Abstract
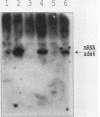
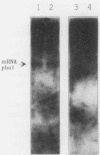
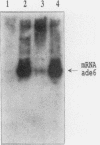
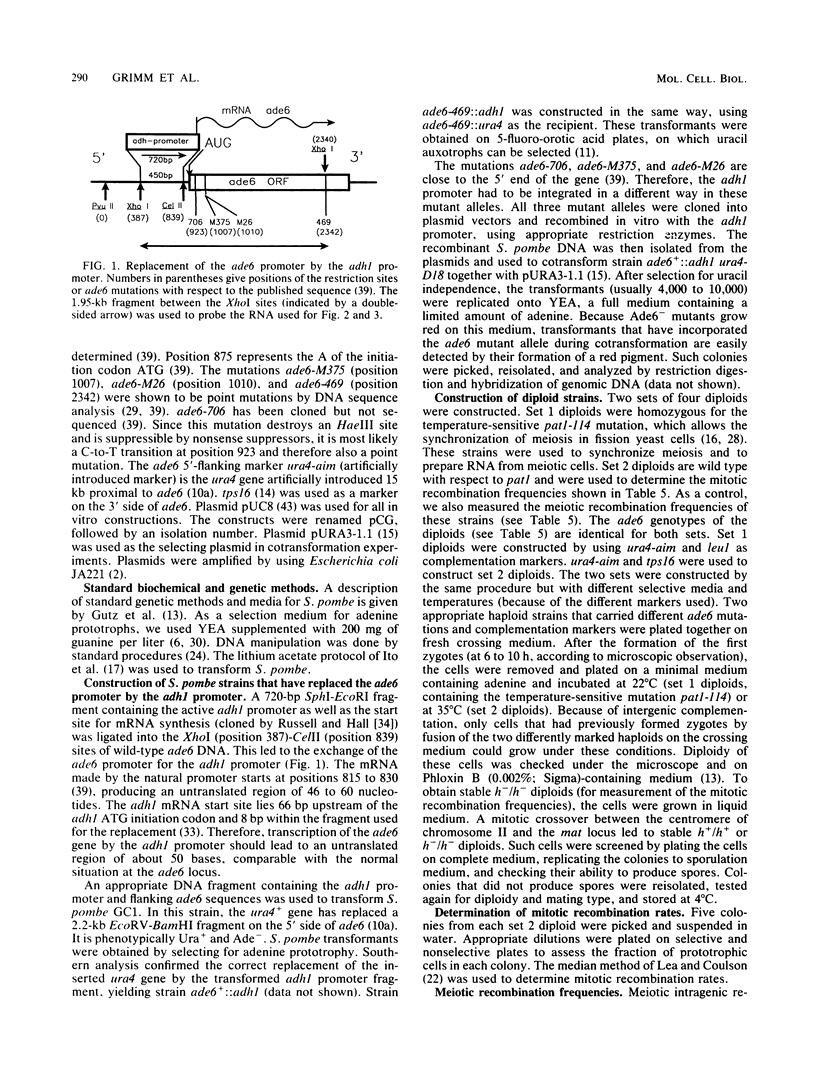
The effect of the strong promoter from the alcohol dehydrogenase gene on mitotic and meiotic intragenic recombination has been studied at the ade6 locus of the fission yeast Schizosaccharomyces pombe. A 700-bp fragment containing the functional adh1 promoter was used to replace the weak wild-type promoter of the ade6 gene. Analysis of mRNA showed that strains with this ade6::adh1 fusion construct had strongly elevated ade6-specific mRNA levels during vegetative growth as well as in meiosis. These increased levels of mRNA correlated with a 20- to 25-fold stimulation of intragenic recombination in meiosis and a 7-fold increased prototroph formation during vegetative growth. Analysis of flanking marker configurations of prototrophic recombinants indicated that simple conversions as well as conversions associated with crossing over were stimulated in meiosis. The strongest stimulation of recombination was observed when the adh1 promoter was homozygous. Studies with heterologous promoter configurations revealed that the highly transcribed allele was the preferred acceptor of genetic information. The effect of the recombinational hot spot mutation ade6-M26 was also investigated in this system. Its effect was only partly additive to the elevated recombination rate generated by the ade6::adh1 fusion construct.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn B. Y., Livingston D. M. Mitotic gene conversion lengths, coconversion patterns, and the incidence of reciprocal recombination in a Saccharomyces cerevisiae plasmid system. Mol Cell Biol. 1986 Nov;6(11):3685–3693. doi: 10.1128/mcb.6.11.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Cummins J. E., Mitchison J. M. Adenine uptake and pool formation in the fission yeast Schizosaccharomyces pombe. Biochim Biophys Acta. 1967 Feb 7;136(1):108–120. doi: 10.1016/0304-4165(67)90326-1. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989 Mar 10;56(5):771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Grimm C., Kohli J., Murray J., Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988 Dec;215(1):81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Gutz H. Site Specific Induction of Gene Conversion in SCHIZOSACCHAROMYCES POMBE. Genetics. 1971 Nov;69(3):317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D., Sipiczki M., Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986 Jan;6(1):80–89. doi: 10.1128/mcb.6.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy J. M., Klar A. J., Hicks J. B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984 Dec;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Abraham J. A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Broach J. R., Hicks J. B. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature. 1981 Jan 22;289(5795):239–244. doi: 10.1038/289239a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K., Nurse P., Schönholzer F., Schweingruber M. E. Cloning and characterization of two genes restoring acid phosphatase activity in pho1- mutants of Schizosaccharomyces pombe. Gene. 1985;39(2-3):223–230. doi: 10.1016/0378-1119(85)90316-6. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Sena E. P., Smith G. R. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics. 1988 Jul;119(3):491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Smith G. R. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989 Sep;123(1):45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M., Flory J., Wu A., Kahn R., DasGupta C., Gonda D., Bianchi M., Tsang S. S. Three phases in homologous pairing: polymerization of recA protein on single-stranded DNA, synapsis, and polar strand exchange. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):821–828. doi: 10.1101/sqb.1983.047.01.094. [DOI] [PubMed] [Google Scholar]
- Ray A., Machin N., Stahl F. W. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. R. Evolutionary divergence of the mRNA transcription initiation mechanism in yeast. Nature. 1983 Jan 13;301(5896):167–169. doi: 10.1038/301167a0. [DOI] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983 Jan 10;258(1):143–149. [PubMed] [Google Scholar]
- Schulz M., Freisem-Rabien U., Jessberger R., Doerfler W. Transcriptional activities of mammalian genomes at sites of recombination with foreign DNA. J Virol. 1987 Feb;61(2):344–353. doi: 10.1128/jvi.61.2.344-353.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J., Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986 Jan;5(1):95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S. E., Roeder G. S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Aug;9(8):3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Szankasi P., Heyer W. D., Schuchert P., Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6-M26. J Mol Biol. 1988 Dec 20;204(4):917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Keil R. L., Roeder G. S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987 Mar 27;48(6):1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985 Feb;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]