ERdj5 triggers BiP to bind to cholera toxin in the endoplasmic reticulum, targeting the toxin to the Hrd1 E3 ubiquitin ligase complex for retrotranslocation.
Abstract
Cholera toxin (CT) traffics from the host cell surface to the endoplasmic reticulum (ER), where the toxin's catalytic CTA1 subunit retrotranslocates to the cytosol to induce toxicity. In the ER, CT is captured by the E3 ubiquitin ligase Hrd1 via an undefined mechanism to prepare for retrotranslocation. Using loss-of-function and gain-of-function approaches, we demonstrate that the ER-resident factor ERdj5 promotes CTA1 retrotranslocation, in part, via its J domain. This Hsp70 cochaperone regulates binding between CTA and the ER Hsp70 BiP, a chaperone previously implicated in toxin retrotranslocation. Importantly, ERdj5 interacts with the Hrd1 adaptor Sel1L directly through Sel1L's N-terminal lumenal domain, thereby linking ERdj5 to the Hrd1 complex. Sel1L itself also binds CTA and facilitates toxin retrotranslocation. By contrast, EDEM1 and OS-9, two established Sel1L binding partners, do not play significant roles in CTA1 retrotranslocation. Our results thus identify two ER factors that promote ER-to-cytosol transport of CTA1. They also indicate that ERdj5, by binding to Sel1L, triggers BiP–toxin interaction proximal to the Hrd1 complex. We postulate this scenario enables the Hrd1-associated retrotranslocation machinery to capture the toxin efficiently once the toxin is released from BiP.
INTRODUCTION
Cholera toxin (CT), secreted by Vibrio cholerae, is the virulence factor responsible for inducing massive secretory diarrhea. Structurally, CT is composed of a catalytic A subunit (CTA) inserted into a pore formed by five receptor-binding B subunits (CTBs; Spangler, 1992). CTA is further divided into two subunits: CTA1 and CTA2. CTA1, which harbors the toxin's catalytic domain, is linked to CTA2 via a disulfide bond. Additional noncovalent interactions between CTA1 and CTB provide structural support for the native holotoxin.
To cause disease, CT binds to the glycolipid ganglioside GM1 receptor on the plasma membrane of intestinal epithelial cells. This interaction induces toxin internalization, enabling the toxin to traffic in a retrograde manner through the Golgi complex to reach the endoplasmic reticulum (ER; Fujinaga et al., 2003). In the ER, CTA is reduced to generate CTA1. CTA1 then retrotranslocates to the cytosol, where it triggers a signaling cascade leading to the opening of a chloride channel; the ensuing chloride ion and water loss by intestinal epithelial cells leads to the cholera disease (Lencer and Tsai, 2003).
In the ER, CT is postulated to hijack the ER-associated degradation (ERAD) machinery to gain access to the cytosol (Hazes and Read, 1997). This machinery normally recognizes and ejects misfolded ER proteins to the cytosol, where they are ubiquitinated and degraded by the proteasome (Smith et al., 2011). The toxin, however, avoids ubiquitination upon reaching the cytosol and evades proteasomal destruction (Rodighiero et al., 2002). While the precise molecular mechanism by which CTA1 retrotranslocates from the ER to the cytosol is unclear, recent findings illuminate some aspects of this decisive intoxication step.
Accumulating evidence pinpoints the central ERAD component as being a membrane-bound E3 ubiquitin ligase (Christianson et al., 2011; Smith et al., 2011). This ligase operates in conjunction with numerous associated adaptors, including integral membrane proteins such as Derlins and Sel1L (Lilley and Ploegh, 2005), the ER lumenal housekeeping chaperones BiP (Hosokawa et al., 2008), and protein disulfide isomerase (PDI; Bernardi et al., 2008), as well as the cytosolic p97 ATPase (Ye et al., 2005). Together they coordinate events on the ER membrane's lumenal and cytosolic sides to promote retrotranslocation of a specific substrate (Gauss et al., 2006).
We previously demonstrated that CTA1 coopts the membrane E3 ubiquitin ligases Hrd1 and gp78 to cross the ER membrane (Bernardi et al., 2010). A few of these ligases’ adaptors have been implicated in toxin retrotranslocation. For example, Derlin-1, but not Derlin-2, facilitates CTA1 retrotranslocation in mammalian cells (Bernardi et al., 2008; Dixit et al., 2008). Derlins, however, may not be involved in promoting toxin transport in zebrafish (Saslowsky et al., 2010). A Derlin-1–interacting, ER-resident ATPase called TorsinA also supports CTA1 ER-to-cytosol transport (Nery et al., 2011). In addition, BiP and PDI have been shown to be critical for mediating CTA1 retrotranslocation (Tsai et al., 2001; Winkeler et al., 2003; Forster et al., 2006; Moore et al., 2010). Finally, two independent reports suggest that p97 plays a modest role in facilitating toxin retrotranslocation (Abujarour et al., 2005; Kothe et al., 2005).
While these findings provide insight into the role of E3 ligases and their associated factors during CTA1 retrotranslocation, how the E3s capture CT once the toxin reaches the ER to initiate retrotranslocation remains unclear. We hypothesize that this reaction is supported by ER factors associated with the Hrd1 complex or any of its adaptor proteins. In this study, using an established cell-based retrotranslocation assay coupled with small interfering RNA (siRNA) and overexpression approaches, we pinpoint Sel1L and ERdj5 in facilitating CTA1 retrotranslocation. ERdj5 is a cochaperone of BiP that contains a J and four redox-active thioredoxin domains implicated in ERAD (Dong et al., 2008; Ushioda et al., 2008). Additionally, we found an intact ERdj5 J domain is critical for this process. Biochemical studies indicate ERdj5 binds to Sel1L directly and promotes BiP–toxin interaction. ERdj5's physical proximity to the Hrd1 complex, via binding to Sel1L, enables ERdj5 to trigger BiP–toxin interaction next to the Hrd1 complex. We propose that this scenario allows the Hrd1-associated retrotranslocation machinery to capture CT efficiently once the toxin is released from BiP.
RESULTS
ERdj5 knockdown decreases CTA1 retrotranslocation
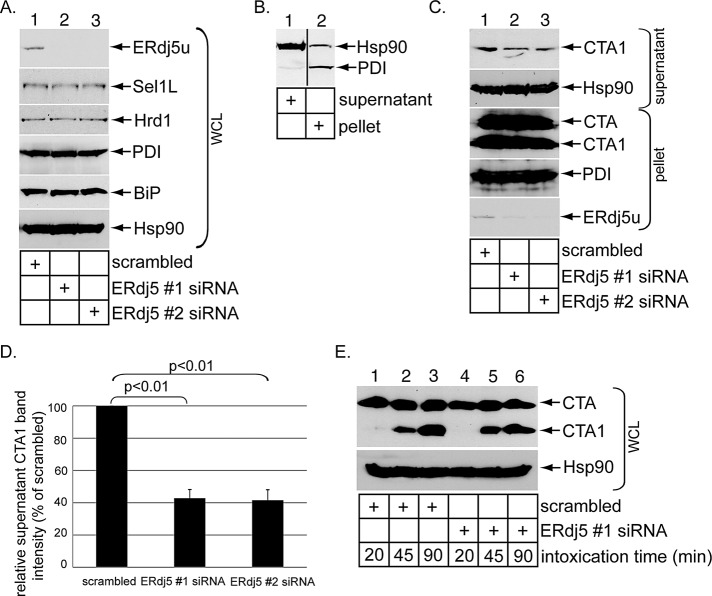
ERdj5 is a bifunctional protein harboring both a J domain and four redox-active thioredoxin domains (Ushioda et al., 2008; Hagiwara et al., 2011). J proteins typically stimulate the ATPase activity of Hsp70 chaperones to regulate chaperone–substrate interactions (Kampinga and Craig, 2010), while redox-active thioredoxin domains are used to oxidize, reduce, and isomerize disulfide bonds (Bulleid and Ellgaard, 2011). We therefore postulate that ERdj5 controls CTA1 retrotranslocation by using either its J domain to stimulate BiP–toxin binding or its thioredoxin domains to reduce CTA and generate CTA1, the substrate for retrotranslocation. For testing this possibility, full-length unspliced ERdj5 (ERdj5u), the major ERdj5 form expressed in 293T cells, was knocked down using two different siRNA oligonucleotides directed specifically against ERdj5 (i.e., ERdj5 #1 and #2 siRNAs) in 293T cells. Cells were transfected with a control siRNA (scrambled), ERdj5 #1 siRNA, or ERdj5 #2 siRNA; harvested; and lysed; and the resulting whole-cell lysate (WCL) was subjected to SDS–PAGE followed by immunoblotting. Compared with a WCL derived from cells transfected with a scrambled siRNA, the ERdj5u level markedly decreased in WCLs derived from cells transfected with the ERdj5-specific siRNAs (Figure 1A, top panel, compare lanes 2 and 3 with lane1). Under the knocked-down conditions, the Hrd1 and Sel1L levels were unaffected (Figure 1A, second and third panels from top). Moreover, ERdj5u knockdown did not up-regulate PDI and BiP, two unfolded protein response (UPR) markers induced upon ER stress (Figure 1A, fourth and fifth panels from top, compare lanes 2 and 3 with lane1), consistent with a previous report (Ushioda et al., 2008). Thus ERdj5u was down-regulated efficiently without triggering massive ER stress.
FIGURE 1:
ERdj5 knockdown decreases CTA1 retrotranslocation. (A) WCLs from 293T cells transfected with a scrambled, ERdj5 #1, or ERdj5 #2 siRNA were analyzed by SDS–PAGE and immunoblotted with the indicated antibodies. The ERdj5 antibody used was from Santa Cruz Biotechnology. (B) Cells were incubated with digitonin and centrifuged. The resulting supernatant and pellet fractions were analyzed for the presence of the cytosolic Hsp90 and ER-resident PDI markers. This protocol represents the fractionation procedure used in the retrotranslocation assay. The vertical black line indicates splicing of the lanes from the same blot. (C) Cells transfected with a scrambled, ERdj5 #1, or ERdj5 #2 siRNA were incubated with CT (10 nM) for 90 min and subjected to the retrotranslocation assay, as in (B). The supernatant and pellet fractions were analyzed by immunoblotting with the indicated antibodies. (D) The CTA1 band intensity generated in (C) was quantified with ImageJ. Mean of four independent experiments. A two-tailed t test was used. Error bars: ±SD. (E) Cells transfected with a scrambled or ERdj5 #1 siRNA were intoxicated with CT (10 nM) for the indicated time and harvested, and the resulting WCLs were analyzed with nonreducing SDS–PAGE followed by immunoblotting with the indicated antibodies.
To determine whether CTA1 retrotranslocation is perturbed when ERdj5u is knocked down, we used a cell-based semipermeabilized CTA1 retrotranslocation assay established by our laboratory (Forster et al., 2006; Bernardi et al., 2008; Moore et al., 2010). This assay has since been used by other laboratories to study CTA1 retrotranslocation (Taylor et al., 2010; Wernick et al., 2010; Nery et al., 2011), as well as by our laboratory and others to examine polyomavirus retrotranslocation (Geiger et al., 2011; Goodwin et al., 2011; Inoue and Tsai, 2011). Briefly, cells are permeabilized with a low concentration (0.04%) of the gentle detergent digitonin. Under this condition, only the plasma membrane, but not internal membranes, is expected to be permeabilized. Subsequent centrifugation of the sample generates a supernatant and pellet fraction. When these samples are subjected to SDS–PAGE followed by immunoblotting, the cytosolic Hsp90 appears mostly in the supernatant fraction (Figure 1B, compare lane 1 with lane 2), while the ER-resident PDI is present essentially in the pellet fraction (Figure 1B, compare lane 2 with lane 1). Thus the supernatant fraction contains the cytosolic material, and the pellet fraction contains membranes, including the ER membrane. CTA1 appearance in the supernatant fraction therefore represents cytosol-localized retrotranslocated toxin.
Accordingly, cells transfected with the scrambled siRNA, ERdj5 #1 siRNA, or ERdj5 #2 siRNA were intoxicated with CT (10 nM) for 90 min and subjected to digitonin permeabilization and centrifugation, with the resulting supernatant and cytosolic fractions analyzed by nonreducing SDS–PAGE followed by immunoblotting. Our data demonstrate that the CTA1 level decreased in the supernatant fraction derived from cells transfected with ERdj5 #1 and ERdj5 #2 siRNAs when compared with scrambled siRNA (Figure 1C, top panel, compare lanes 2 and 3 with lane 1). As only a small fraction of total CTA1 appeared in the supernatant fraction (Figure 1C, compare first and third panels from top), any expected corresponding changes of the CTA1 level in the pellet fraction were not obvious, consistent with our previous observations (Bernardi et al., 2008, 2010; Moore et al., 2010). Quantification of the CTA1 band intensity demonstrated that an approximately 60% decrease in toxin retrotranslocation when ERdj5u was down-regulated using either siRNAs (Figure 1D). We conclude that ERdj5u facilitates CTA1 retrotranslocation.
One potential mechanism by which ERdj5u affects CTA1 retrotranslocation is by using its redox-active thioredoxin domains to reduce CTA to generate CTA1. For assessing whether CTA1 formation was affected by ERdj5u down-regulation, cells transfected with scrambled or ERdj5 #1 siRNA were intoxicated with CT (10 nM) for 20, 45, or 90 min. The resulting WCLs were subjected to nonreducing SDS–PAGE followed by immunoblotting. In cells transfected with scrambled siRNA, CTA1 appeared at both the 45- and 90-min, but not the 20 min, postintoxication time points (Figure 1E, top panel, compare lanes 2 and 3 with lane 1). Thirty minutes postintoxication is approximately the time in which some CT first reaches the ER, where CTA is reduced to CTA1. In cells transfected with ERdj5 #1 siRNA, the CTA1 level generated at both the 45- and 90-min time points was similar to that seen in cells transfected with scrambled siRNA (Figure 1E, top panel, compare lanes 5 and 6 with lanes 2 and 3). Therefore ERdj5u mediates CTA1 retrotranslocation without affecting CTA reduction.
WT but not mutant ERdj5 overexpression stimulates CTA1 retrotranslocation
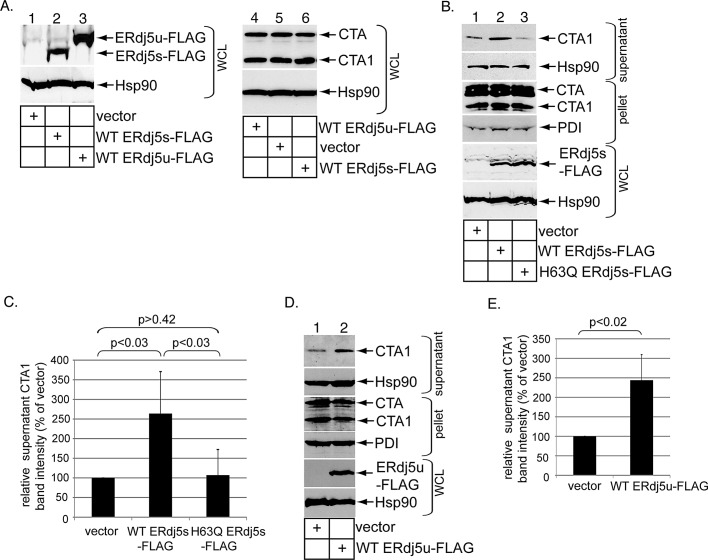
To complement the loss-of-function approach, we used a gain-of-function strategy to further support ERdj5's role in promoting CTA1 retrotranslocation. In addition to full-length ERdj5u, a cDNA encoding an ERdj5 alternatively spliced form was reported that lacks 46 amino acids located within a redox-inactive thioredoxin domain of ERdj5u (Gu et al., 2003); we refer to this spliced form as ERdj5s. Although an ERdj5s cDNA was derived from mRNA isolated from 293T cells, the ERdj5s protein is below the sensitivity threshold of the antibody used in our study, as only ERdj5u can be detected (Figure 1A, top panel). Nonetheless, 293T cells were transfected with either human wild-type (WT) ERdj5s containing a C-terminal FLAG epitope (WT ERdj5s-FLAG) or human WT ERdj5u containing a C-terminal FLAG epitope (WT ERdj5u-FLAG) and harvested, and the resulting WCLs were subjected to SDS–PAGE followed by immunoblotting with a FLAG antibody. We found WT ERdj5s-FLAG migrated faster than WT ERdj5u-FLAG, as expected (Figure 2A, top panel, compare lane 2 with lane 3). When these cells were incubated with CT (10 nM) for 90 min and the resulting WCLs were analyzed, the data indicate that overexpressing WT ERdj5s-FLAG or WT ERdj5u-FLAG did not increase CTA1 formation (Figure 2A, top panel, compare lane 5 with lanes 4 and 6). We conclude that overexpressing either ERdj5 variant does not promote CTA reduction to generate CTA1.
FIGURE 2:
ERdj5 overexpression stimulates CTA1 retrotranslocation. (A) WCLs derived from 293T cells transfected with vector, WT ERdj5s-FLAG, or WT ERdj5u-FLAG and intoxicated with CT (10 nM) were analyzed by immunoblotting using the indicated antibodies. (B) Cells transfected with vector, WT ERdj5s-FLAG, or H63Q ERdj5s-FLAG were incubated with CT (10 nM) for 90 min and subjected to the retrotranslocation assay, as in Figure 1C. The supernatant and pellet fractions and the WCLs were analyzed by immunoblotting with the indicated antibodies. (C) The CTA1 band intensity in (B) was quantified as in Figure 1D. Mean of five independent experiments. A two-tailed t test was used. Error bars: ± SD. (D) As in (B), except only vector and WT ERdj5u-FLAG were transfected. (E) The CTA1 band intensity in (D) was analyzed as in (C). Mean of seven independent experiments. A two-tailed t test was used. Error bars: ±SD.
For determination of whether CTA1 retrotranslocation was affected by ERdj5 overexpression, cells transfected with vector or WT ERdj5s-FLAG were intoxicated with CT (10 nM) for 90 min and subjected to the semipermeabilized retrotranslocation assay as before. Indeed, WT ERdj5s-FLAG overexpression caused a 2.5-fold increase in the supernatant CTA1 level (Figure 2B, top panel, compare lane 2 with lane 1; quantified in Figure 2C). To assess whether WT ERdj5s's J domain is responsible for this effect, we expressed a mutant version of ERdj5s in which a critical histidine residue located in its J domain responsible for stimulating BiP ATPase activity is mutated to glutamine (H63Q ERdj5s-FLAG); the full-length version of this mutant protein failed to stimulate BiP ATPase activity and accelerate degradation of an ERAD substrate when compared with the corresponding WT protein (Ushioda et al., 2008). In contrast with WT ERdj5s-FLAG, H63Q ERdj5s-FLAG overexpression failed to stimulate CTA1 retrotranslocation (Figure 2B, top panel, compare lane 3 with lane 1; quantified in Figure 2C). This observation is not due to lower H63Q ERdj5s-FLAG expression when compared with WT ERdj5s-FLAG (Figure 2B, fifth panel from top, compare lane 3 with lane 2). Similar to WT ERdj5s-FLAG, WT ERdj5u-FLAG overexpression also increased CTA1 retrotranslocation by 2.5 fold (Figure 2D, top panel, compare lane 2 with lane 1; quantified in Figure 2E). We conclude that ERdj5 uses its J domain to stimulate CTA1 retrotranslocation, and that this reaction represents a rate-limiting step.
ERdj5 binds to BiP and regulates BiP–CTA interaction
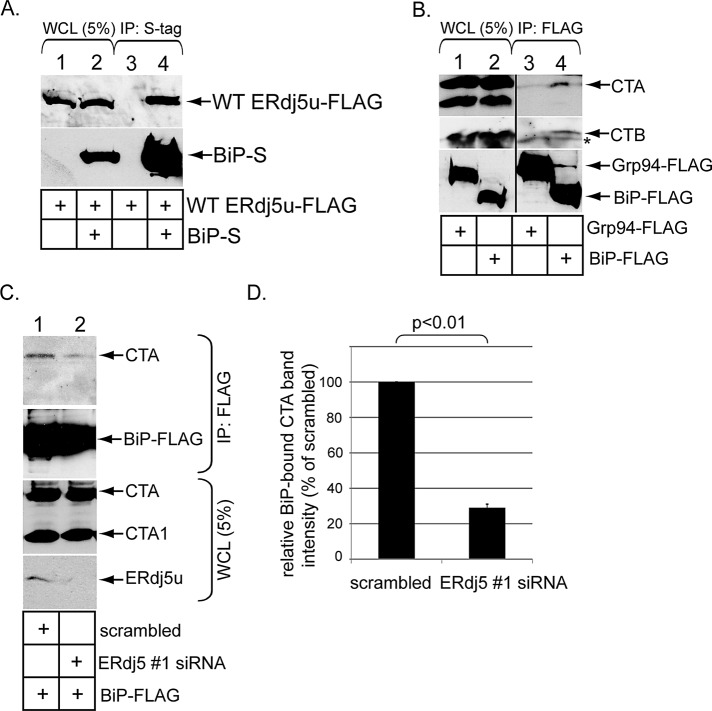
ERdj5u was shown previously to interact with BiP (Ushioda et al., 2008). To verify this interaction in our system, we transfected WT ERdj5u-FLAG alone or in combination with C-terminally S-tagged human BiP (BiP-S). When the resulting WCLs were subjected to precipitation using S-antibody–conjugated beads, WT ERdj5u-FLAG precipitated only when BiP-S was coexpressed (Figure 3A, top panel, compare lane 4 with lane 3). Similar results were seen using WT ERdj5s-FLAG rather than WT ERdj5u-FLAG (unpublished data). Thus ERdj5 binds to BiP.
FIGURE 3:
ERdj5 binds to BiP and regulates BiP–CTA interaction. (A) Cells were transfected with WT ERdj5u-FLAG alone or in combination with BiP-S, and lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were incubated with S-antibody–conjugated beads, and the immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with the indicated antibodies. (B) Cells transfected with the control Grp94-FLAG or BiP-FLAG were incubated with CT (10 nM) for 90 min and lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were incubated with FLAG antibody–conjugated beads, and the immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with CTA, CTB, or FLAG antibodies. The vertical black line indicates antibody splicing of the lanes from the same blot. The asterisk denotes a nonspecific protein that interacts with the CTB antibody. (C) Cells transfected with BiP-FLAG and cotransfected with either a scrambled or ERdj5 #1 siRNA were incubated with CT (10 nM) for 90 min and lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were incubated with FLAG antibody–conjugated beads, and the immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with the indicated antibodies. WCL samples were also analyzed by immunoblotting with an antibody against CTA and ERdj5. (D) The CTA1 band intensity in (C) was analyzed as in Figure 1D. Mean of three independent experiments. A two-tailed t test was used. Error bars: ±SD.
BiP was previously implicated in binding to CTA in vitro (Winkeler et al., 2003). For evaluating whether this interaction occurs in cells, cells expressing either human BiP containing a C-terminal FLAG epitope (BiP-FLAG) or the ER lumenal chaperone Grp94 containing a C-terminal FLAG epitope (Grp94-FLAG). When WCLs derived from these cells were subjected to precipitation using FLAG antibody–conjugated beads, CTA and CTB (but not CTA1) coprecipitated only with BiP-FLAG but not with Grp94-FLAG (Figure 3B, top and second panels, compare lane 4 with lane 3). Importantly, BiP–CTA interaction decreased significantly when ERdj5u was knocked down (Figure 3C, top panel, compare lane 2 with lane 1; quantified in Figure 3D). We conclude that ERdj5 promotes BiP–CTA interaction in cells.
ERdj5 binds to Sel1L
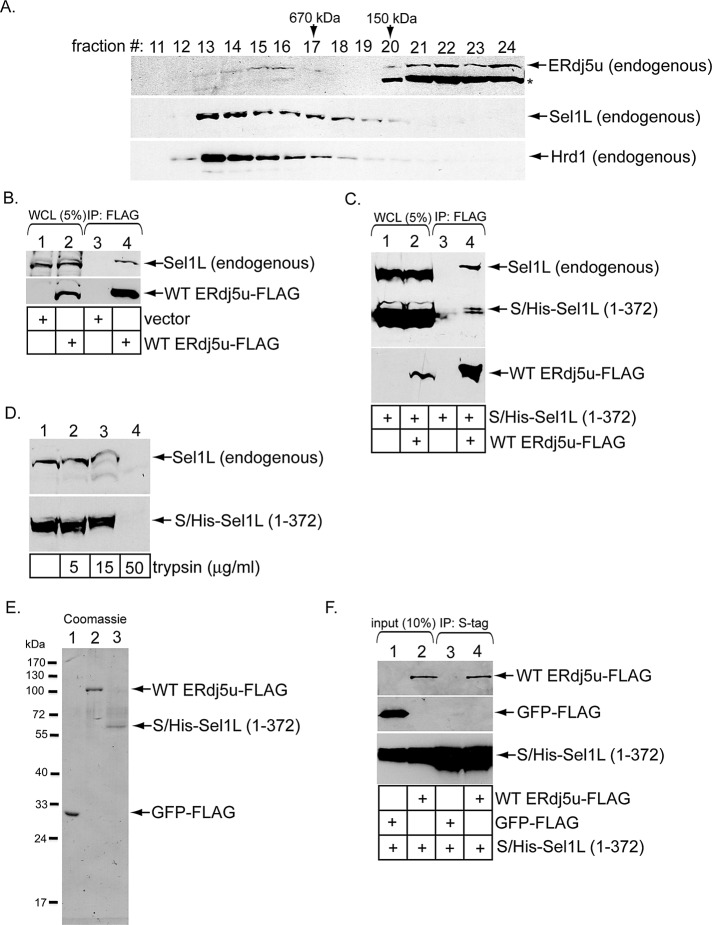
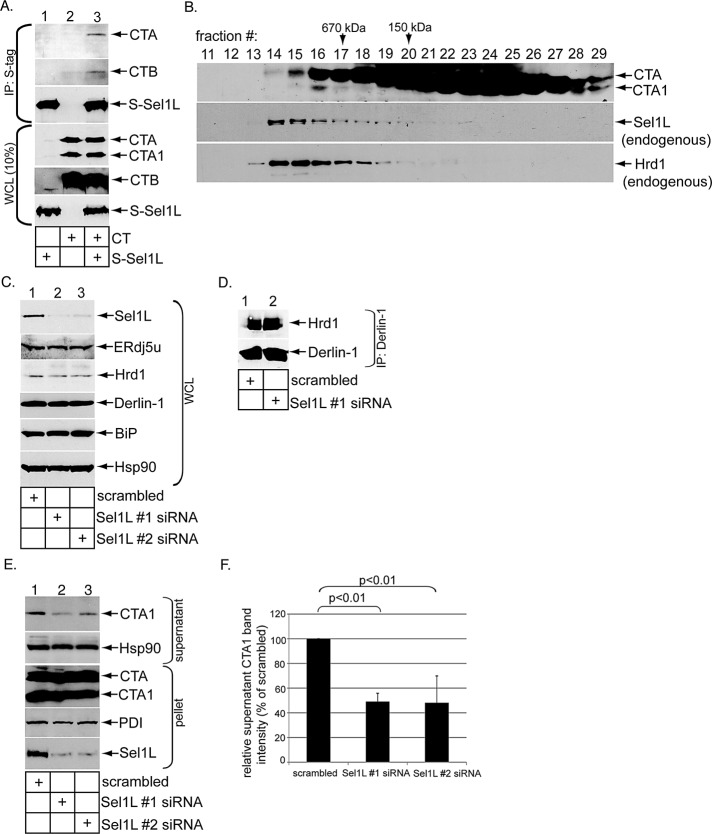
As ERdj5 stimulates BiP-toxin binding, establishing ERdj5's physical proximity to the Hrd1 complex would provide a means for the retrotranslocation machinery to capture the toxin. Therefore we asked whether ERdj5 binds to the Hrd1 interaction partner Sel1L (Lilley and Ploegh, 2005) because Sel1L is known to engage BiP in mammalian cells (Hosokawa et al., 2008). Additionally, the yeast Sel1L homologue, Hrd3p, is also an established binding partner of Kar2p, the yeast BiP homologue (Denic et al., 2006). When a WCL derived from 293T cells was subjected to gel-filtration analysis, a small portion of endogenous ERdj5 cofractionated with endogenous Hrd1 and Sel1L in fractions corresponding to protein complexes larger than 670 kDa (Figure 4A, compare top, middle, and bottom panels, fractions 13–16). (A different ERdj5 antibody was used in this experiment, as it displays a higher affinity for endogenous ERdj5u, although this antibody cross-reacts with the 72-kDa protein ERp72, indicated with an asterisk.) Because monomeric ERdj5 is <100 kDa, this finding suggests that ERdj5 complexes with other ER proteins. To directly test ERdj5–Sel1L interaction, we subjected a WCL derived from cells transfected with vector or WT ERdj5u-FLAG to precipitation with FLAG antibody–conjugated beads. Endogenous Sel1L precipitated only in cells transfected with WT ERdj5s-FLAG, but not vector (Figure 4B, top panel, compare lane 4 with lane 3). A similar result was found using WT ERdj5s-FLAG (Supplemental Figure S1, top panel, compare lane 4 with lane 3). Both WT ERdj5u-FLAG and WT ERdj5s-FLAG bind to Sel1L with equal affinity (unpublished data). We conclude that transfected ERdj5 binds to Sel1L to associate with the Hrd1 complex.
FIGURE 4:
ERdj5u binds to Sel1L. (A) A WCL derived from 293T cells was subjected to gel filtration. Individual fractions were subjected to SDS–PAGE and analyzed by immunoblotting using the indicated antibodies. The asterisk denotes ERp72 that cross-reacts with an ERdj5 antibody (Proteintech Group). (B) Cells were transfected with vector or WT ERdj5u-FLAG and lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were incubated with FLAG antibody–conjugated beads, and the immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with the indicated antibodies. (C) Cells transfected with S/His-Sel1L (1–372) alone or S/His-Sel1L (1–372) and WT ERdj5u-FLAG were lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were processed as in (B). (D) A WCL expressing S/His-Sel1L (1–372) was incubated with the indicated trypsin concentration, subjected to SDS–PAGE, and immunoblotted using an antibody against Sel1L. (E) GFP-FLAG, WT ERdj5u-FLAG, and S/His-Sel1L (1–372) were expressed in 293T cells, and the purified proteins were subjected to SDS–PAGE followed by Coomassie blue staining. Fraction 27 during purification of S/His-Sel1L (1–372) is shown. (F) S/His-Sel1L (1–372) was incubated with either GFP-FLAG or WT ERdj5u-FLAG, and the samples were precipitated using S-antibody–conjugated beads. The immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with the indicated antibodies. The input samples were also analyzed by immunoblotting with the appropriate antibodies.
We next sought to identify regions in Sel1L that interact with ERdj5. Sel1L is predicted to be a type I transmembrane protein with most of the protein found in the ER lumen (Mueller et al., 2006). Its lumenal portion contains 11 “Sel1-repeats” or tetratricopeptide repeat (TPR) domains (Christianson et al., 2008). A construct containing an S-tag fused to Sel1L's first 372 amino acids that harbors four TPR domains (S-Sel1L (1–372)) was previously found not to bind to Hrd1, in contrast with the corresponding full-length S-Sel1L construct (Christianson et al., 2008). We inserted a 6× histidine tag (His) between the S-tag and the Sel1L (1–372) sequence to generate the S/His-Sel1L (1–372) construct. Cells were transfected with S/His-Sel1L (1–372) alone or S/His-Sel1L (1–372) and WT ERdj5u-FLAG. The resulting WCLs were subjected to precipitation with FLAG antibody–conjugated beads. We found that both endogenous Sel1L and S/His-Sel1L (1–372) coprecipitated with WT ERdj5u-FLAG (Figure 4C, top and bottom panels, lane 4), perhaps with S/His-Sel1L (1–372) displaying a slightly lower affinity to ERdj5 than full-length Sel1L. These findings implicate Sel1L's N-terminal 372 amino acids in binding to ERdj5. S/His-Sel1L (1–372) and endogenous Sel1L display similar sensitivity to digestion by increasing trypsin concentration (Figure 4D, compare top and bottom panels, lanes 1–4), suggesting that the truncated Sel1L is not globally misfolded and therefore is unlikely to engage ERdj5 as a substrate. To determine whether Sel1L (1–372) binds to ERdj5 directly, we purified WT ERdj5u-FLAG and S/His-Sel1L (1–372), as well as the control protein green fluorescent protein (GFP)-FLAG from 293T cells (Figure 4E, lanes 1–3). When S/His-Sel1L (1–372) was incubated with either GFP-FLAG or WT ERdj5u-FLAG, and the samples were subjected to precipitation with S-antibody–conjugated beads, we found only WT ERdj5u-FLAG, not GFP-FLAG, coprecipitated with S/His-Sel1L (1–372) (Figure 4F, top panel, compare lane 4 with lane 3). We conclude that Sel1L's N-terminal domain binds directly to ERdj5.
Our results demonstrating that ERdj5 interacts with Sel1L, coupled with the previous finding that BiP binds to Sel1L (Hosokawa et al., 2008), raise the possibility that BiP's interaction with Sel1L might require ERdj5. However, during purification of S/His-Sel1L (1–372), we noted that a pool of S/His-Sel1L (1–372) copurified only with BiP (Figure S2A, fraction 32), while another pool did not (Figure S2A, fraction 27). This observation suggests that Sel1L's N-terminal domain can bind BiP without ERdj5's assistance. Intriguingly, glycosylated Sel1L (1–372) was also found in fraction 32, implicating glycosylation of Sel1L in mediating its interaction with BiP. (S/His-Sel1L (1–372) from fraction 27 was used in Figure 4, E and F, because this fraction lacked BiP). To further substantiate that BiP's binding to Sel1L does not require ERdj5, we found ERdj5 knockdown does not affect the ability of transfected S/His-Sel1L (1–372) to coprecipitate endogenous BiP (Figure S2B, compare lane 8 with lane 7). We conclude that the BiP–Sel1L interaction does not require ERdj5.
Sel1L binds to CT and mediates CTA1 retrotranslocation
Because ERAD substrates can use Sel1L-independent mechanisms for ER membrane transport (Bernasconi et al., 2010; Stanley et al., 2011), we next asked whether Sel1L functions in CTA1 retrotranslocation. We first sought to establish a physical interaction between Sel1L and CT. Cells transfected with or without S-Sel1L were incubated with or without CT (10 nM) for 90 min. The resulting WCLs were incubated with S-antibody–conjugated beads to precipitate S-Sel1L. We found that CTA and CTB precipitated only when S-Sel1L is expressed (Figure 5A, top two panels, compare lane 3 with lane 2), indicating Sel1L binds to the holotoxin. That Sel1L binds to CT and our earlier result demonstrating the Hrd1 complex also interacts with CT (Bernardi et al., 2010) prompted us to test whether the toxin cofractionates with Sel1L and Hrd1 in a gel-filtration analysis. Indeed, we found a small CTA pool cofractionates with endogenous Sel1L and Hrd1 (Figure 5B, fractions 14–18). Thus a pool of CT forms a complex with the Hrd1-associated machinery, consistent with the coimmunoprecipitation data.
FIGURE 5:
Sel1L binds to CT and mediates CTA1 retrotranslocation. (A) Cells transfected with or without S-Sel1L were intoxicated with or without CT (10 nM) for 90 min and lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were incubated with S-antibody–conjugated beads, and the immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with the indicated antibodies. WCL samples were also analyzed by immunoblotting with the appropriate antibodies. (B) As in Figure 4A, except cells were intoxicated with CT (10 nM), semipermeabilized with 0.01% digitonin, and centrifuged to remove toxin in the cytosol; this was followed by solubilization of the resulting pellet with 1% digitonin. (C) As in Figure 1A, except two different Sel1L-directed siRNAs (Sel1 #1 and #2 siRNAs) were used. (D) Cells transfected with a scrambled or Sel1L #1 siRNA were harvested and lysed in a buffer containing 1% deoxy Big CHAP, and the resulting WCLs were subjected to immunoprecipitation with a Derlin-1−specific antibody. The immunoprecipitates were analyzed by immunoblotting using the indicated antibodies. (E) Cells transfected with a scrambled, Sel1L #1 siRNA, or Sel1L #2 siRNA were intoxicated with CT (10 nM) for 90 min. Cells were subjected to the retrotranslocation assay, and the supernatant and pellet fractions were analyzed as in Figure 1C. (F) The CTA1 band intensity in (D) was analyzed as in Figure 1D. Mean of three independent experiments. A two-tailed t test was used. Error bars: ±SD.
For addressing whether Sel1L functionally regulates CTA1 retrotranslocation, cells were transfected with a scrambled siRNA or either of two siRNAs directed specifically against Sel1L (i.e., Sel1L #1 and #2 siRNAs) and harvested, and the resulting WCL was subjected to SDS–PAGE and immunoblotting. An efficient Sel1L knockdown was achieved using either Sel1L-specific siRNAs (Figure 5C, top panel, compare lanes 2 and 3 with lane 1). Under the knockdown conditions, neither the ERdj5, Hrd1, nor Derlin1 levels were affected (Figure 5C, second, third, and fourth panels from top). Furthermore, the UPR marker BiP was also unaffected (Figure 5C, fifth panel from top). A coimmunoprecipitation experiment revealed that Sel1L knockdown (using Sel1L #1 siRNA) does not affect Hrd1–Derlin-1 interaction (Figure 5D, compare lane 2 with lane 1), suggesting that Sel1L down-regulation does not disrupt the Hrd1 complex globally. However, when cells were subjected to the semipermeabilized retrotranslocation assay, CTA1 ER-to-cytosol transport was attenuated when Sel1L was knocked down using either siRNA (Figure 5E, top panel, compare lane 2 and 3 with lane 1; quantified in Figure 5F). We conclude that Sel1L plays a functional role during CTA1 retrotranslocation.
ERdj5 knockdown decreases Hrd1–CTA interaction
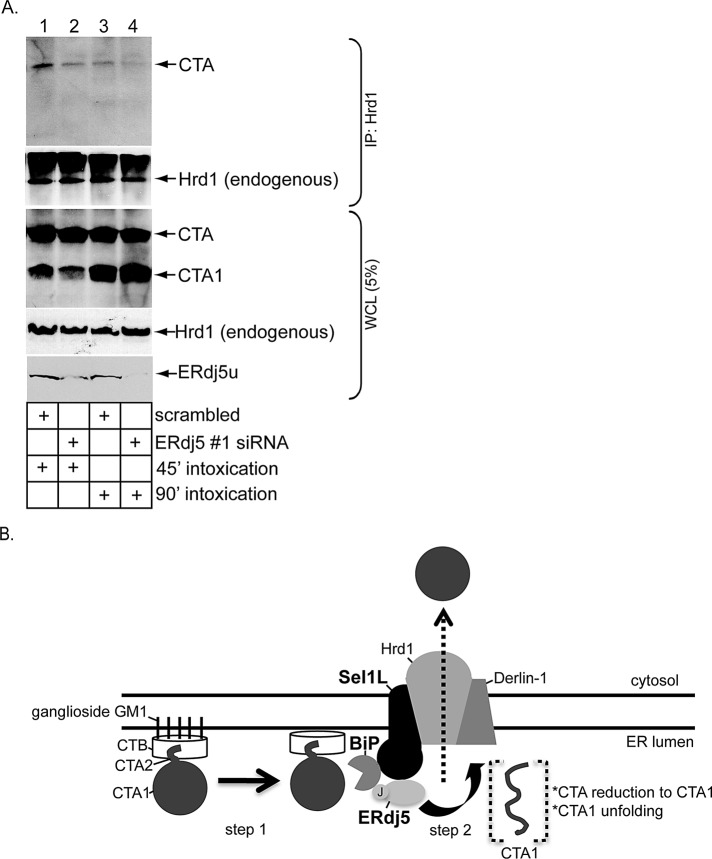
We demonstrated previously that the Hrd1-associated complex binds to and facilitates toxin retrotranslocation (Bernardi et al., 2010). We reasoned that, should ERdj5's regulation of the CTA–BiP interaction precede binding of CTA to Hrd1, silencing ERdj5 ought to block CTA–Hrd1 interaction. Indeed, in cells incubated with CT for either 45 min or 90 min, coimmunoprecipitation experiments revealed that endogenous Hrd1 binds CTA less efficiently at both time points when ERdj5 is knocked down using ERdj5 #1 siRNA (Figure 6A, top panel, compare lane 2 with lane 1 and lane 4 with lane 3). Under the scrambled condition, less CTA bound to Hrd1 at the 90-min time point than at the 45-min time point (Figure 6A, top panel, compare lane 3 with lane 1), perhaps reflecting either less toxin binding to Hrd1 or more efficient toxin release from Hrd1 at the later time point. Regardless, these findings demonstrate that ERdj5 regulates a critical upstream step that delivers CTA to Hrd1, likely due to ERdj5's ability to promote CTA-BiP binding.
FIGURE 6:
ERdj5 knockdown reduces Hrd1–CTA interaction. (A) Cells transfected with either scrambled or ERdj5 #1 siRNA were incubated with CT (10 nM) for 45 min or 90 min and lysed in a buffer containing 1% deoxy Big CHAP. The resulting WCLs were incubated with a Hrd1 antibody, and the immunoprecipitates were subjected to SDS–PAGE followed by immunoblotting with the indicated antibodies. WCL samples were also analyzed by immunoblotting with the indicated antibodies. (B) Model of ERdj5- and Sel1L-dependent CTA1 retrotranslocation. Using its J domain, ERdj5 stimulates BiP's ATPase activity, enabling BiP to engage the toxin (step 1). Because ERdj5 interacts with the Hrd1 adaptor Sel1L, this reaction occurs proximal to the Hrd1-associated complex. On release from BiP, CTA is reduced and unfolded and is competent for retrotranslocation across the Hrd1 membrane complex (step 2). See the text for more detailed discussion.
EDEM1 and OS-9 do not play important roles in CTA1 retrotranslocation
We identified ERdj5 as a direct Sel1L-binding partner that regulates CTA1 retrotranslocation. EDEM1 and OS-9 are two ER-resident, Sel1L-interacting proteins that execute important roles during ERAD (Christianson et al., 2008; Cormier et al., 2009). Accordingly, we tested whether EDEM1 and OS-9 control CTA1 retrotranslocation. When EDEM1 and OS-9 were knocked down in cells using their respective oligonucleotide-specific siRNAs (Figure S3A), the CTA1 level in the supernatant fraction derived from the semipermeabilized retrotranslocation assay was unaffected when compared with scrambled control (Figure S3B, top panel, compare lanes 2 and 3 with lane 1; quantified in Figure S3C). These findings indicate that EDEM1 and OS-9 do not play significant roles in facilitating ER-to-cytosol transport of CTA1.
DISCUSSION
Retrotranslocation of the enzymatic CTA1 subunit from the ER into the cytosol represents a decisive CT intoxication step. The toxin is postulated to hijack the ERAD machinery to gain entry into the cytosol (Hazes and Read, 1997). The central ERAD components are transmembrane E3 ubiquitin ligases (Christianson et al., 2011; Smith et al., 2011). In this context, our laboratory previously demonstrated an important role for the Hrd1 and gp78 E3 ubiquitin ligases in catalyzing CTA1 retrotranslocation (Bernardi et al., 2010). We also showed that Derlin-1, an adaptor of these ligases, delivers the toxin to Hrd1 and gp78 (Bernardi et al., 2010). Nonetheless, how CT is captured by these E3 membrane complexes to prepare for retrotranslocation is unclear. Using loss-of-function and gain-of-function approaches in a cell-based semipermeabilized system coupled with biochemical interaction studies, we found the ER-resident ERdj5 and Sel1L proteins promote CTA1 retrotranslocation. Our results depict a scenario in which ERdj5 localizes to the Hrd1 membrane complex by binding to the adaptor Sel1L (Figure 6B, step 1). Here, via its J domain, ERdj5 stimulates BiP's ATPase activity. This reaction enables BiP to bind the toxin, effectively delivering the toxin to the Hrd1 complex. Once the toxin is released from BiP, CTA is reduced to generate CTA1 via the action of an unidentified reductase. PDI then unfolds CTA1 (Tsai et al., 2001; Forster et al., 2006), priming the toxin for retrotranslocation across the Hrd1 complex (Figure 6B, step 2). How CTA is released from BiP to enable its subsequent reduction and unfolding remains unclear.
Specifically, our data demonstrate that ERdj5 knockdown decreases CTA1 ER-to-cytosol transport, indicating ERdj5 plays a critical role in CTA1 retrotranslocation. However, that ERdj5 knockdown did not block CTA1 retrotranslocation completely suggests that either any residual ERdj5 level is sufficient to support toxin retrotranslocation or that other ER-resident J proteins play a complementary role in driving toxin translocation. Indeed, a report suggests that the ER-resident J protein ERdj3 mediates CTA1 retrotranslocation (Massey et al., 2011), although it is unclear whether ERdj3 physically interacts with the Hrd1 membrane complex. This J protein was previously implicated in shiga toxin retrotranslocation (Yu and Haslam, 2005). In addition to ERdj3, the ER-resident ERdj4 J protein was recently shown to interact with the Hrd1 adaptor Derlin-1 (Lai et al., 2012) to control degradation of different ERAD substrates (Dong et al., 2008; Buck et al., 2010). Thus ERdj4 may potentially impact CTA1 ER-to-cytosol transport as well.
How might ERdj5 support toxin retrotranslocation? There are at least two possibilities. First, ERdj5's four thioredoxin domains may reduce CTA's single disulfide bond to generate the retrotranslocation CTA1 substrate. However, we found that ERdj5 knockdown did not affect CTA1 formation in cells, suggesting that ERdj5's thioredoxin domains are not likely to be responsible for reducing CTA's disulfide bond. Whether ERdj5's reductase activity impacts other ERAD components to indirectly control CTA1 retrotranslocation requires further experimentation. In this context, recent reports identified an ER flavoprotein called ERFAD (Riemer et al., 2009) bound to the PDI family member ERp90 (Riemer et al., 2011). As ERFAD interacts with both Sel1L and ERdj5 (Riemer et al., 2009), the ERFAD-ERp90 complex may provide the source of reductase activity to generate CTA1.
Another possibility by which ERdj5 promotes CTA1 retrotranslocation is through engaging the ER Hsp70 BiP. As BiP was shown previously to control CTA1 retrotranslocation in vitro by holding the toxin in a soluble, transport-competent state (Winkeler et al., 2003), it is possible that ERdj5's J domain stimulates BiP's ATPase activity to induce toxin binding. The importance of ER-resident J proteins and BiP in maintaining ERAD substrates in a soluble state competent for retrotranslocation has also been observed in yeast (Nishikawa et al., 2001). Indeed, we found BiP binds to CTA in cells, and this interaction is markedly decreased in ERdj5's absence, suggesting that ERdj5's J domain controls BiP–toxin interaction. Consistent with this result, overexpression of WT ERdj5, but not an ERdj5 J domain mutant, stimulated CTA1 retrotranslocation. Thus our evidence demonstrates that ERdj5's J domain plays a critical role during toxin retrotranslocation. That BiP binds to CTA and not CTA1 indicates that this chaperone interacts with the toxin prior to its reduction. Although perturbing BiP activity in cells leads to ER stress (Lee, 2005), thereby precluding an accurate assessment of BiP's role in mediating CTA1 retrotranslocation, our observations that ERdj5's J domain is both critical for promoting BiP–toxin binding and toxin retrotranslocation strongly suggest that BiP executes an important role in toxin ER-to-cytosol transport in cells.
As BiP binds to Sel1L in mammalian cells (Hosokawa et al., 2008), and Kar2p interacts with Hrd3p in yeast (Denic et al., 2006), we next explored the possibility that BiP's cochaperone ERdj5 might also bind to Sel1L in mammalian cells. Gel-filtration analysis suggests that a small portion of ERdj5 exists in a large-molecular-weight complex that cofractionates with Sel1L and Hrd1. Coimmunoprecipitation experiments, as well as in vitro binding studies using purified components, established a direct physical interaction between ERdj5 and Sel1L's N-terminal region (i.e., aa 1–372). Whether the four TPR domains contained within this region are responsible for contacting ERdj5 is unknown. Interestingly, a previous study found Sel1L's N-terminal region did not bind to its other chaperone adaptors, OS-9 and XTP3-B (Christianson et al., 2008), suggesting that Se11L's C-terminal lumenal domain proximal to the ER membrane mediates binding to these adaptors. We do not know whether Sel1L's N- or C-terminal domain engages EDEM1, another Sel1L adaptor. Regardless, it appears that different regions of Se11L's lumenal domain are geared to interact with different adaptors. This is likely a critical feature of Sel1L's role as a molecular scaffold, providing an ERAD substrate an opportunity to select and engage only a subset of Sel1L adaptors prior to transport across the Hrd1 complex; that CTA1 uses only ERdj5, but not EDEM1 nor OS-9, underscores this selectivity. Our binding studies also demonstrate that spliced and unspliced ERdj5 displayed a similar affinity to Sel1L (unpublished data). As spliced ERdj5 lacks 46 amino acids within a redox-inactive domain present in unspliced ERdj5 (i.e., the so-called Trxb1 domain; Hagiwara et al., 2011), this part is not likely to be responsible for engaging Sel1L.
Identifying an ERdj5-Sel1L complex prompted us to examine whether Sel1L functions in CTA1 retrotranslocation, especially considering that ERAD substrate degradation can occur using a Sel1L-independent mechanism (Mueller et al., 2006; Bernasconi et al., 2010; Ninagawa et al., 2011; Stanley et al., 2011). CT coprecipitated with Sel1L, suggesting a physical interaction between them. Moreover, a pool of CT cofractionated with Sel1L and Hrd1 in gel-filtration analyses. Although these findings do not indicate whether Sel1L or Hrd1 directly bind to the toxin, they are consistent with the premise that CT forms a complex with the Hrd1-associated retrotranslocation machinery, which includes Sel1L and Hrd1. Next our functional results demonstrate that, when Sel1L is knocked down, CTA1 ER-to-cytosol transport decreases by ∼50%, indicating Sel1L plays a role in this process. This effect is unlikely due to Hrd1 destabilization, because Hrd1's steady-state level is unchanged under the Sel1L knockdown condition, consistent with a recent report (Iida et al., 2011). In yeast, Sel1L's homologue Hrd3p stabilizes Hrd1p (Plemper et al., 1999; Gardner et al., 2000), whereas in mammals, Hrd1 stabilizes Sel1L (Iida et al., 2011). In our experiments, Hrd1 interacts efficiently with its adaptor, Derlin-1, even when Sel1L is silenced, suggesting that the Hrd1 membrane complex is largely intact in Sel1L's absence. This observation is also consistent with the finding that silencing Sel1L marginally affected the Hrd1 complex (Iida et al., 2011). One possible explanation for why down-regulating Sel1L only led to a 50% reduction in CTA1 retrotranslocation is that gp78, which also mediates CTA1 retrotranslocation (Bernardi et al., 2010), does not bind Sel1L (Hosokawa et al., 2008; Christianson et al., 2011). Therefore CTA1 translocation across the gp78 complex would not require Sel1L. This explanation raises the question as to how gp78, lacking Sel1L as a binding partner, is linked physically to the BiP-ERdj5 chaperones. Perhaps unidentified gp78 adaptors provide this link, or gp78 uses a different machinery to capture the toxin. Interestingly, Sel1L's role in CTA1 membrane transport is not restricted to this toxin, as a recent report suggests that Sel1L promotes ricin toxin A chain retrotranslocation (Redmann et al., 2011).
Finally, we found that silencing ERdj5 decreases CTA–Hrd1 interaction, indicating ERdj5 executes a role in CTA1 retrotranslocation upstream of Hrd1. This result further supports our model (Figure 6B), wherein Sel1L forms a scaffold on which the ERdj5-BiP chaperones dock and capture CT (and presumably other ERAD substrates). Because the BiP–Sel1L interaction does not require ERdj5, the CTA bound to BiP is likely targeted to the Sel1L/Hrd1 complex because of BiP's interaction with Sel1L, but not due to ERdj5's binding to Sel1L. Regardless, once CTA is released from BiP, it is eventually transferred to Hrd1 via Derlin-1 (Bernardi et al., 2010). As we are unable to establish any physical interaction between CTA and ERdj5 (unpublished data), we believe ERdj5 does not serve as a chaperone to transfer the released toxin to Hrd1. After reduction and unfolding, CTA1 translocates across the Hrd1 complex to reach the cytosol. We note that, in addition to acting as a scaffold to enable binding of other adaptors, the yeast Sel1L homologue, Hrd3p, itself can engage ERAD substrates directly (Denic et al., 2006; Gauss et al., 2006; Stanley et al., 2011). These two possibilities may operate simultaneously to expand the repertoire of substrates that can be captured by the Hrd1 membrane complex to initiate retrotranslocation. Clearly, substrate recognition during ERAD is a complex process, regulated by different E3 ligase adaptors in a substrate-dependent manner.
MATERIALS AND METHODS
Materials
Polyclonal antibodies against PDI and Hsp90 and an ERdj5 antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); another ERdj5 and Hrd1 antibodies were purchased from Proteintech Group (Chicago, IL); the Sel1L antibody was from Enzo Life Sciences (Farmingdale, NY); the FLAG and EDEM1 antibodies were from Sigma-Aldrich (St. Louis, MO); the S-tag and OS-9 antibodies were from Abcam (Cambridge, MA); and the polyclonal and the monoclonal BiP antibodies were from BD Biosciences (San Jose, CA). The polyclonal CTA antibody was produced against denatured CTA purchased from EMD Biosciences (San Diego, CA). The polyclonal Derlin-1 antibody was a gift from T. Rapoport (Harvard University), and the S-Sel1L (1–372) construct was a gift from J. Christianson (Oxford University). Purified CT was purchased from EMD Biosciences.
Generation of ERdj5, BiP, Grp94, and Sel1L constructs
BiP, Grp94, and WT ERdj5s cDNAs were isolated from 293T cells total RNA. For generating BiP-S, BiP-FLAG, and WT ERdj5s-FLAG, the C-terminal S- or FLAG-tag sequences were inserted before each ER retention KDEL sequence using primers containing the respective tag sequences. The resulting PCR products were inserted into a pcDNA3.1(−) vector using a standard cloning method. For generation of WT ERdj5u-FLAG, the spliced region was amplified using overlapping PCR, and the resulting PCR product was inserted into WT ERdj5s-FLAG using a standard cloning method. For generation of H63Q ERdj5s-FLAG, histidine at the residue 63 was mutated to glutamine with overlapping PCR. The resulting PCR product was inserted into pcDNA3.1(−) vector using a standard cloning method. For generation of Grp94-FLAG, the BiP coding sequence of BiP-FLAG was replaced with the Grp94 coding sequence. For generation of S/His-Sel1L (1–372), the histidine tag sequence was inserted between the S tag and Sel1L coding sequences of the S-Sel1L (1–372) expression vector using an overlapping PCR method. All vectors were sequenced prior to experimentation.
Retrotranslocation assay
As described previously in Bernardi et al. (2008), except after digitonin treatment, cells were centrifuged at 100,000 rpm for 10 min in a TLA 100 rotor (Beckman Coulter, Brea, CA).
Cell transfection
The 293T cells were grown to 30% confluency on a 10-cm dish prior to transfection with the Effectene system (Qiagen, Chatsworth, CA). The cells were grown for an additional 48 h prior to experimentation.
siRNA knockdown of ERdj5, Sel1L, EDEM1, and OS-9
The sequences of the siRNAs used in this study are: ERdj5 #1 siRNA (5′-GGACAAGGAACCAAAGAAT-3′; Invitrogen, Carlsbad, CA); ERdj5 #2 siRNA (5′-GCTCTTGGCTAGGATGATT-3′; Invitrogen); Sel1L #1 siRNA (5′-GGCTATACTGTGGCTAGAA-3′; Invitrogen); Sel1l #2 siRNA: (5′-GCUCAGUAGUACAGAGAAUUU-3′; Dharmacon (Thermo Fisher Scientific, Lafayette, CO); EDEM1 siRNA: (5′-GAGCAAUGAUACAGGAUUAUU-3′; Dharmacon); OS-9 #1 siRNA: (5′-AUCCCUGAGUUGUUGAGCCCAAU-3′; Invitrogen); and OS-9 #2 siRNA: (5′-UAACAAACUGGACAGCAGCGUUUCC-3′; Invitrogen). Duplex siRNA (20 nM) was transfected into 293T cells for 24 h according to the manufacturer's protocol.
Coimmunoprecipitation
The 293T cells were incubated with or without CT (10 or 100 nM) for 90 min. Cells were harvested and lysed in buffer containing KOAc (150 mM) Tris (pH 7.5, 30 mM), MgCl2 (4 mM), NEM (10 mM), and protease inhibitors with 1% deoxy Big CHAP (Sigma-Aldrich, St. Louis, MO) for 30 min on ice. Cells were centrifuged at 16,000 × g for 10 min, and the supernatant was used for immunoprecipitation experiments. Where indicated, S-antibody–conjugated beads (EMB Millipore, Darmstadt, Germany) or FLAG-antibody–conjugated beads (Sigma-Aldrich) were added to the WCL and incubated at 4°C for 2–4 h. The immune complex was sedimented, washed, and subjected to SDS–PAGE followed by immunoblotting with the appropriate antibody. For the S/His-Sel1L (1–372)–BiP interaction experiment, cells were first treated with 0.02% digitonin and centrifuged. The resulting pellet was solubilized with 1% Triton X-100 to generate the lysate used for immunoprecipitation.
Trypsin digestion
A WCL expressing S/His-Sel1L (1–372) was incubated with 5, 15, or 50 μg/ml trypsin at 4°C for 30 min. Samples were subjected to SDS–PAGE followed by immunoblotting using an antibody against Sel1L.
Gel filtration
The 293T cells were solubilized in 1% digitonin, and the resulting WCL was loaded onto a Bio-Sil SEC 400 gel-filtration column (Bio-Rad, Hercules, CA) and separated with a buffer containing 0.05% digitonin. For the CT-intoxicated sample, 293T cells were first semipermeabilized with 0.01% digitonin and centrifuged to remove any toxin that arrived in the cytosol. The resulting pellet fraction was further solubilized in 1% digitonin and loaded onto the column. Forty fractions (0.5 ml each) were collected, and 0.05 ml of fractions 11–24 was subjected to SDS–PAGE followed by immunoblotting with CTA, ERdj5, Sel1L, and Hrd1 antibodies. For the ERdj5 or CTA immunoblot, 0.3 ml of fractions 11–24 was concentrated and subjected to SDS–PAGE.
Purification of GFP-FLAG, WT ERdj5u-FLAG, and S/His-Sel1L (1–372)
For purification of GFP-FLAG and WT ERdj5u-FLAG, ∼5 × 107 293T cells were transfected with the DNA constructs. At 48 h posttransfection, cells were harvested; lysed in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, and a protease inhibitor cocktail; and centrifuged at 16,100 × g for 10 min. The resulting supernatant was mixed with anti-FLAG M2 agarose beads (Sigma-Aldrich) and incubated at 4ºC for 2 h. After extensive washing with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, and 0.1% Triton X-100, bound proteins were eluted with 0.25 mg/ml FLAG peptide (Sigma-Aldrich). For purification of S/His-Sel1L (1–372), ∼1.5 × 108 293T cells were transfected with the S/His-Sel1L (1–372) construct using Polyethylenimine “Max” (Polysciences, Warrington, PA). At 48 h posttransfection, cells were harvested; semipermeabilized in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.02% digitonin, and a protease inhibitor cocktail (Roche, Indianapolis, IN); and centrifuged at 16,100 × g for 10 min. The resulting pellet fraction was further lysed with a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, and a protease inhibitor cocktail, and centrifuged at 50,000 × g for 20 min in the TLA 100.3. The resulting lysate was mixed with a stock imidazole solution to generate a final 30-mM imidazole sample solution and applied to a HisTrap HP column (GE Healthcare, Waukesha, WI) in a fast protein liquid chromatography (FPLC) system (Bio-Rad). After the column was extensively washed with a buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 0.1% Triton X-100, and 30 mM imidazole, bound proteins were eluted with a 30–500 mM imidazole gradient. The peak fractions of S/His-Sel1L (1–372) were pooled and desalted with a spin desalting column (Thermo Fisher Scientific). The desalted sample was applied to a UNO Q column in the FPLC system and eluted with a 50–500 mM NaCl gradient. The peak fractions were pooled, concentrated with a Microcon 30,000 MWCO (Millipore, Billerica, MA), and dialyzed against a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, and 0.1% Triton X-100.
Supplementary Material
Acknowledgments
We thank Christopher Walczak (Michigan) for critical review of the manuscript. B.T. holds an Investigator in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund and is funded by the National Institutes of Health (R01 083252-04). L.B. is a member of the Cellular and Molecular Biology program at the University of Michigan.
Abbreviations used:
- CT
cholera toxin
- CTA
cholera toxin catalytic A subunit
- CTB
cholera toxin catalytic B subunit
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FPLC
fast protein liquid chromatography
- GFP
green fluorescent protein
- PDI
protein disulfide isomerase
- siRNA
small interfering RNA
- TPR
tetratricopeptide repeat
- UPR
unfolded protein response
- WT
wild-type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0522) on January 30, 2013.
REFERENCES
- Abujarour RJ, Dalal S, Hanson PI, Draper RK. p97 is in a complex with cholera toxin and influences the transport of cholera toxin and related toxins to the cytoplasm. J Biol Chem. 2005;280:15865–15871. doi: 10.1074/jbc.M406316200. [DOI] [PubMed] [Google Scholar]
- Bernardi KM, Forster ML, Lencer WI, Tsai B. Derlin-1 facilitates the retrotranslocation of cholera toxin. Mol Biol Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye Y, Tsai B. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retrotranslocation. Mol Biol Cell. 2010;21:140–151. doi: 10.1091/mbc.E09-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid NJ, Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–492. doi: 10.1016/j.tibs.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2011;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier JH, Tamura T, Sunryd JC, Hebert DN. EDEM1 recognition and delivery of misfolded proteins to the Sel1L-containing ERAD complex. Mol Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dixit G, Mikoryak C, Hayslett T, Bhat A, Draper RK. Cholera toxin up-regulates endoplasmic reticulum proteins that correlate with sensitivity to the toxin. Exp Biol Med (Maywood) 2008;233:163–175. doi: 10.3181/0705-RM-132. [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell. 2008;19:2620–2630. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster ML, Sivick K, Park YN, Arvan P, Lencer WI, Tsai B. Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J Cell Biol. 2006;173:853–859. doi: 10.1083/jcb.200602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga Y, Wolf AA, Rodighiero C, Wheeler H, Tsai B, Allen L, Jobling MG, Rapoport T, Holmes RK, Lencer WI. Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulum. Mol Biol Cell. 2003;14:4783–4793. doi: 10.1091/mbc.E03-06-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol. 2011;13:1305–1314. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- Goodwin EC, et al. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. MBio. 2011;2:e00101. doi: 10.1128/mBio.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SH, Chen JZ, Ying K, Wang S, Jin W, Qian J, Zhao EP, Xie Y, Mao YM. Cloning and identification of a novel cDNA which encodes a putative protein with a DnaJ domain and a thioredoxin active motif, human macrothioredoxin. Biochem Genet. 2003;41:245–253. doi: 10.1023/a:1025510502147. [DOI] [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Hazes B, Read RJ. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 1997;36:11051–11054. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida Y, Fujimori T, Okawa K, Nagata K, Wada I, Hosokawa N. SEL1L protein critically determines the stability of the HRD1-SEL1L endoplasmic reticulum-associated degradation (ERAD) complex to optimize the degradation kinetics of ERAD substrates. J Biol Chem. 2011;286:16929–16939. doi: 10.1074/jbc.M110.215871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Tsai B. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog. 2011;7:e1002037. doi: 10.1371/journal.ppat.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe M, Ye Y, Wagner JS, De Luca HE, Kern E, Rapoport TA, Lencer WI. Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J Biol Chem. 2005;280:28127–28132. doi: 10.1074/jbc.M503138200. [DOI] [PubMed] [Google Scholar]
- Lai CW, Otero JH, Hendershot LM, Snapp E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem. 2012;287:7969–7978. doi: 10.1074/jbc.M111.311290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Lencer WI, Tsai B. The intracellular voyage of cholera toxin: going retro. Trends Biochem Sci. 2003;28:639–645. doi: 10.1016/j.tibs.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey S, Burress H, Taylor M, Nemec KN, Ray S, Haslam DB, Teter K. Structural and functional interactions between the cholera toxin A1 subunit and ERdj3/HEDJ, a chaperone of the endoplasmic reticulum. Infect Immun. 2011;79:4739–4747. doi: 10.1128/IAI.05503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P, Bernardi KM, Tsai B. The Ero1α-PDI redox cycle regulates retrotranslocation of cholera toxin. Mol Biol Cell. 2010;21:1305–1313. doi: 10.1091/mbc.E09-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninagawa S, Okada T, Takeda S, Mori K. SEL1L is required for endoplasmic reticulum-associated degradation of misfolded luminal proteins but not transmembrane proteins in chicken DT40 cell line. Cell Struct Funct. 2011;36:187–195. doi: 10.1247/csf.11018. [DOI] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retrotranslocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112:4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- Redmann V, Oresic K, Tortorella LL, Cook JP, Lord M, Tortorella D. Dislocation of ricin toxin A chains in human cells utilizes selective cellular factors. J Biol Chem. 2011;286:21231–21238. doi: 10.1074/jbc.M111.234708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J, Appenzeller-Herzog C, Johansson L, Bodenmiller B, Hartmann-Petersen R, Ellgaard L. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2009;106:14831–14836. doi: 10.1073/pnas.0900742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J, Hansen HG, Appenzeller-Herzog C, Johansson L, Ellgaard L. Identification of the PDI-family member ERp90 as an interaction partner of ERFAD. PLoS One. 2011;6:e17037. doi: 10.1371/journal.pone.0017037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodighiero C, Tsai B, Rapoport TA, Lencer WI. Role of ubiquitination in retrotranslocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 2002;3:1222–1227. doi: 10.1093/embo-reports/kvf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky DE, Cho JA, Chinnapen H, Massol RH, Chinnapen DJ, Wagner JS, De Luca HE, Kam W, Paw BH, Lencer WI. Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J Clin Invest. 2010;120:4399–4409. doi: 10.1172/JCI42958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler BD. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol Rev. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley AM, Carvalho P, Rapoport T. Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking. FEBS Lett. 2011;585:1281–1286. doi: 10.1016/j.febslet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Navarro-Garcia F, Huerta J, Burress H, Massey S, Ireton K, Teter K. Hsp90 is required for transfer of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J Biol Chem. 2010;285:31261–31267. doi: 10.1074/jbc.M110.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Wernick NL, De Luca H, Kam WR, Lencer WI. N-terminal extension of the cholera toxin A1-chain causes rapid degradation after retrotranslocation from endoplasmic reticulum to cytosol. J Biol Chem. 2010;285:6145–6152. doi: 10.1074/jbc.M109.062067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkeler A, Gödderz D, Herzog V, Schmitz A. BiP-dependent export of cholera toxin from endoplasmic reticulum-derived microsomes. FEBS Lett. 2003;554:439–442. doi: 10.1016/s0014-5793(03)01217-1. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Haslam DB. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect Immun. 2005;73:2524–2532. doi: 10.1128/IAI.73.4.2524-2532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.