = multi-ethnic study of atherosclerosis.
Description of each study is provided in the Supplementary Materials. For all studies, each participant self-identified as either White (European, European American) or African American and provided written informed consent. The Institutional Review Board at the parent institution for each respective study approved the study protocols.
The characteristics of the discovery study samples at the time of ABI measurement are presented in Tables 1A and 1B by ethnic group. More than half were women and the mean age ranged from 44 years (CFS) to 73 years (CHS) in samples of European ancestry, and from 41 years (CFS) to 73 years (CHS) in African Americans. The mean ABI was 1.10 and the prevalence of PAD (ABI < 0.90) varied across studies, ranging from 4% to 12% in European Americans and from 5% to 21% in African Americans. Risk factor burden appeared greater in African Americans, as demonstrated by higher prevalence of hypertension, type 2 diabetes, and obesity compared to European ancestry participants.
2.2. ABI phenotypes
The details of the ABI protocol for each study are provided in Supplementary Materials, Supplementary Methods Table 1. For each leg, the systolic blood pressure at each ankle was divided by the systolic blood pressure in the arm. In the ARIC study, ABI was measured in only one leg and one arm chosen at random. The lower of the two ABIs calculated with each leg was used for analyses with the exception of the Amish study which used the average of an individual’s two ABIs in the analyses.
We defined two PAD phenotypes for genetic association analyses. First, we used the continuous range of ABI ≤ 1.40. Next, we defined PAD as ABI < 0.90 and conducted a case (ABI < 0.9)/control comparison (ABI ≥ 0.90 and < 1.40) analysis. Participants with an ABI > 1.40 were excluded as these subjects likely had medial artery calcification and therefore the artery would not be compressible to allow for determination of pressure in the artery. Excluding participants with ABI did not truncate the distribution substantially, and ABI was still normally distributed.
2.3. Genotyping
Genotyping in the CARe cohorts and PDHS was conducted at the Broad Institute using the ITMAT-Broad-CARe (IBC) Illumina iSELECT custom array [14]. The Old Order Amish Study genotyping was also performed using the ITMAT IBC Illumina iSELECT custom array. For KORA F3 (Discovery stage) genotyping with the same array was performed in the Genome Analysis Centre, Helmholtz Zentrum München.
The IBC array was designed to capture genetic variation in loci known or postulated to be associated with cardiovascular disease, metabolic disease and inflammatory diseases [14]. Specifically, a cosmopolitan tagging approach was used to capture genetic diversity across ~2100 candidate genes [14]. Loci were primarily chosen in three groups as follows: (1) 435 loci were chosen areas with a high probability of functional significance, (2) 1349 loci were chosen as having involvement in phenotypes of interest (i.e. cardiovascular disease or cardiovascular disease-related traits such as inflammation, hemostasis, obesity, diabetes) or were well-established loci requiring a number of SNPs for coverage, and (3) 232 lower priority loci were chosen, which also included larger genes [14]. Further details on the IBC array can be found elsewhere [14]. Details of the genotyping and quality control procedures are provided in Supplementary Methods Table 2.
2.4. Statistical analysis
For each study, residuals of ABI stratified by gender and race were created from linear regression models and used as phenotypes in the association analysis; results of gender were pooled but all analyses were stratified by race. The ABI residuals were adjusted for age, clinic site for multi-site studies, principal components (participants of European ancestry) or global European ancestry (African American participants), ever smoking, type 2 diabetes, hypertension (>140/90 or use of anti-hypertensive medication), LDL cholesterol, HDL cholesterol, and body mass index (BMI). In each ethnic group, SNPs were modeled additively, and the association of each SNP with ABI was tested using linear regression. The PDHS study did not use diabetes as a covariate as all subjects were diagnosed with type 2 diabetes. For CFS and FHS, linear mixed effects (LME) models were used to account for familial correlations. Multivariable logistic regression was used to test for the association of each SNP with PAD. For CFS and FHS, generalized estimating equations (GEE) were used to account for familial correlations. The covariate adjustment for PAD was the same as used for the ABI phenotype.
A fixed effects meta-analysis with inverse-variance weighting was then conducted in PLINK V 1.0.6 [18] and Stata V 9.0 (College Station, TX) to combine the results for all studies. The association of each additional copy of the minor allele with ABI was quantified by the regression slope (β), its standard error [SE(β)] and the corresponding p-value. We calculated a meta-analysis odds ratio (OR) for each of the most significant SNP associations for PAD. The meta-analysis OR represents the increase/decrease in odds of PAD for each additional copy of the minor allele of the SNP. We also tested for heterogeneity of study-specific regression parameters using the Cochran’s Q statistic in Stata V9.0, and report the p-values for heterogeneity. Associations were considered to be significant on an experiment-wide level at a p-value ≤ 2 × 10−6 which was determined based on the estimate of the number of independent tests [19]. SNPs with MAF < 0.01 were excluded.
A gene-based test of association using the meta-analyzed p-values at the discovery stage was performed using the program Versatile Gene-Based Association Study (VEGAS) [20];http://gump.qimr.edu.au/VEGAS/). The SNPs are matched to genes using the UCSC Genome Browser hg18 assembly with the gene region defined by ±50 kb up-and downstream of the gene. The test is based on the sum of chi-square-statistics and the linkage disequilibrium (LD) of these SNPs is taken into account according to the correlation structure in the HapMap CEU samples. An empirical p-value is provided based on all SNPs, as well as based on the SNPs within the top 20% with regard to their p-value. Since roughly 2100 genes are covered by the IBC chip, a p-value < 2.4 × 10−5 is considered significant.
2.5. Replication
Given that contemporary genetics consortia and results indicate that very large replication samples are needed to successfully replicate SNPs, and that false positives are an ongoing issue in studies such as these, we attempted to replicate only the two SNPs that met experiment-wide significance for ABI in European Americans in an additional 13,524 individuals of European ancestry from six population-based studies (Copenhagen city heart study, n = 5182; genetic study of aspirin responsiveness (GeneS-TAR), n = 618; KORA F3 (independent of KORA F3 participants in the discovery sample), n = 1440; KORA F4, n = 411; national health and nutrition examination survey (NHANES), n = 2358, and prevention of renal and vascular end-stage disease (PREVEND), n = 3515) and 1916 individuals of European ancestry from clinically based samples (cardiovascular disease in intermittent claudication (CAVASIC), n = 434; genetic determinants of peripheral arterial disease (GenePAD), n = 811; and Linz peripheral arterial disease (LIPAD), n = 671). GeneSTAR provided in silico genotyping (genotyped participants with the same candidate gene chip used in the discovery cohorts) while in the remaining studies genotyped the 2 SNPs de novo using Taqman or Sequenom genotyping platforms. Description of the replication studies, ABI protocol and calculation, and participant characteristics are provided in Supplementary Methods, Supplementary Method Table 1 and Supplementary Results Table 1.
2.6. Power and sample size for discovery and replication stages
Participants of European ancestry
With a minor allele frequency (MAF) of 0.10, additive SNP modeling, and experiment-wide significance level of 2 × 10−6, for each additional copy of the risk allele, we have 80% power to detect a beta coefficient for ABI of 0.0092 and an OR for PAD of 1.45 in the discovery stage (n = 21,547). For European ancestry individuals in the replication stage for ABI (n = 15,440) using the more stringent significance level of 2 × 10−6 we have 80% power to detect a beta coefficient of 0.0106; using a less stringent level (i.e. α = 0.025 based on carrying forward 2 SNPs for replication), we have 80% power to detect a beta coefficient of 0.0058. In the discovery plus replication stage for ABI (n = 36, 987), we have 80% power to detect a beta coefficient of 0.0065 using 2 × 10−6 as the type 1 error rate.
Participants of African-American ancestry
With a minor allele frequency (MAF) of 0.10, additive SNP modeling, and experiment-wide significance level of 2 × 10−6, for each additional copy of the risk allele, we have 80% power to detect a beta coefficient for ABI of 0.0155 and an OR for PAD of 1.67 in the sample of 7267 individuals in the discovery stage. No replication sample was available for this ethnicity.
3. Results
3.1. European ancestry studies: meta-analysis of ABI and PAD
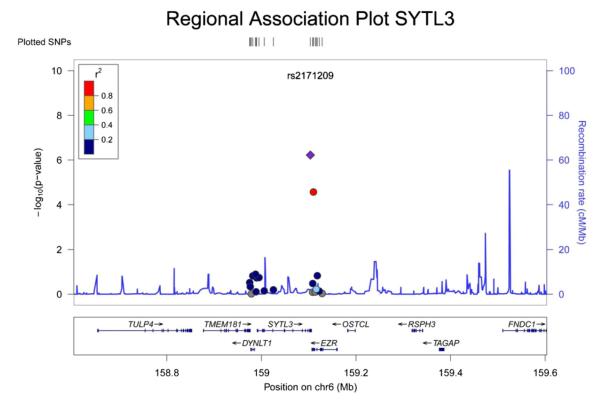
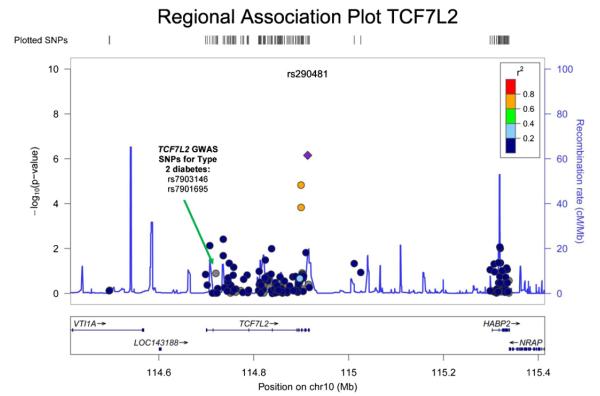
In European ancestry discovery samples, two SNPs were significantly associated with ABI (Table 2A, Figs. 1 and 2): each additional copy of the minor allele of rs2171209 in SYTL3 was associated with a 0.007 lower ABI (95% CI −0.010, 0.004, p = 6.02 × 10−7, p for heterogeneity = 0.55) and each additional copy of the minor allele of rs290481 in TCF7L2 was associated with a 0.008 lower ABI (95% CI −0.011, −0.005, p = 7.01 × 10−7, p for heterogeneity = 0.08). Rs290481 is located in intron 14 within the 3′ region of the TCF7L2 gene on chromosome 10 is distinct from a cluster of SNPs in the 5′ region of TCF7L2 (represented by rs7903146), previously reported to be associated with type 2 diabetes in genome-wide association studies (r2 = 0.001 between rs290481 and rs7903146) [21]. Among those of European ancestry, rs2171209 was not significantly associated with the categorical PAD diagnosis made by the ABI threshold of 0.90 – each additional copy of the minor allele was associated with just a 1.09-fold greater odds of PAD (95% CI: 0.97, 1.22, p = 0.14); however, rs290481 was associated with PAD (OR = 1.20, 95% CI: 1.06, 1.35, p = 0.004), although this association did not meet the experiment-wide significance level. There was no association between rs290481 and ABI (β = 0.001 95% CI: −0.006, 0.005, p = 0.80) or rs2171209 and ABI (β =−0.002 95% CI −0.009, 0.004 p = 0.46) detected in African Americans. One additional SNP was nominally associated with ABI in individuals of European ancestry at p < 10−4 (Table 2A). The most significant SNP associations for ABI in models minimally adjusted for age, sex, and study site only remained similar to the fully adjusted models. When we removed all participants with type 2 diabetes from the analysis of the CARe studies (ARIC, CFS, CHS, FHS and MESA), the beta coefficients were essentially unchanged from those in Table 2A. Similarly, when we removed participants less than 60 years of age from the analysis in the CARe studies, beta coefficients were essentially unchanged from those in Table 2A.
Table 2A.
Candidate gene SNP associations for ankle-brachial index: discovery meta-analysis, p < 10−4. European and European Americans, n = 21,547.
| SNP | Chr | Physical position | Nearest gene | Feature | Major/minor allele |
MAFa | Beta | 95% CI | p Value | phetb |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2171209 | 6 | 159,103,550 | SYTL3 | Intron | C/T | 0.22 | −0.007 | −0.010,−0.004 | 6.02×10−7 | 0.55 |
| rs290481 | 10 | 114,913,815 | TCF7L2 | Intron | C/T | 0.17 | −0.008 | −0.011,−0.005 | 7.01×10−7 | 0.08 |
| rs11061318 | 12 | 130,135,058 | GPR133 | Missense Ser → Leu | C/T | 0.03 | −0.016− | −0.023,−0.009 | 4.46×10−6 | 0.91 |
MAF = minor allele frequency.
p-for-heterogeneity by Cochran’s Q.
Fig. 1.
This plot shows the p-values for rs2171209, as well as for SNPs in the region of rs2171209, with ABI in a meta-analysis of the discovery studies. The x-axis shows chromosomal location in Mb (chromosome 10), as well as genes residing in this region. The y-axis on the left displays the −log 10(p-value) for each SNP, and the y-axis on the right shows the recombination rate in this region. The top SNP, rs2171209 is represented as a purple diamond, while supporting SNPs and other SNPs in the area are color-coded by linkage disequilibrium with rs2171209 (see r2 linkage disequilibrium legend on the plot). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 2.
This plot shows the p-values for rs290481, as well as for SNPs in the region of rs290481, with ABI in a meta-analysis of the discovery studies. The x-axis shows chromosomal location in Mb (chromosome 10), as well as genes residing in this region. The y-axis on the left displays the −log 10(p-value) for each SNP, and the y-axis on the right shows the recombination rate in this region. The top SNP, rs290481 is represented as a purple diamond, while supporting SNPs and other SNPs in the area are color-coded by linkage disequilibrium with rs290481 (see r2 linkage disequilibrium legend on the plot). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Because rs290481 is located in a gene previously strongly associated with type 2 diabetes, we conducted a test for interaction of this genotype with type 2 diabetes for ABI within the CARe discovery cohorts (ARIC, CFS, CHS, FHS, MESA). The test for interaction was performed on an additive scale using linear regression within each CARe cohort, and then combining the results in the diabetes and no diabetes strata using fixed effect inverse-variance weighting meta-analysis. The magnitude of the effect with ABI was greater in diabetics (p for interaction 0.04, Supplementary Results Table 2, with each copy of the minor allele in participants with diabetes conferring a lower level of ABI (n = 1896, β = −0.03, p < 0.001) compared to participants without diabetes (n = 16,685, β = −0.007, p = 0.007).
The gene-based analysis using VEGAS at the discovery stage did not reveal any different significantly associated genes with continuous ABI from our individual SNP analysis (Supplementary Results Table 3). SYTL3 and TCF7L2 were among the most significant genes for both analyses using all SNPs and the top 20% of SNPs (SYTL3: p(all SNPs) = 0.00216, p(top20% of SNPs) = 0.00007; TCF7L2: p(all SNPs) = 0.03589 p(top20% of SNPs) = 0.00161).
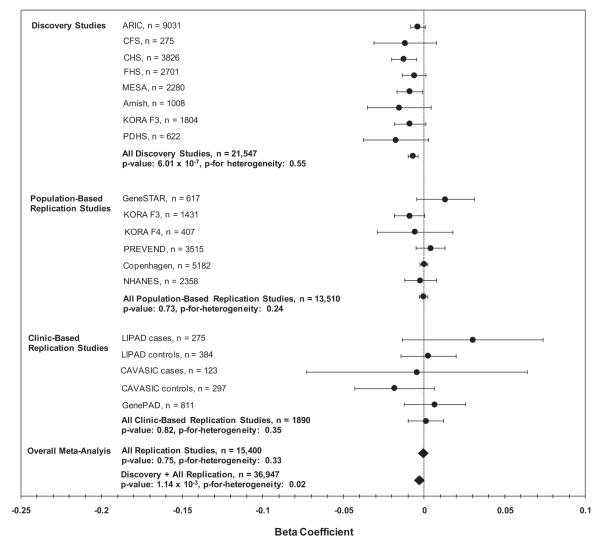
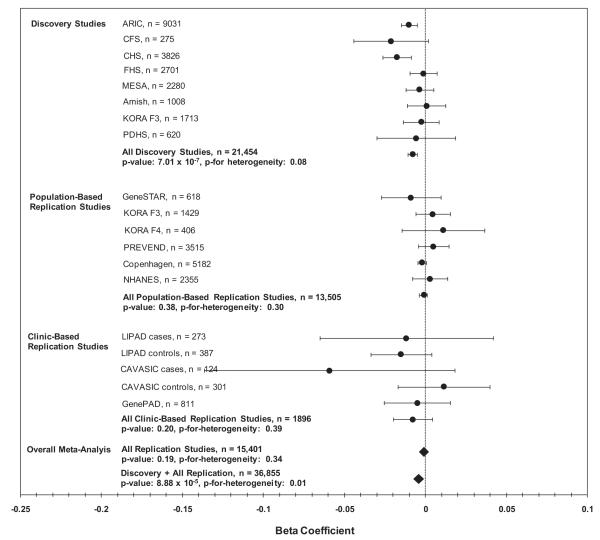
In the replication stage, the association between ABI and rs2171209 in SYTL3 was not significant in the population-based (β = −0.0004, n = 13,510, p = 0.73) and clinically based replication samples (β = 0.001, n = 1890, p = 0.82) (Fig. 3). Consequently, in the combined discovery plus replication meta-analysis the association was no longer significant (n = 36,947, β −0.003, p = 1.14 × 10−3) (Fig. 3). rs290481 in TCF7L2 also failed to replicate in the population-based replication studies (β = −0.001, n = 13,505, p = 0.38) and in the clinically based replication studies (β = 0.008, n = 1896, p = 0.20) (Fig. 4). In the combined discovery and replication meta-analysis the association between rs290481 and ABI no longer met experiment-wide significance (n = 36,855, β 0.004, p = 8.88 × 10−5) (Fig. 4).
Fig. 3.
This plot shows the association of rs2171209 with ABI for each of the discovery and replication studies, and then results with these studies combined by meta-analysis. The x-axis contains beta coefficients for the association of rs2171209 with ABI in the discovery and replication studies, and the y-axis shows studies or groups of studies. Circles represent the beta coefficient (except for overall replication and discovery + replication meta-analysis results, where beta coefficients are designated by a diamond), and error bars are 95% confidence intervals. p-Values for heterogeneity are by Cochran’s Q. Study abbreviations are as follows: ARIC = atherosclerosis risk in communities, CFS = Cleveland family study, CHS = cardiovascular health study, FHS = Framingham heart study, MESA = multi-ethnic study of atherosclerosis, KORA F3 and KORA F4 = cooperative research in the region of Augsburg, PDHS = Penn diabetes heart study, GeneSTAR = genetic study of aspirin responsiveness, PREVEND = prevention of renal and vascular end-stage disease, Copenhagen = Copenhagen city heart study, NHANES = national health and nutrition examination survey, LIPAD = Linz peripheral arterial disease, CAVASIC = cardiovascular disease in intermittent claudication, and GenePAD = genetic determinants of peripheral arterial disease.
Fig. 4.
This plot shows the association of rs290481 with ABI for each of the discovery and replication studies, and then results with these studies combined by meta-analysis. The x-axis contains beta coefficients for the association of rs290481 with ABI in the discovery and replication studies, and the y-axis shows studies or groups of studies. Circles represent the beta coefficient (except for overall replication and discovery + replication meta-analysis results, where beta coefficients are designated by a diamond), and error bars are 95% confidence intervals. p-Values for heterogeneity are by Cochran’s Q. Study abbreviations are as follows: ARIC = atherosclerosis risk in communities, CFS = Cleveland family study, CHS = cardiovascular health study, FHS = Framingham heart study, MESA = multi-ethnic study of atherosclerosis, KORA F3 and KORA F4 = cooperative research in the region of Augsburg, PDHS = Penn diabetes heart study, GeneSTAR = genetic study of aspirin responsiveness, PREVEND = prevention of renal and vascular end-stage disease, Copenhagen = Copenhagen city heart study, NHANES = national health and nutrition examination survey, LIPAD = Linz peripheral arterial disease, CAVASIC = cardiovascular disease in intermittent claudication, and GenePAD = genetic determinants of peripheral arterial disease.
None of the SNP associations in individuals of European ancestry achieved experiment-wide significance for PAD (Table 2B). One of the most significant associations for PAD was in a coding, non-synonymous SNP rs3745274 on chromosome 19 in CYP2B6 (OR 1.24, p = 4.99 × 10−5).
Table 2B.
Candidate gene SNP associations for PAD (ABI < 0.9): discovery meta-analysis, p < 10−4. European and European Americans, n = 20,539.
| SNP | Chr | Physical position | Nearest gene | Feature | Major/minor allele |
MAFa | Odds ratio | 95% CI | p Value | phetb |
|---|---|---|---|---|---|---|---|---|---|---|
| rs11088283 | 21 | 34,745,649 | KCNE1 | Intron | A/G | 0.47 | 0.85 | (0.78, 0.93) | 4.88×10−5 | 0.09 |
| rs3745274 | 19 | 46,204,681 | CYP2B6 | Coding, non-synonymous | G/T | 0.24 | 1.24 | (1.12, 1.38) | 4.99×10−5 | 0.01 |
| rs12428227 | 13 | 109,700,293 | COL4A1 | Intron | A/G | 0.17 | 1.23 | (1.10, 1.38) | 5.20×10−5 | 0.14 |
| rs17151901 | 8 | 10,290,865 | MSRA | Intron | C/T | 0.02 | 1.36 | (1.09, 1.69) | 6.53×10−5 | 0.01 |
MAF = minor allele frequency.
p-for-heterogeneity by Cochran’s Q.
3.2. African-Americans: meta-analysis of ABI and PAD
In African Americans none of the SNP associations with ABI and PAD were statistically significant (Tables 3A and 3B). The strongest association for ABI was rs2243100 on chromosome 17 in SLC25A11 (β 0.011 95% CI 0.006, 0.017, p = 5 × 10−5) and for PAD was rs4987756 on chromosome 18 in BCL2 (OR 2.99, 95% CI 1.88, 4.76, p = 3.78 × 10−6).
Table 3A.
Candidate gene SNP associations for ankle-brachial index: discovery meta-analysis, p < 10−4. African Americans, n = 7267.
| SNP | Chr | Physical position | Nearest gene | Feature | Major/minor allele | MAFa | Beta | 95% CI | p Value | phetb |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2243100 | 17 | 4,779,777 | SCL25A11 | Locus-region | C/T | 0.20 | 0.011 | 0.006, 0.017 | 5.00×10−5 | 0.32 |
| rs2243093 | 17 | 4,776,675 | GP1BA | 5′ UTR | T/C | 0.22 | 0.012 | 0.007, 0.017 | 5.94×10−5 | 0.42 |
| rs2660896 | 12 | 94,947,913 | LTA4H | Intron | C/T | 0.25 | −0.010 | −0.015,−0.005 | 7.94×10−5 | 0.96 |
| rs2242406 | 16 | 74,131,531 | CHST5 | Intron | C/T | 0.01 | −0.034 | −0.051,−0.017 | 9.46×10−5 | 0.18 |
MAF = minor allele frequency.
p-for-heterogeneity by Cochran’s Q.
Table 3B.
Candidate gene SNP associations for PAD (ABI < 0.9): discovery meta-analysis, p < 10−4. African Americans, n = 7267.
| SNP | Chr | Physical position | Nearest gene | Feature | Major/minor allele | MAFa | Odds ratio | 95% CI | p Value | phetb |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4987756 | 18 | 59,060,091 | BCL2 | Intron | A/G | 0.01 | 2.99 | 1.88, 4.76 | 3.78×10−6 | 0.63 |
| rs1256143 | 14 | 63,981,380 | MTHFD1 | Intron | C/T | 0.19 | 1.72 | 1.21, 1.66 | 1.43×10−5 | 0.05 |
| rs13004470 | 2 | 242,159,756 | BOK | Intron | C/T | 0.18 | 1.39 | 1.19, 1.62 | 4.69×10−5 | 0.81 |
| rs9830448 | 3 | 154,349,978 | RAPB2 | Locus region | C/A | 0.07 | 1.58 | 1.28, 2.02 | 4.79×10−5 | 0.73 |
MAF = minor allele frequency.
p-for-heterogeneity by Cochran’s Q.
4. Discussion
We conducted a large candidate gene association study of ~2100 cardiovascular candidate genes for ABI and PAD in over 21,000 individuals of European ancestry and over 7000 African Americans. In individuals of European ancestry, a SNP in the TCF7L2 gene (rs290481) and a SNP in the SYTL3 gene (rs2171209) were significantly associated in the discovery stage with variation in ABI measurements and a suggestive association was identified in a SNP in CYP2B6 for PAD. These findings are intriguing as the genes are linked to key PAD risk factors. TCF7L2 is the strongest genetic risk factor for susceptibility to type 2 diabetes [22–25] and CYP2B6 affects smoking behavior [26] and thus may be important in tobacco-related diseases such as PAD. However, we were unable to replicate the SNP–ABI associations in additional samples from population-based studies or clinically based samples. Furthermore, the associations were not detected in African Americans. We did not observe any significant associations for ABI or PAD in African Americans, possibly due to the relatively small sample size limiting our power to detect associations.
4.1. In the context of the current literature
Genetic factors leading to susceptibility to PAD remain largely unknown but are likely to be attributed to variants in many genes, each with small effects [13] or possibly from rare variants (minor allele frequency < 1%) with larger effects. While many of these variants may lead to risk for PAD through effects on established risk factors or shared effects with CAD and other atherosclerotic diseases [27], other variants may uniquely influence development of arterial disease in the lower extremities. Although our findings after the discovery stage did not bear out in the replication samples, the two genes are interesting candidates for ABI in light of the current literature and deserve some discussion. Genome-wide association studies of individuals of European ancestry have consistently reported an association between genetic variants of TCF7L2 and type 2 diabetes that has been confirmed in Japanese and African American samples [19,22–24,28–30]. However, our SNP in TCF7L2, rs290481 was not significantly associated with type 2 diabetes in large scale association analysis [21]. The replicated index SNP in TCF7L2 associated with type 2 diabetes is rs7902146. Further, rs290481 is not in linkage disequilibrium with rs7902146 (r2 = 0.001) [21]. TCF7L2 encodes a high mobility group (HMG) box-containing transcription factor that is involved in the Wnt signaling pathway [31] and is associated with impaired beta cell function, impaired insulin secretion and increased hepatic glucose production. Therefore, the TCF7L2 association in our discovery cohorts that presented even after adjusting for type 2 diabetes might deserve further attention in functional studies to elucidate its role in atherosclerosis.
The association between SYTL3 and ABI may be mediated by lipoprotein(a) (Lp(a)). A genome-wide association study in a small founder population of 386 Hutterites identified an association between the extended LPA gene region on chromosome 6q26–q27 including SYTL3 SNPs with Lp(a) levels[32]. Genetic variation within the LPA gene region including a very common copy number variation and other genetic variants explain up to 90% of Lp(a) concentrations[33]. Since Lp(a) concentrations and genetic variants within the LPA region are a strong risk factor for cardiovascular disease[34], it might well be that a SNP in the SYTL3 gene reflects a signal from LPA. Polymorphisms within the LPA gene region were associated with PAD in a past study [35]. Lp(a) may be an independent risk factor for PAD [36] but results are conflicting [37,38].
4.2. Strengths and limitations
To our knowledge this study is the largest candidate gene association study concerning ABI conducted in both individuals of European ancestry and African Americans and includes the most extensive number of candidate genes investigated. In the CARe consortium, imputed GWAS data is available on the African-American participants; however, given our relatively modest African-American sample size, we chose to perform analysis of the IBC chip first. We have also chosen the IBC chip because it was specifically designed as a large scale cardiovascular-centric candidate gene array, and the genetic variants on the chip were informed by GWAS for vascular and inflammatory diseases as well as expression QTLs for atherosclerosis.
Several limitations of our candidate gene meta-analysis merit comment: (i) The ankle-brachial blood pressure measurement protocols used in the studies were heterogeneous. Hence, phenotype heterogeneity may have impacted our ability to detect associations. (ii) The ARIC study contributed over 40% of the European ancestry sample and measured ABI in only one leg which may have led to phenotype misclassification most problematic for the PAD phenotype. The mean ABI did differ significantly between European Americans in ARIC and European Americans from the other CARe cohorts (all p < 0.05) with mean ± SD of the ARIC ABI 1.12 ± 0.13, CHS ABI 1.06 ± 0.15, CFS ABI 1.08 ± 0.10, FHS ABI 1.13 ± 0.12, and MESA ABI 1.11 ± 0.12. However, a sensitivity analysis excluding the ARIC samples showed parameter estimates of similar size. (iii) Not all studies had information on lower extremity revascularization, which may also have contributed to PAD misclassification. In general, these misclassifications should cause bias toward the null. (iv) Control selection bias could have affected our PAD results in some way, although given that all of our studies except one (the PDHS) contributing to the PAD analysis were prospective cohort studies where knowledge of PAD would not affect exposure (i.e. genotype status) and the genotype precedes prevalent PAD, this is of lesser concern. For ABI analyses, we also analyzed our clinical replication samples separately by case-control status to avoid additional bias or heterogeneity. (v) Although we adjusted for population stratification using principal components in the European ancestry analysis and global ancestry in the African-American analysis, residual confounding could still be present. (vi) Our sample of African Americans was modest in size and likely limited our power to detect associations. For example, given the sample size of African-Americans we included and a risk allele frequency of 0.10, we only had 80% power to detect an increment in ABI of approximately 0.02 or greater per each copy of the risk allele. The observed effect size of the experiment-wide significant SNPs in European ancestry participants was much smaller than this value. According to our calculations in the methods section, for the European ancestry analyses, we can detect modest differences in ABI (similar to the ones we observed in this study), but are likely underpowered for PAD.
Some of the mentioned limitations might have contributed to the observation that the most important findings from the discovery phase could not be confirmed in the replication phase. However, it is unlikely that this fully explains the differences between the two study stages which necessitate additional large study samples.
5. Conclusions
The search for genes influencing ABI and PAD remains challenging. Although we cannot claim new findings in our study, two associations at the discovery stage for ABI (SYTL3, TCF7L2) may deserve further attention in other populations and functional studies. Further study of the genes identified in this study for ABI (SYTL3, TCF7L2) and PAD (CYP2B6) is warranted in other populations as further investigation of the function of these loci may uncover important biological insights into the pathogenesis of PAD. Identification of main effects may have been difficult in our study due to the presence of interactions and heterogeneity across participating studies. New and more powerful approaches to PAD gene discovery are sorely needed.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the participants of all studies in the manuscript.
Funding The Candidate Gene Association Resource (CARe) is supported by contract number HHSN268200625226C from the National Institutes of Health (NIH)/National Heart Lung and Blood Institute (NHLBI), and subcontract number 5215810-55000000041 to C.L.W. A full listing of the grants and contracts that have supported CARe is provided at http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx. Please see information in supplementary information file for a complete list of funding information for each study participating in this manuscript.
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.atherosclerosis.2012.01.039.
References
- [1].Ankle Brachial Index Collaboration FG. Fowkes GD, Murray I, et al. Ankle brachial index combined with Framingham Risk Score to predict cardiovascular events and mortality: a meta-analysis. JAMA. 2008;300:197–208. doi: 10.1001/jama.300.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Steg PG, Bhatt DL, Wilson PW, et al. One-year cardiovascular event rates in outpatients with atherothrombosis. JAMA. 2007;297:1197–206. doi: 10.1001/jama.297.11.1197. [DOI] [PubMed] [Google Scholar]
- [3].Bhatt DL, Steg PG, Ohman EM, et al. International prevalence, recognition, and treatment of cardiovascular risk factors in outpatients with atherothrombosis. JAMA. 2006;295:180–9. doi: 10.1001/jama.295.2.180. [DOI] [PubMed] [Google Scholar]
- [4].Hirsch AT, Haskal ZJ, Hertzer NR, et al. ACC/AHA 2005 Practice Guidelines for the management of patients with peripheral arterial disease (lower extremity, renal, mesenteric, and abdominal aortic): a collaborative report from the American Association for Vascular Surgery/Society for Vascular Surgery, Society for Cardiovascular Angiography and Interventions, Society for Vascular Medicine and Biology, Society of Interventional Radiology, and the ACC/AHA Task Force on Practice Guidelines (Writing Committee to Develop Guidelines for the Management of Patients With Peripheral Arterial Disease): endorsed by the American Association of Cardiovascular and Pulmonary Rehabilitation; National Heart, Lung, and Blood Institute; Society for Vascular Nursing; TransAtlantic Inter-Society Consensus; and Vascular Disease Foundation. Circulation. 2006;113:e463–654. doi: 10.1161/CIRCULATIONAHA.106.174526. [DOI] [PubMed] [Google Scholar]
- [5].Valentine RJ, Guerra R, Stephan P, et al. Family history is a major determinant of subclinical peripheral arterial disease in young adults. J Vasc Surg. 2004;39:351–6. doi: 10.1016/j.jvs.2003.07.011. [DOI] [PubMed] [Google Scholar]
- [6].Valentine RJ, Verstraete R, Clagett GP, Cohen JC. Premature cardiovascular disease is common in relatives of patients with premature peripheral atherosclerosis. Arch Intern Med. 2000;160:1343–8. doi: 10.1001/archinte.160.9.1343. [DOI] [PubMed] [Google Scholar]
- [7].Carmelli D, Fabsitz RR, Swan GE, et al. Contribution of genetic and environmental influences to ankle-brachial blood pressure index in the NHLBI Twin Study. National Heart, Lung, and Blood Institute. Am J Epidemiol. 2000;151:452–8. doi: 10.1093/oxfordjournals.aje.a010230. [DOI] [PubMed] [Google Scholar]
- [8].Kullo IJ, Turner ST, Kardia SL, et al. A genome-wide linkage scan for ankle-brachial index in African American and non-Hispanic white subjects participating in the GENOA study. Atherosclerosis. 2006;187:433–8. doi: 10.1016/j.atherosclerosis.2005.10.003. [DOI] [PubMed] [Google Scholar]
- [9].Murabito JM, Guo CY, Fox CS, D’Agostino RB. Heritability of the ankle-brachial index: the Framingham Offspring study. Am J Epidemiol. 2006;164:963–8. doi: 10.1093/aje/kwj295. [DOI] [PubMed] [Google Scholar]
- [10].Wassel CL, Loomba R, Ix JH, et al. Family history of peripheral artery disease is associated with prevalence and severity of peripheral artery disease: the San Diego population study. J Am Coll Cardiol. 2011;58:1386–92. doi: 10.1016/j.jacc.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Newman AB, Shemanski L, Manolio TA, et al. Ankle-arm index as a predictor of cardiovascular disease and mortality in the Cardiovascular Health Study. The Cardiovascular Health Study Group. Arterioscler Thromb Vasc Biol. 1999;19:538–45. doi: 10.1161/01.atv.19.3.538. [DOI] [PubMed] [Google Scholar]
- [12].Murabito JM, White CC, Kavousi M, et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ Cardiovasc Genet. 2011 doi: 10.1161/CIRCGENETICS.111.961292. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Knowles JW, Assimes TL, Li J, Quertermous T, Cooke JP. Genetic susceptibility to peripheral arterial disease: a dark corner in vascular biology. Arterioscler Thromb Vasc Biol. 2007;27:2068–78. doi: 10.1161/01.ATV.0000282199.66398.8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50K SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- [16].De Caterina R, Talmud PJ, Merlini PA, et al. Strong association of the APOA5-1131T>C gene variant and early-onset acute myocardial infarction. Atherosclerosis. 2011;214:397–403. doi: 10.1016/j.atherosclerosis.2010.11.011. [DOI] [PubMed] [Google Scholar]
- [17].Musunuru K, Lettre G, Young T, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–75. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lettre G, Palmer CD, Young T, et al. Genome-wide association study of coronary heart disease and its risk factors in 8090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu JZ, McRae AF, Nyholt DR, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saxena R, Gianniny L, Burtt NP, et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–5. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- [25].Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, Novartis Institutes of BioMedical Research et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- [26].Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cluett C, McDermott MM, Guralnik J, et al. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ Cardiovasc Genet. 2009;2:347–53. doi: 10.1161/CIRCGENETICS.108.825935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- [29].Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- [30].Takeuchi F, Serizawa M, Yamamoto K, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58:1690–9. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jin T, Liu L. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol. 2008;22:2383–92. doi: 10.1210/me.2008-0135. [DOI] [PubMed] [Google Scholar]
- [32].Ober C, Nord AS, Thompson EE, et al. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J Lipid Res. 2009;50:798–806. doi: 10.1194/jlr.M800515-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Utermann G. Genetic architecture and evolution of the lipoprotein(a) trait. Curr Opin Lipidol. 1999;10:133–41. doi: 10.1097/00041433-199904000-00007. [DOI] [PubMed] [Google Scholar]
- [34].Erqou S, Thompson A, Di Angelantonio E, et al. Apolipoprotein(a) isoforms and the risk of vascular disease: systematic review of 40 studies involving 58 000 participants. J Am Coll Cardiol. 2010;55:2160–7. doi: 10.1016/j.jacc.2009.10.080. [DOI] [PubMed] [Google Scholar]
- [35].Dieplinger B, Lingenhel A, Baumgartner N, et al. Increased serum lipoprotein(a) concentrations and low molecular weight phenotypes of apolipoprotein(a) are associated with symptomatic peripheral arterial disease. Clin Chem. 2007;53:1298–305. doi: 10.1373/clinchem.2007.088013. [DOI] [PubMed] [Google Scholar]
- [36].Volpato S, Vigna GB, McDermott MM, et al. Lipoprotein(a), inflammation, and peripheral arterial disease in a community-based sample of older men and women (the InCHIANTI study) Am J Cardiol. 2010;105:1825–30. doi: 10.1016/j.amjcard.2010.01.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pradhan AD, Shrivastava S, Cook NR, et al. Symptomatic peripheral arterial disease in women: nontraditional biomarkers of elevated risk. Circulation. 2008;117:823–31. doi: 10.1161/CIRCULATIONAHA.107.719369. [DOI] [PubMed] [Google Scholar]
- [38].Molgaard J, Klausen IC, Lassvik C, et al. Significant association between low-molecular-weight apolipoprotein(a) isoforms and intermittent claudication. Arterioscler Thromb. 1992;12:895–901. doi: 10.1161/01.atv.12.8.895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.