Abstract
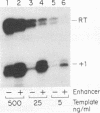
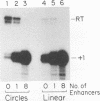
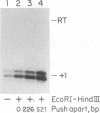
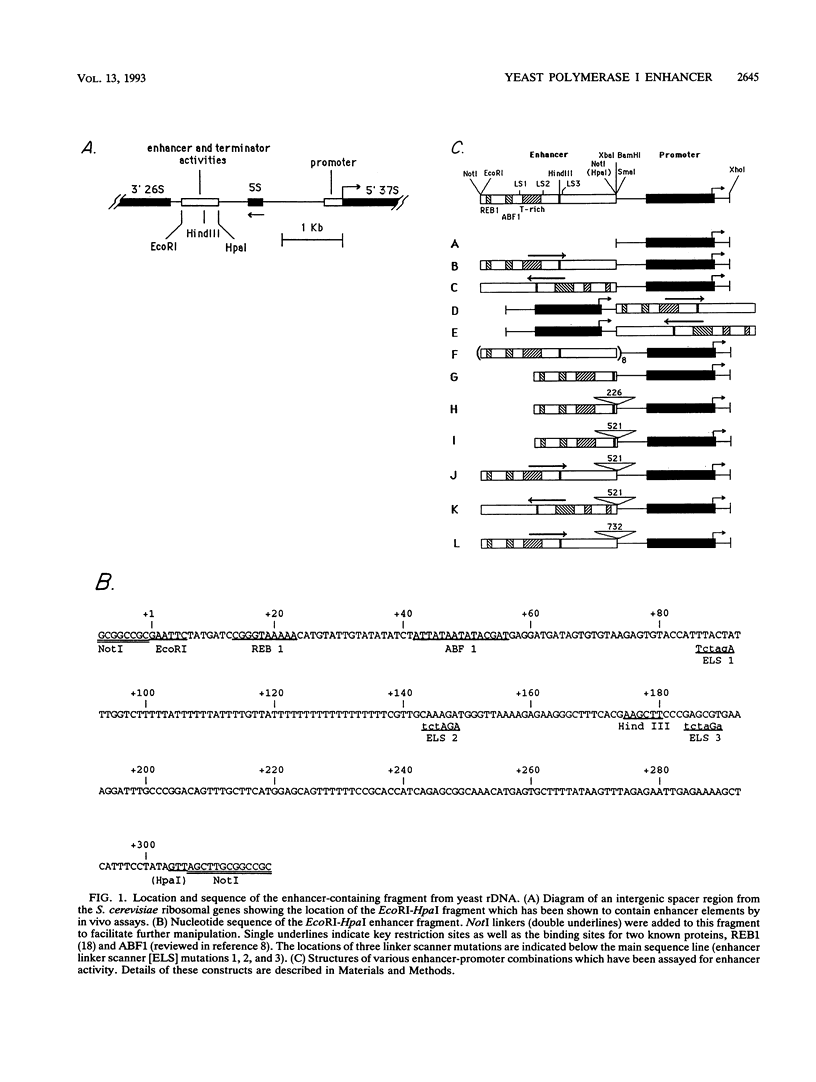
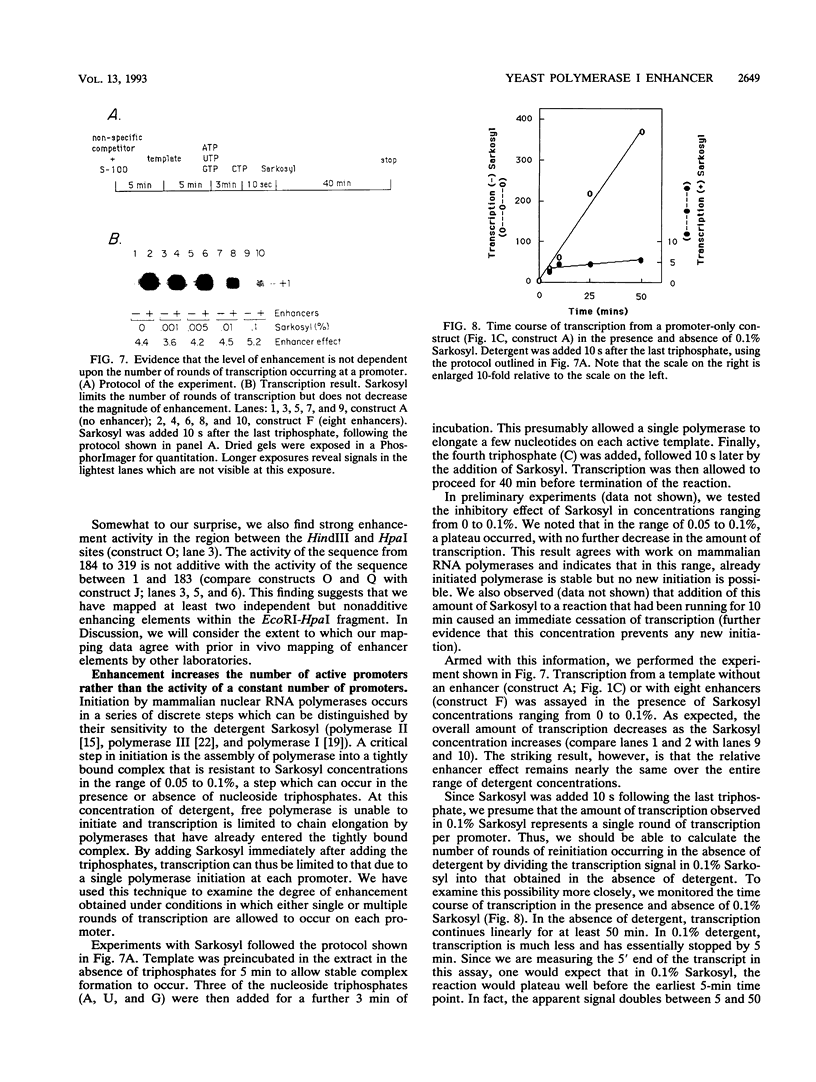
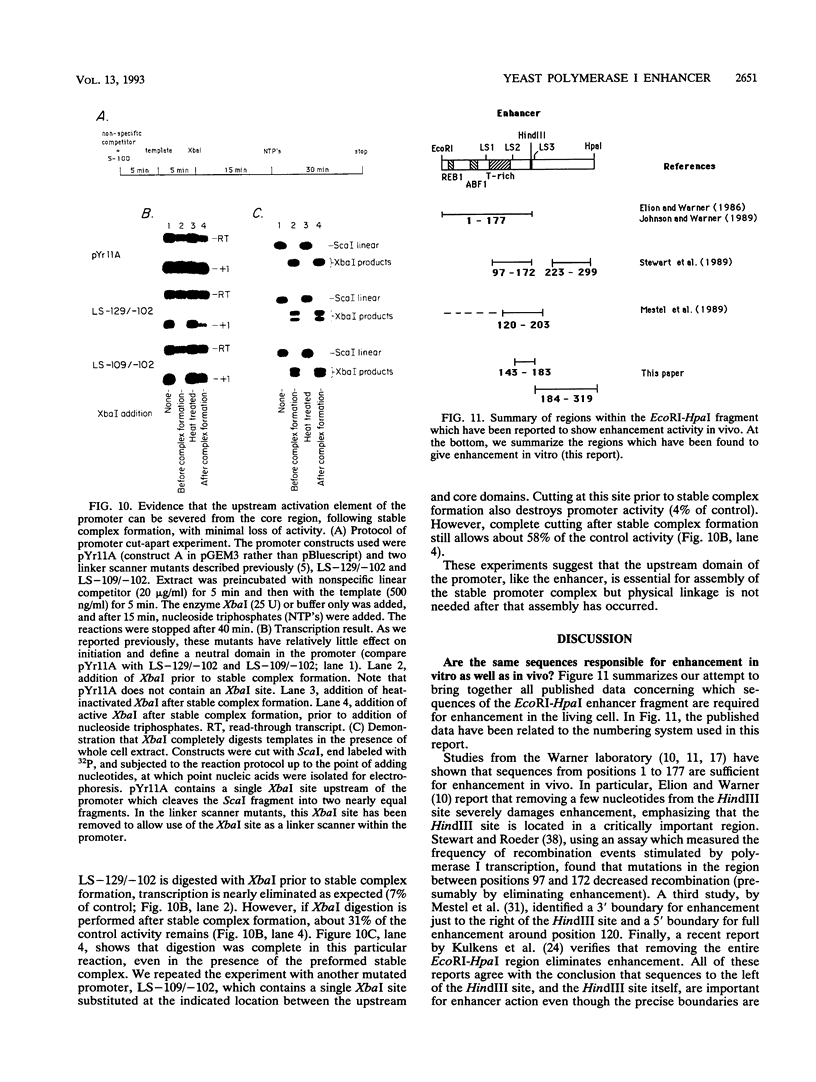
In vitro conditions are reported under which an EcoRI-HpaI fragment of the Saccharomyces cerevisiae ribosomal gene spacer will enhance transcription from an adjacent RNA polymerase I promoter. Enhancement is largely independent of orientation and distance and is proportional to copy number. Mapping experiments reveal that two separate regions of the EcoRI-HpaI fragment are independently capable of promoter stimulation. These regions appear to correspond to elements which have been shown by previous workers to cause enhancement in vivo. Using the detergent Sarkosyl to limit the number of rounds of transcription from each promoter, we found that the degree of enhancement is similar whether one or many rounds of transcription occur. This finding supports a model in which the enhancer increases the number of stable promoter complexes but does not alter the loading of polymerase on an active promoter. Once the stable promoter complex is formed, the enhancer can be physically severed from the promoter with no loss of enhancement. Likewise, the upstream activation region of the promoter can be severed from the core promoter domain once the stable complex has been formed. These results are interpreted to mean that the enhancer functions only to assist stable complex formation and, once that is accomplished, the enhancer is dispensable.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Bellomy G. R., Record M. T., Jr Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog Nucleic Acid Res Mol Biol. 1990;39:81–128. doi: 10.1016/s0079-6603(08)60624-8. [DOI] [PubMed] [Google Scholar]
- Butlin M., Quincey R. The yeast rRNA gene enhancer does not function by recycling RNA polymerase I and cannot act as a UAS. Curr Genet. 1991 Jul;20(1-2):9–16. doi: 10.1007/BF00312759. [DOI] [PubMed] [Google Scholar]
- Carey M., Leatherwood J., Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990 Feb 9;247(4943):710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Choe S. Y., Schultz M. C., Reeder R. H. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1992 Jan 25;20(2):279–285. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992 Mar 6;68(5):965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992 May 15;69(4):685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Doorenbosch T., Mager W. H., Planta R. J. Multifunctional DNA-binding proteins in yeast. Gene Expr. 1992;2(3):193–201. [PMC free article] [PubMed] [Google Scholar]
- Dunaway M., Dröge P. Transactivation of the Xenopus rRNA gene promoter by its enhancer. Nature. 1989 Oct 19;341(6243):657–659. doi: 10.1038/341657a0. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Kelly J. L., Lehman I. R. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Horikoshi M., Roeder R. G., Green M. R. Analysis of the role of the transcription factor ATF in the assembly of a functional preinitiation complex. Cell. 1988 Sep 23;54(7):1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Horikoshi M., Hai T., Lin Y. S., Green M. R., Roeder R. G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988 Sep 23;54(7):1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- Johnson S. P., Warner J. R. Unusual enhancer function in yeast rRNA transcription. Mol Cell Biol. 1989 Nov;9(11):4986–4993. doi: 10.1128/mcb.9.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Q. D., Morrow B. E., Warner J. R. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990 Oct;10(10):5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Nagamine M., Kominami R., Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986 Oct;6(10):3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Hidaka M., Nishizawa M., Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Jun;233(3):355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- Kovelman R., Roeder R. G. Sarkosyl defines three intermediate steps in transcription initiation by RNA polymerase III: application to stimulation of transcription by E1A. Genes Dev. 1990 Apr;4(4):646–658. doi: 10.1101/gad.4.4.646. [DOI] [PubMed] [Google Scholar]
- Kulkens T., Riggs D. L., Heck J. D., Planta R. J., Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991 Oct 11;19(19):5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T., van der Sande C. A., Dekker A. F., van Heerikhuizen H., Planta R. J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992 Dec;11(12):4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell. 1984 May;37(1):285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Lang W. H., Reeder R. H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science. 1992 Sep 18;257(5077):1682–1685. doi: 10.1126/science.1388287. [DOI] [PubMed] [Google Scholar]
- Lue N. F., Kornberg R. D. Accurately initiated, enhancer-dependent transcription by RNA polymerase I in yeast extracts. J Biol Chem. 1990 Oct 25;265(30):18091–18094. [PubMed] [Google Scholar]
- McStay B., Hu C. H., Pikaard C. S., Reeder R. H. xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X. laevis. EMBO J. 1991 Aug;10(8):2297–2303. doi: 10.1002/j.1460-2075.1991.tb07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestel R., Yip M., Holland J. P., Wang E., Kang J., Holland M. J. Sequences within the spacer region of yeast rRNA cistrons that stimulate 35S rRNA synthesis in vivo mediate RNA polymerase I-dependent promoter and terminator activities. Mol Cell Biol. 1989 Mar;9(3):1243–1254. doi: 10.1128/mcb.9.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow B. E., Johnson S. P., Warner J. R. The rRNA enhancer regulates rRNA transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1283–1289. doi: 10.1128/mcb.13.2.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A. F., van Heerikhuizen H., Planta R. J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989 Dec 11;17(23):9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Roan J. G., Dunaway M. Spacer regulation of Xenopus ribosomal gene transcription: competition in oocytes. Cell. 1983 Dec;35(2 Pt 1):449–456. doi: 10.1016/0092-8674(83)90178-2. [DOI] [PubMed] [Google Scholar]
- Schultz M. C., Choe S. Y., Reeder R. H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. C., Reeder R. H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992 May 15;69(4):697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Stewart S. E., Roeder G. S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Aug;9(8):3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Treisman R., Maniatis T. Simian virus 40 enhancer increases number of RNA polymerase II molecules on linked DNA. Nature. 1985 May 2;315(6014):73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- Weber F., Schaffner W. Simian virus 40 enhancer increases RNA polymerase density within the linked gene. Nature. 1985 May 2;315(6014):75–77. doi: 10.1038/315075a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Mager W. H., Nieuwint R. T., Wassenaar G. M., van der Kuyl A. C., Murre J. J., Hoekman M. F., Brockhoff P. G., Planta R. J. Analysis of upstream activation sites of yeast ribosomal protein genes. Nucleic Acids Res. 1987 Aug 11;15(15):6037–6048. doi: 10.1093/nar/15.15.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sande C. A., Kulkens T., Kramer A. B., de Wijs I. J., van Heerikhuizen H., Klootwijk J., Planta R. J. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989 Nov 25;17(22):9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]