Abstract
The presence of persister cells has been proposed as a factor in biofilm resilience. In the present study we investigated whether persister cells are present in Burkholderia cepacia complex (Bcc) biofilms, what the molecular basis of antimicrobial tolerance in Bcc persisters is, and how persisters can be eradicated from Bcc biofilms. After treatment of Bcc biofilms with high concentrations of various antibiotics often a small subpopulation survived. To investigate the molecular mechanism of tolerance in this subpopulation, Burkholderia cenocepacia biofilms were treated with 1024 µg/ml of tobramycin. Using ROS-specific staining and flow cytometry, we showed that tobramycin increased ROS production in treated sessile cells. However, approximately 0.1% of all sessile cells survived the treatment. A transcriptome analysis showed that several genes from the tricarboxylic acid cycle and genes involved in the electron transport chain were downregulated. In contrast, genes from the glyoxylate shunt were upregulated. These data indicate that protection against ROS is important for the survival of persisters. To confirm this, we determined the number of persisters in biofilms formed by catalase mutants. The persister fraction in ΔkatA and ΔkatB biofilms was significantly reduced, confirming the role of ROS detoxification in persister survival. Pretreatment of B. cenocepacia biofilms with itaconate, an inhibitor of isocitrate lyase (ICL), the first enzyme in the glyoxylate shunt, reduced the persister fraction approx. 10-fold when the biofilms were subsequently treated with tobramycin. In conclusion, most Bcc biofilms contain a significant fraction of persisters that survive treatment with high doses of tobramycin. The surviving persister cells downregulate the TCA cycle to avoid production of ROS and at the same time activate an alternative pathway, the glyoxylate shunt. This pathway may present a novel target for combination therapy.
Introduction
Burkholderia cenocepacia is a member of the Burkholderia cepacia complex (Bcc), a group of 17 closely related and phenotypically similar species [1]. These bacteria are opportunistic pathogens that can cause severe lung infections in immunocompromised people, including cystic fibrosis (CF) patients [2]. The prevalence and outcome of infections appear to be species dependent and infections with Burkholderia multivorans and B. cenocepacia are associated with high rates of transmission and high mortality [3]. B. cenocepacia J2315 belongs to the highly transmissible ET12 lineage which has infected many CF patients in Canada, the UK and various European countries [4]. Unfortunately, Bcc organisms are difficult to eradicate because of their innate resistance to a wide range of antibiotics and their capacity to form biofilms [5]. Mechanisms of resistance include changes in lipopolysaccharide structure, the presence of several multidrug efflux pumps, inducible chromosomal β-lactamases and altered penicillin-binding proteins [6]. In addition, biofilm-associated cells are often more tolerant to antimicrobials than planktonic cells [7] due to reduced drug penetration in the biofilm, the lower growth rate of sessile cells and/or the expression of specific resistance genes [8], [9]. Even when patients are treated with high doses of antibiotics known to be effective against Bcc species in vitro, it is often impossible to clear the infection [10].
Persister cells, cells that have entered a dormant multidrug-tolerant state, are thought to be involved in the recalcitrance of biofilm related infections [11]. These persisters are not mutants but phenotypic variants of the wild type [11]. Upon treatment of a biofilm with high doses of an antibiotic most of the bacteria are killed, but some of them neither grow nor die. After removal of the antibiotic, these surviving cells will start to grow and will give rise to a new infection. The resulting biofilm will again only contain a very small fraction of persister cells [12]. The mechanisms leading to persistence are not well understood. Screening knockout mutant libraries has not led to the identification of mutants completely lacking persister cell formation, suggesting that dormancy mechanisms are redundant [11]. Currently, it is assumed that toxin/antitoxin modules are involved in persister formation [12]. Toxins are proteins that inhibit important cellular functions such as translation or replication. This condition can be reversed by the expression of the corresponding antitoxin which can form an inactive complex with the toxin. By causing a reversible dormant state, toxins protect bacteria against antibiotics which require active targets in order to be effective [11]. The phenomenon of persister formation in exponentially growing planktonic cultures has been demonstrated for several micro-organisms (including Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus and Mycobacterium tuberculosis) [11], but has so far not been investigated in the Bcc.
Recently, Kohanski et al. demonstrated that the production of reactive oxygen species (ROS) contributes to the antimicrobial activity of bactericidal antibiotics. The primary drug-target interactions stimulate the oxidation of NADH via the electron transport chain, which itself is dependent on the tricarboxylic acid cycle (TCA). A hyperactivation of the electron transport chain results in increased superoxide formation, leading to damage to iron-sulfur clusters in proteins with the release of ferrous iron. This ferrous iron can be oxidised in the Fenton reaction with the production of hydroxyl radicals capable of damaging proteins, DNA and lipids and ultimately leading to cell death [13].
The glyoxylate cycle is an anaplerotic pathway of the TCA cycle, which bypasses the decarboxylation steps in which NADH is produced. This pathway allows microorganisms to utilize simple carbon compounds as a carbon source [14]. The glyoxylate cycle is absent in humans, making it an interesting drug target [14]. For example, Van Schaik et al [15] found that inhibition of isocitrate lyase (ICL, the first enzyme of this shunt), by itaconate during experimental chronic Burkholderia pseudomallei lung infections forces the infection into an acute state, which can then be treated with antibiotics.
In the present study we wanted to investigate whether persister cells are present in Bcc biofilms, what the molecular basis of antimicrobial tolerance in Bcc persisters is, and how persisters can be eradicated from Bcc biofilms.
Materials and Methods
Strains and Culture Conditions
The strains used in the present study are shown in Tables 1 and 2 [16], [17]. All strains were cultured at 37°C on Luria-Bertani agar (LBA, Oxoid, Hampshire, UK) or on Mueller Hinton agar (MHA, Oxoid). Overnight cultures were diluted in Luria-Bertani broth (LBB, Oxoid) and incubated aerobically at 37°C. The succinate dehydrogenase antisense overexpression mutants were grown on LBA supplemented with 800 µg/ml trimethoprim (Tp) (Ludeco, Brussels, Belgium) or in LBB supplemented with 800 µg/ml Tp and 0.2% rhamnose (Sigma Aldrich, Bornem, Belgium).
Table 1. Strains used in the present study.
| Strain | LMG number | Strain information | Source (reference) |
| B. cenocepacia J2315 | LMG 16656 | CF patient, UK, ET12 strain | BCCM/LMG Bacteria Collection |
| B. cenocepacia K56-2 | LMG 18863 | CF patient, Canada | BCCM/LMG Bacteria Collection |
| B. cenocepacia C5424 | LMG 18827 | CF patient, Canada | Miguel Valvano [16] |
| B. cenocepacia MDL1 | C5424 ΔkatA mutant strain | Miguel Valvano [16] | |
| B. cenocepacia MDL2 | C5424 ΔkatB mutant strain | Miguel Valvano [16] | |
| B. cenocepacia D2 | ΔBCAL2118ΔBCAM1588 | This study | |
| B. cenocepacia SDH7 | Overexpression of BCAM0967 | This study | |
| B. cenocepacia SDH8 | Overexpression of BCAM0968 | This study | |
| B. ambifaria AMMD | LMG 19182 | Pea rhizosphere, USA | BCCM/LMG Bacteria Collection |
| B. cenocepacia HI2424 | LMG 24507 | Soil, USA, PHDC strain | John LiPuma [17] |
| B. lata ATCC 17769 | LMG 6992 | Soil, Trinidad and Tobago | BCCM/LMG Bacteria Collection |
| B. multivorans ATCC 17616 | LMG 17588 | Soil, USA | BCCM/LMG Bacteria Collection |
| B. vietnamiensis ATCC 53617 | LMG 22486 | Wastewater, USA | BCCM/LMG Bacteria Collection |
| B. dolosa R-5670 | LMG 18943 | CF patient, US | BCCM/LMG Bacteria Collection |
| B. contaminans CCUG 34411 | LMG 16227 | CF patient, Sweden | BCCM/LMG Bacteria Collection |
| B. ubonensis NCTC 13147 | LMG 20358 | Surface soil, Thailand | BCCM/LMG Bacteria Collection |
| B. cepacia ATCC 25416 | LMG 1222 | Allium cepa, US | BCCM/LMG Bacteria Collection |
| B. pyrrocinia ATCC 15958 | LMG 14191 | Soil, US | BCCM/LMG Bacteria Collection |
Table 2. The MIC and fraction of persisters for early and late Bcc clonal isolates obtained from infected CF patients.
| Species | Strain | Time interval (years) | MIC | Average % persisters(n = 3) | SD | |
| Clonal isolates from Canada | ||||||
| B. multivorans | C8298 | 8 | Early | 64 | 0.1551 | 0.2352 |
| D2156 | Late | 64 | 0.0068 | 0.0076 | ||
| B. multivorans | C8814 | 5 | Early | 32 | 0.3411 | 0.3080 |
| D0999 | Late | 128 | 0.0012 | 0.0008 | ||
| B. multivorans | C6396 | 9 | Early | 64 | 0.0031 | 0.0004 |
| D0913 | Late | 128 | 0.0005 | 0.0003 | ||
| B. cenocepacia | C4053 | 6 | Early | 128 | 0.0004 | 0.0004 |
| C6121 | Late | 64 | 0.0019 | 0.0009 | ||
| B. cenocepacia | C3921 | 10 | Early | 1024 | 0.0249 | 0.0270 |
| C9343 | Late | 1024 | 0.0051 | 0.0044 | ||
| B. cenocepacia | C6483 | 4 | Early | 256 | 0.0019 | 0.0031 |
| C8474 | Late | 256 | 0.0214 | 0.0363 | ||
| B. cenocepacia | C4629 | 7 | Early | 128 | 0.0226 | 0.0326 |
| C8482 | Late | 128 | 0.0036 | 0.0007 | ||
| B. cenocepacia | C5876 | 8 | Early | 128 | 0.0020 | 0.0004 |
| D0465 | Late | 256 | 0.0000 | 0.0001 | ||
| B. cenocepacia | C5424 | 3 | Early | 256 | 0.0527 | 0.0753 |
| C7376 | Late | 128 | 0.0114 | 0.0152 | ||
| Clonal isolates from the US | ||||||
| B. cenocepacia | AU0326 | 11 | Early | 256 | 0.0066 | 0.0095 |
| AU18962 | Late | 512 | 0.0276 | 0.0441 | ||
| B. cenocepacia | AU0734 | 13 | Early | 256 | 0.0010 | 0.0007 |
| AU21801 | Late | 256 | 0.0001 | 0.0001 | ||
| B. vietnamiensis | AU0808 | 12 | Early | 16 | 0.0019 | 0.0025 |
| AU21645 | Late | 8 | 0.0092 | 0.0116 | ||
| B. dolosa | AU3503 | 10 | Early | 32 | 0.0040 | 0.0044 |
| AU21993 | Late | 32 | 0.0821 | 0.0702 | ||
| B. dolosa | AU0265 | 14 | Early | 128 | 0.9246 | 0.7618 |
| AU21961 | Late | 128 | 0.1230 | 0.1847 | ||
Minimal Inhibitory Concentration (MIC)
MICs were determined in duplicate according to the EUCAST broth microdilution protocol using flat-bottom 96-well microtiter plates (TPP, Trasadingen, Switzerland) [18]. Tobramycin and ciprofloxacin (Sigma Aldrich) concentrations tested ranged from 0.25 to 1024 µg/ml and from 0.25 to 128 µg/ml, respectively. Itaconate and 3-nitropropionate (3-NP) (Sigma Aldrich) concentrations ranged from 0.20 to 100 mM. 2-Thenoyltrifluoroacetone (Sigma Aldrich) concentrations ranged from 5 to 625 nM. The MIC was defined as the lowest concentration for which no significant difference in optical density (λ = 590 nm) was observed between the inoculated and blank wells after 24 h incubation. Differences in absorbance were considered significant when the 95%-confidence interval (calculated using Microsoft Excel) did not contain zero. All MIC determinations were performed in duplicate and replicates never differed more than two fold. When a two fold difference was observed between replicates, the lowest concentration was recorded as the MIC.
Quantification of Persister Cells in Biofilms and Planktonic Cultures
To determine the number of surviving cells, 24 h old biofilms or planktonic cultures were exposed to tobramycin, an aminoglycoside antibiotic, or ciprofloxacin, a fluoroquinolone, in concentrations ranging from 0.5 to 64× the MIC for 24 h. Biofilms were grown in the wells of a round-bottom 96-well microtiter plate (TPP), as described previously [19]. After 24 h of growth, the supernatant was removed and 120 µl of an antibiotic solution in physiological saline (PS) or 120 µl PS ( = control) was added and the plates were further incubated at 37°C. Twelve wells were used for each condition. After 24 h of treatment, cells were harvested by vortexing and sonication (2×5 min) (Branson 3510, Branson Ultrasonics Corp, Danbury, CT) and quantified by plating on LB (n ≥2 for all experiments). For the planktonic experiments an overnight culture was diluted to an optical density of 0.1 (approximately 108 cells/ml). After an additional 24 h growth in a shaking warm water bath, cell suspensions with an optical density of 1 (approximately 109 cells/ml) were transferred to falcon tubes and centrifuged for 9 min at 5000 rpm. Cells were resuspended in an antibiotic solution in PS, or in PS and further incubated at 37°C. After 24 h of treatment the tubes were centrifuged, resuspended in PS and quantified by plating on LB (n ≥2 for all experiments).
RNA Extraction and Microarray Analysis
Biofilms were grown as described above and exposed to an antibiotic (concentration of 4×MIC) or a 0.9% NaCl solution (untreated controls) for 24 h. Treated and untreated B. cenocepacia J2315 sessile cells were harvested by vortexing (5 min) and sonication (5 min) and transferred to sterile tubes. RNA was extracted immediately following harvesting of the cells, using the Ambion RiboPure Bacteria Kit (Ambion, Austin, TX) according to the manufacturer’s instructions and the procedure included a DNase I treatment for 1 h at 37°C. After extraction, RNA of both treated and untreated samples was concentrated using Microcon YM-50 filter devices (Milipore, Billerica, MA) and linearly amplified using the MessageAmp II-Bacteria Kit (Ambion) prior to the microarray analysis. The method was first optimised with the use of standard Escherichia coli RNA. Assessment of RNA yield and quality, cDNA synthesis and the hybridization and washing of the custom made 4×44K arrays (Agilent, Santa Clara, CA) was performed as described previously [20]. The gene expression analysis was performed using GeneSpring GX 7.3 (Agilent) and data were normalized using the procedure recommended for two-colour Agilent microarrays. Only features dedicated to B. cenocepacia J2315 that were labelled “present” or “marginal” were included in the analysis. After this initial filtering, a student’s T-test analysis was performed (p<0.05). The experimental protocols and the raw microarray data can be found in ArrayExpress under the accession number E-MEXP-3532.
Quantitative RT-PCR
In order to validate the microarray results, the expression of 11 selected genes was examined using RT-qPCR. Biofilms were treated and RNA was extracted as described above. cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Forward and reverse primers were developed using tools available on the NCBI website and they were compared with the B. cenocepacia J2315 genome sequence using BLAST to determine their specificity (Table 3). The primer concentration used was 600 nM (300 nM for BCAM1588). All qPCR experiments were performed on a Bio-Rad CFX96 Real-Time System C1000 Thermal Cycler as described previously [20]. Each sample was spotted in duplicate and control samples without added cDNA were included in each experiment. The initial 3 min denaturation step at 95°C was followed by 40 amplification cycles, consisting of 15 s at 95°C and 60 s at 60°C. A melting curve analysis was included at the end of each run. To allow accurate normalization of our data, we also included three reference genes (BCAL2694, BCAS0175, BCAM2784) for which we confirmed expression stability using GeNorm [21] prior to the actual analyses (Figure S1).
Table 3. Primer sequences.
| Gene | Annotation | FW primer | RV primer |
| Glyoxylate shunt | |||
| BCAL2122 | Malate synthase | GGCCGACCGCTCGAAGATCA | CGCCTTGTCGTTCGCTTCCG |
| BCAL2118 | Isocitrate lyase | GCACCGTACGCCGACCTGAT | GCTTGCCCGGGAACTGCTTG |
| BCAM1588 | Isocitrate lyase | TCCTTGCCCGCTGCCTTCAT | ATCCCGAACGCGAAGCTGGT |
| Tricarboxylic acid cycle | |||
| BCAM0961 | Aconitate hydratase | CTCGGCCAGCCGGTGTACTT | CAGCGACTTCGTGCCTTCGC |
| BCAM2701 | Aconitate hydratase | CGGTGATCCAGGCGCAGGAT | CAGGCGCATCCACGATGCAC |
| BCAM1833 | Aconitate hydratase/methylisocitrate dehydratase | GCATGCTGACGGTCGCCAAG | GGTCGGGCAGGCCTTCGATT |
| BCAL2735 | Isocitrate dehydrogenase | CGCTGAAGCCGGAAGCGAAC | ACCGCGCTGCCCTTGATCTT |
| BCAL2736 | Isocitrate dehydrogenase | ATCAAGGGGCCGCTCACGAC | GGCGCAGGCACACGTAGAGA |
| BCAL1517 | Dihydrolipoamide dehydrogenase | GAAGTGAGCGGCGAGGGTGA | TCGGCACCGAGTCGAACGTC |
| BCAL0957 | Succinyl-CoA synthethase subunit alpha | ACGCAGGGCATTACCGGCAA | CGCGGCCGTTTGCGTATTCA |
| BCAM0970 | Succinate dehydrogenase iron-sulfur subunit | CCTCCAGTCGCCGGAAGAGC | CACCAGAAGCTCGGGCACGA |
| BCAM0965 | Malate dehydrogenase | CCGGTCGCGTCGATCGAGAA | GCGAAGCGGAAGTCCGGGTA |
| Other genes | |||
| BCAM2318 | Putative ferredoxin oxidoreductase | TGAAGCTGATCGCGCCGGAT | GCGCACCGCCTTCATGTACG |
| Reference genes | |||
| BCAL0036 | ATP synthase beta chain | CAAGACCGTCAACATGATGGA | TCGAGTCCTTCATTTCGTGGTA |
| BCAL0289 | Glutamate synthase | ATCATCCAGCAGGGTCTGAAGA | GCCATTTCCTCGCGATAGAA |
| BCAL0421 | DNA gyrase B subunit | GTTCCACTGCATCGCGACTT | GGGCTTCGTCGAATTCATCA |
| BCAL1459 | Calcineurin-like phosphoesterase | ATCCCTTGAAATCGAGCATCA | TACGCTCGACAACGTGCATTA |
| BCAL1659 | Ribose transport permease | ACCGTGTTCGGACGCTATCT | CATCAGTGCGAATACGAGAATTTT |
| BCAL1861 | Acetoacetyl-CoA reductase | GATCACCTGCTTCGTGACGTT | GACGTCGTGTTCCGCAAGAT |
| BCAL2694 | Dehydrogenase | CTTGCCGTGATCCTCGAGAT | GAGATCAGCGAGGCCGAGTA |
| BCAM0991 | Tryptophane synthase beta chain | GCCAACGTCTACCGGATGAA | GACCGTGCCGATGATGTAGAA |
| BCAM2784 | Aminotransferase | CCCCGTTCTCGCTCTACGT | GTGTCGCCGAGGCAGAAAT |
| BCAS0175 | Hydrolase | ATGGCCAGTTCGCTCATCA | ACGCGATGTCGATACTCGAAT |
Flow Cytometry
The induction of ROS by tobramycin was confirmed by staining of treated (4×MIC tobramycin, 24 h) and untreated biofilms with the ROS-specific fluorescent dye 2′,7′-dichlorodihydro-fluorescein diacetate (DCFHDA, Sigma Aldrich), followed by quantification of labeled cells by flow cytometry. After 24 h of treatment, as described above, 22 µl DCFHDA (0.1 mM) was added to each well. After a 30 min incubation at 37°C in the dark, the wells were rinsed and 100 µl PS was added. Tobramycin-treated and untreated sessile cells were harvested by scraping and then transferred to sterile tubes. The tubes were centrifuged (for 6 min, at 9000 rpm), the cells were resuspended and 100 fold diluted in PS. All solutions were prepared using MQ water (Millipore, Billerica, MA) and were filter-sterilized before use (Puradisk FP30; Whatman, Middlesex, UK). Labeled cells were quantified using a Cyan ADP flow cytometer (Beckman Coulter, Suarlée, Belgium) with a 488 nm argon laser and a 530–540 nm emission filter. Data of at least 50000 cells were collected for each sample.
Effect of Superoxide Dismutase (SOD), Isocitrate Lyase (ICL) or Succinate Dehydrogenase (SDH) Inhibition on Survival of Bcc Persisters
To confirm the importance of protection against ROS for the survival of persister cells, the number of surviving cells was quantified in biofilms treated with tobramycin (4×MIC, 24 h) in combination with the SOD inhibitor diethyldithiocarbamate (0.05 mM) (Sigma Aldrich). To confirm the importance of ICL for persistence, the number of surviving cells was quantified in treated (4×MIC, 24 h) and untreated biofilms grown in LBB supplemented with 50 mM itaconate or 10 mM 3-NP, two ICL inhibitors. These concentrations were below the MIC of these inhibitors for the strains tested (data not shown). Itaconate and 3-NP solutions were neutralised with NaOH in LBB and sterilized by filtration. To evaluate the role of SDH in persistence, the number of surviving cells was determined in biofilms grown in LBB supplemented with 250 nM 2-thenoyltrifluoroacetone, a concentration well below the MIC (data not shown).
Construction of ICL Mutant
The genome of B. cenocepacia J2315 contains two ICL encoding genes, BCAL2118 and BCAM1588. To confirm their importance, we constructed a double ICL mutant. All B. cenocepacia mutant strains were constructed following the protocol described by Hamad et al., which allows for the creation of unmarked nonpolar gene deletions [22]. Briefly, this mutagenesis procedure requires the upstream and downstream regions flanking the target gene to be cloned into pGPI-SceI-XCm plasmid. The PCR amplifications of these regions (about 500 bp each) were performed with the HotStar HiFidelity polymerase kit (QIAGEN, Milan, Italy) according to the manufacturer’s instructions. The following primer pairs were used for the deletion of BCAL2118 (aceA): KOaceXL (5′-TTTCTAGAATTTCACGATACGGGA-3′ XbaI site is underlined), KOaceBL (5′-TTGGATCCCGGACAGGTAGATGGC-3′ BamHI site is underlined), KOaceBR (5′-TTGGATCCACAAGGGCTTCACGGC-3′ BamHI site is underlined), and KOaceKR (5′-TTGGTACCCTATCTCGGCATCATC-3′ KpnI site is underlined). The following primer pairs were used for the deletion of BCAM1588: KO1588XL (5′-TTTCTAGATGACCGATCCGCACGC-3′ XbaI site is underlined), KO1588BL (5′-TTGGATCCCGACCGACAACCTGGC-3′ BamHI site is underlined), KO1588BR (5′-TTGGATCCCTTGTTCTGGGCACGC-3′ BamHI site is underlined), and KO1588KR (5′-TTGGTACCATCAAAGAATTGTCC-3′ KpnI site is underlined). The mutagenesis plasmids were conjugated into B. cenocepacia J2315 and also into the ΔBCAL2118 mutant strain, in order to create the ΔBCAL2118ΔBCAM1588 double mutant (designated as D2). Exconjugants were selected in the presence of Tp (200 µg/ml), chloramphenicol (400 µg/ml)and ampicillin (200 µg/ml). The insertion of the mutagenesis plasmid into B. cenocepacia genome was confirmed by spraying catechol on the cells, which turn yellow in the presence of this compound and 2,3-catechol-dioxygenase, encoded by xylE (located in pGPI-SceI-XCm plasmid). Subsequently, pDAI-SceI-SacB plasmid (encoding the I-SceI endonuclease), was mobilized by conjugation. Site-specific double-strand breaks take place in the chromosome at the I-SceI recognition site, which is present in pGPI-SceI-XCm plasmid, resulting in tetracycline-resistant (due to the presence of pDAI-SceI-SacB) and Tp-susceptible (indicating the loss of the integrated plasmid) exconjugants. The gene deletions were confirmed by PCR reaction and sequencing of the amplification products. The isolation of deletion mutants cured from pDAI-SceI-SacB was achieved by growing B. cenocepacia cells on LB medium without antibiotics and then screening the resulting colonies for loss of tetracycline resistance.
Construction of SDH Antisense Overexpression Mutants
To confirm the importance of SDH we constructed an antisense overexpression mutant. Succinate dehydrogenase contains 4 subunits. Gene encoding for the two hydrophobic subunits sdhC and sdhD (BCAM0967 and BCAM0968) were amplified by PCR using a PhusionR high Fidelity PCR Kit (Bioké NEB, Leiden, the Nederlands). The following primers were used for amplifying BCAM0967: forward primer GTACAAGCATATGTTAGAATGCTCCGAACA (NdeI restriction site is underlined) and reverse primer ATTCTAGAATGACTGACGCAGTAAGAAAGC (XbaI restriction site is underlined). The following primers were used for amplifying BCAM0968: forward primer GTACAAGCATATGTTACACTCTCCAGAGAA (NdeI restriction site is underlined) and reverse primer AATCTAGAATGGCAGCCAACAACCGAATC (XbaI restrictions site is underlined). Cycling conditions were 30s 98°C, 30 cycli of 10s 98°C, 30s 60°C, and 24s 72°C, and 10 min 72°C. PCR products were purified with a High Pure PCR product purification kit (Roche, Vilvoorde, Belgium), digested and subsequently ligated into a plasmid described by Cardona et al. [23], with a rhamnose-inducible promotor and a Tp selection marker. DNA ligations, restriction endonuclease digestions, and agarose gel electrophoresis were performed according to standard techniques [24]. Restriction enzymes and T4 DNA ligase were purchased from Bioké NEB. Plasmid transformation experiments with E. coli DH5α were carried out by the calcium chloride method [25]. Resistant colonies were isolated and screened for the presence of the construct. Plasmids were transferred into B. cenocepacia by triparental mating [26] using pRK2013 as a helper plasmid [27]. Exconjugants were selected on LB agar plates supplemented with 800 µg/ml Tp and 50 µg/ml gentamicin.
Results and Discussion
Presence of Persister Cells
The presence of persister cells has been proposed as an important factor in biofilm resilience [8]. To investigate the presence of these dormant, multidrug-tolerant phenotypic variants in Bcc biofilms, mature biofilms were treated with tobramycin, an aminoglycoside antibiotic, frequently used in the treatment of infected CF patients. The experiments were carried out using B. cenocepacia J2315, a member of the epidemic ET12 lineage of which the genome has been sequenced. The MIC was previously determined to be 256 µg/ml [18]. Biofilms were treated with various concentrations of tobramycin, ranging from 0.5 to 64×MIC for 24 h.
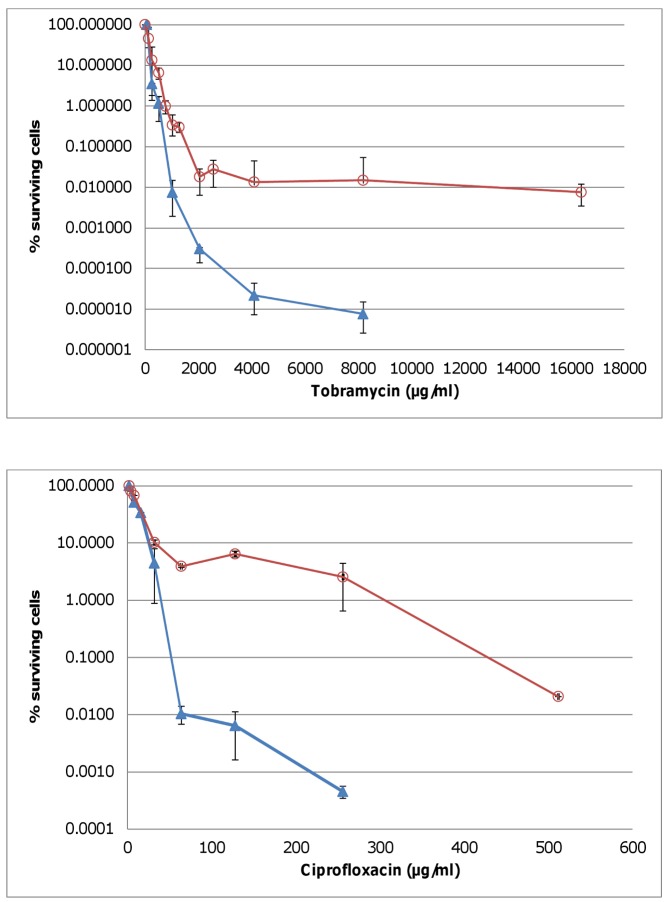
We found that most cells were killed when J2315 biofilms were treated with tobramycin in concentrations ranging from 1 to 4×MIC, but even at higher concentrations (up to 64×MIC), some of the cells survived, indicating the presence of persisters (Fig. 1). Similar results were obtained for other Bcc strains (Table 4). Treatment with ciprofloxacin, a fluoroquinolone, resulted in an even higher fraction of surviving cells (Fig. 1). The presence of persisters is not unique to biofilms as we found similarly-shaped survival curves in planktonic cultures, although the fraction of surviving cells was considerably lower (Fig. 1).
Figure 1. The percentage surviving cells after treatment of B. cenocepacia J2315 with tobramycin or ciprofloxacin.
The results from the biofilm experiments are indicated with open circles and those from the planktonic cultures with triangles. Error bars represent min-max (n ≥2).
Table 4. The average percentage surviving cells for different Bcc strains after treatment of biofilms with high concentration of tobramycin or ciprofloxacin (n ≥3).
| Tobramycin (4×MIC) | Ciprofloxacin (4×MIC) | |||
| Strain | % surviving cells | SD | % surviving cells | SD |
| B. cenocepacia J2315 | 0.3409 | 0.1877 | 10.2425 | 1.4721 |
| B. cenocepacia K56-2 | 0.0064 | 0.0072 | 3.7081 | 3.5486 |
| B. cenocepacia C5424 | 0.0319 | 0.0038 | 37.9861 | 15.6789 |
| B. cenocepacia HI2424 | 0.0007 | 0.0007 | 3.6601 | 0.6902 |
| B. ambifaria AMMD | 0.0096 | 0.01424 | 14.6604 | 11.3466 |
| B. lata ATCC 17769 | 0.8581 | 1.5047 | 4.6846 | 2.3581 |
| B. multivorans ATCC17616 | 0.0011 | 0.0012 | 26.1484 | 21.8644 |
| B. ubonensis LMG 20358 | 0.0548 | 0.0867 | 0.6486 | 1.1232 |
| B. contaminans LMG 16227 | 0.0183 | 0.0268 | 10.5396 | 9.1080 |
| B. dolosa LMG 18943 | 0.1165 | 0.0839 | 18.6143 | 6.3400 |
| B. cepacia LMG 1222 | 0.2369 | 0.4611 | 4.3806 | 5.0242 |
| B. vietnamiensis LMG 22486 | 0.8721 | 1.1513 | 1.8217 | 1.8081 |
| B. pyrrocinia LMG 14191 | 2.4287 | 3.4043 | 6.7670 | 6.6825 |
Mulcahy et al. [28] analyzed clonal pairs of early and late P. aeruginosa isolates from single CF patients and found that in the majority of these patients, cultures of late isolates contained increased numbers of drug-tolerant persister cells. We analyzed clonal pairs of early and late Bcc isolates, obtained from 14 CF patients. The MICs and the fraction of persister cells are shown in Table 2. In general, the MICs were very similar (i.e. within a two-fold dilution), indicating that the strains did not acquire resistance during the course of the infection. Only in four patients the persister fraction was higher in the late isolates. In three patients there was no difference between the early and the late isolates and in eight patients the number of surviving cells was lower in the late than in the early isolate. Our observations suggest that, unlike in P. aeruginosa, the fraction of persisters does not increase with the duration of colonisation.
Induction of ROS by Tobramycin
Recently, it was shown that bactericidal antibiotics induce the production of reactive oxygen species (ROS) [13]. Tobramycin is known to corrupt protein synthesis and by using ROS-specific staining and flow cytometry, we confirmed that tobramycin also drastically increased ROS production in treated sessile cells. Most of the cells in treated, unlike untreated biofilms showed a high fluorescent signal (Fig. 2).
Figure 2. The number of cells in function of the DCHFDA-derived fluorescent signal for untreated and treated biofilms.
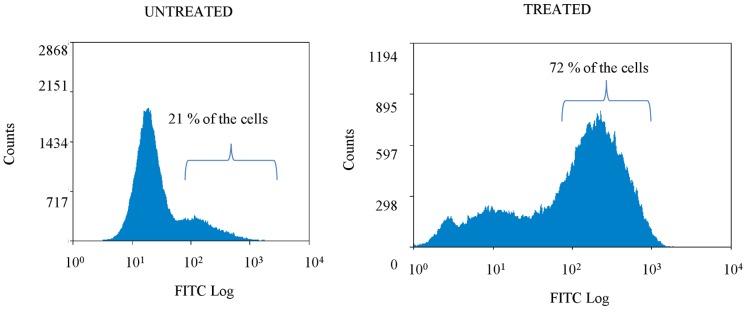
Treated biofilms were exposed to tobramycin in a concentration of 4×MIC (1024 µg/ml) (24 h). The mean percentage of cells with a high fluorescent signal (>30) is indicated (n = 7). Differences between treated and untreated biofilms are statistically significant (p<0.05).
Gene Expression in Cells Treated with High Doses of Tobramycin
A transcriptome analysis showed a considerable change in gene expression in sessile cells treated with high doses of tobramycin relative to untreated sessile cells. Of the 8729 B. cenocepacia J2315 sequences represented on the array, 2688 (30.8%) were significantly (p<0.05) upregulated, while the expression of 2413 sequences (27.6%) was significantly (p<0.05) downregulated. 669 (9.4%) protein-coding genes were significantly (p<0.05) upregulated by a factor of at least two, while the expression of 651 protein-coding genes (9.1%) was significantly (p<0.05) downregulated by a factor of at least two. In addition, a significant differential expression of two or more was noted for 270 intergenic regions (161 upregulated and 109 downregulated) and for 37 rRNA genes (all downregulated). Only a single tRNA gene was upregulated, but 31 tRNA genes were downregulated. A breakdown of differentially expressed protein-encoding genes by functional category is presented in supplementary data (Fig. S2) [29], [30].
In this study we focused on genes involved in the production of ROS and we found that the major pathways were all downregulated in cells surviving high doses of tobramycin treatment (Table 5). We observed that genes from the TCA cycle, including the majority of genes resulting in the production of NADH or FADH2, genes involved in the electron transport chain and the gene coding for ferredoxin reductase were downregulated in persister cells. On the other hand genes involved in production of NADPH, which plays a pivotal role in maintaining the proper cellular redox balance, were upregulated [31]. For example BCAL3276, the gene coding for NAD-kinase, an enzyme that mediates the formation of NADP (a key coenzym known to tilt cellular metabolism towards the synthesis of NADPH and away from the formation of NADH) was upregulated. Similarly, BCAL3395, the gene coding for malic enzyme, which is one of the NADPH producing enzymes, was also upregulated. Isocitrate dehydrogenase kinase-phosphatase, a bifunctional enzyme that can inactivate isocitrate dehydrogenase, thereby directing isocitrate to the glyoxylate shunt, as well as both isocitrate lyase (ICL) encodig genes were also found to be upregulated. In line with these findings, BCAL0813, encoding a RNA polymerase factor sigma 54 which represses the glyoxylate pathway [32] was downregulated. Glutamate dehydrogenase (BCAL3359) was upregulated and α-ketoglutarate dehydrogenase (BCAL1515) was downregulated indicating an increased production of α-ketoglutarate, a ROS scavenger. Other proteins involved in response to oxidative stress which were significantly upregulated include proteins involved in glutathione biosynthesis, thioredoxine, peroxiredoxine and glutathione peroxidase. The expression levels of selected genes were confirmed by qPCR (Table 5).
Table 5. Differences in gene expression expressed as fold changes in treated vs untreated biofilms.
| Gene number | Annotation | Microarray | qPCR |
| Glyoxylate shunt | |||
| BCAL2122 | Malate synthase | 1.4* | −3.3* |
| BCAL2118 | Isocitrate lyase AceA | 2.3* | 1.9 |
| BCAM1588 | Isocitrate lyase | 3.1* | 1.6 |
| BCAL0813 | RNA polymerase factor sigma 54 | −1.5* | − |
| BCAL1949 | Glyoxylate carboligase | −1.5 | − |
| Tricarboxylic acid cycle | |||
| BCAM0961 | Aconitate hydratase | −1.1 | − |
| BCAM2701 | Aconitate hydratase | −1.3* | −1.3 |
| BCAM1833 | Aconitate hydratase/methylisocitrate dehydratase | 1.5* | 1.1 |
| BCAL2735 | Isocitrate dehydrogenase | 1.3* | −2.5* |
| BCAL2736 | Isocitrate dehydrogenase | 1.4 | 1.5 |
| BCAL1515 | α-ketoglutarate dehydrogenase E1 | −2.0* | − |
| BCAL1516 | Dihydrolipoamide succinyltransferase | −3.3* | − |
| BCAL1517 | Dihydrolipoamide dehydrogenase | −2.5* | − |
| BCAL2207 | Putative dihydrolipoamide dehydrogenase | −1.3* | − |
| BCAL1215 | Dihydrolipoamide dehydrogenase | −1.4* | − |
| BCAL0956 | Succinyl-CoA synthetase beta chain | −2.0* | − |
| BCAL0957 | Succinyl-CoA synthetase subunit alpha | −3.3* | −10.0* |
| BCAM0967 | Putative succinate dehydrogenase | −1.7* | − |
| BCAM0968 | Putative succinate dehydrogenase | −2.5* | − |
| BCAM0969 | Succinate dehydrogenase flavoprotein | −2.5* | − |
| BCAM0970 | Succinate dehydrogenase iron-sulfur subunit | −5.0* | −25.0* |
| BCAL2908 | Fumarate hydratase | −1.3* | 1.9* |
| BCAL2287 | Putative fumarate dehydrogenase | 1.0 | − |
| BCAM0965 | Malate dehydrogenase | 1.0 | −2.0* |
| BCAL2746 | Putative citrate synthase | −1.3 | − |
| BCAM0964 | Putative lyase | −1.4* | − |
| BCAS0023 | HpcH/HpaI aldolase/citrate lyase family | −2.5* | − |
| BCAM0972 | Type II citrate synthase | −5.0* | − |
| Oxidative phosphorylation | |||
| BCAL2142 | Cytochrome o ubiquinol oxidase subunit III | −2.0* | − |
| BCAL2143 | Ubiquinol oxidase polypeptide I | −2.0* | − |
| BCAL0750 | Cytochrome c oxidase polypeptide I | −1.7* | − |
| BCAL0752 | Cytochrome c oxidase assembly protein | −2.5* | − |
| BCAL0753 | Hypothetical protein | −2.5* | − |
| BCAL0754 | Putative cytochrome c oxidase subunit III | −2.0* | − |
| BCAL0030–0037 | F0F1 ATP synthase subunit A-ε | −1.7*–−2.5*° | − |
| BCAL2331–2343 | NADH dehydrogenase subunit B-N | −2.5–−10.0*° | − |
| NAD(P)H production | |||
| BCAL3276 | NAD-kinase | 1.4* | − |
| BCAL0672 | Isocitrate dehydrogenase kinase/phosphatase | 1.3* | − |
| BCAL3359 | Glutamate dehydrogenase | 4.2* | − |
| BCAL3395 | Malic enzyme | 1.7* | − |
| Response to oxidative stress | |||
| BCAL1250 | Putative glutathione S-transferase | 1.6* | − |
| BCAL3331 | Putative glutathione S-transferase | 3.4* | − |
| BCAL0463 | Putative thioredoxin | 1.6* | − |
| BCAL2013 | AhpC/TSA family protein | 1.9* | − |
| BCAL2106 | Glutathion peroxidase | 1.6* | − |
| BCAM2318 | Putative ferredoxin oxidoreductase | −10.0* | −33.3* |
| Fe-storage | |||
| BCAM2627 | Putative hemin ABC transporter protein | 5.2* | − |
| BCAM2630 | Hemin importer ATP binding subunit | 2.8* | − |
| BCAM2224 | Putative pyochelin receptor protein FptA | 2.7* | − |
| BCAL1790 | Putative iron-transport protein | 2.5* | − |
| BCAL1347 | Putative Fe uptake system extracellular binding protein | 2.5* | − |
| BCAM2228 | Putative pyochelin synthetase PchF | 2.1* | − |
| BCAL1789 | Putative iron-transport protein | 2.0* | − |
| BCAL1371 | Putative TonB-dependent siderophore receptor | 2.0* | − |
| BCAL1702 | Putative ornibactin biosynthesis protein | −2.2* | − |
-: no qPCR experiments were performed. *: significant change in expression between the treated and the untreated biofilms (n = 3, p<0.05). °: range of fold changes for the various subunits.
Eradication of Persister Cells
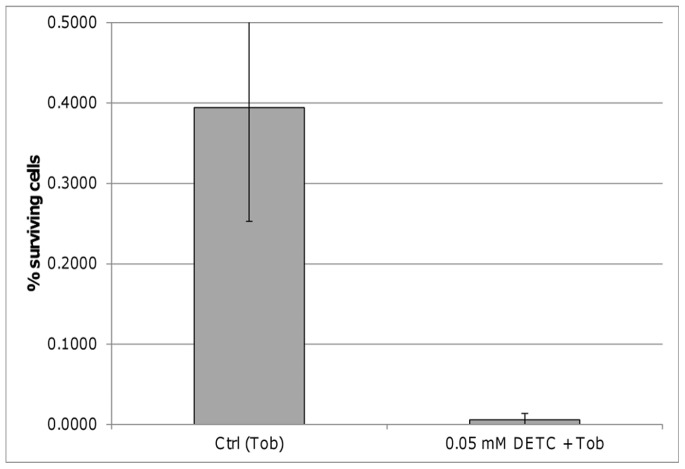
Our transcriptome analysis suggested that protection against ROS is important in survival of persister cells. To confirm this we determined the number of persisters in biofilms formed by catalase mutants. B. cenocepacia contains two catalases of which one (KatA) is a specialized catalase/peroxidase which helps maintaining the normal activity of the TCA cycle, while KatB is a classical catalase/peroxidase which plays a global role in cellular protection against oxidative stress by converting toxic H2O2 into H2O and O2 [20]. We found the persister fraction to be slightly reduced in biofilms formed by the ΔkatA mutant but almost 40-fold reduced in biofilms formed by the ΔkatB mutant (Fig. 3). Furthermore, the number of surviving cells was almost 100 times lower after addition of the superoxide dismutase inhibitor diethyldithiocarbamate (DETC) (Fig. 4). Superoxide dismutases are antioxidant enzymes that detoxify O2 −. by a dismutation reaction generating H2O2 and O2 [16] . The inhibitor had no effect on untreated biofilms in the concentrations tested, but reduced the number of surviving cells in a concentration dependent manner when combined with tobramycin. Together, these results indicate that protection against ROS is indeed important for the survival of persister cells.
Figure 3. Effect of knocking out catalase function on survival of peristers.
The number of surviving cells in biofilms formed by B. cenocepacia C5424 and two catalase mutants after treatment with tobramycin in a concentration of 4×MIC (512 µg/ml). Error bars represent standard deviation (n = 3).
Figure 4. Effect of superoxide dismutase inhibition on survival of persisters.
The number of surviving cells in B. cenocepacia J2315 biofilms after treatment with tobramycin alone (4×MIC, 24 h) or in combination with diethyldithiocarbamate (DETC 0.05 mM). Error bars represent standard deviation (n = 3, p<0.05).
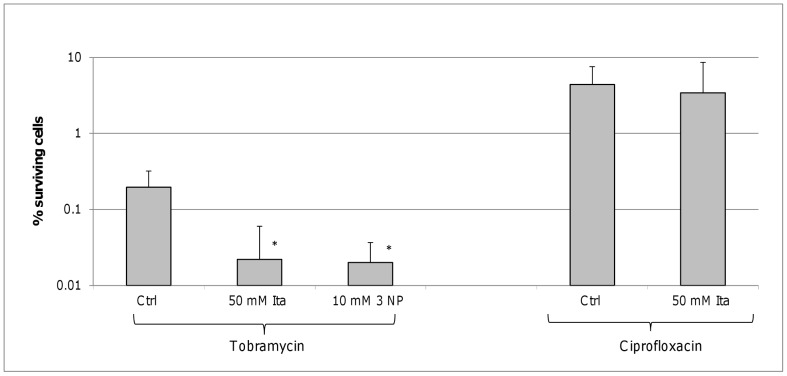
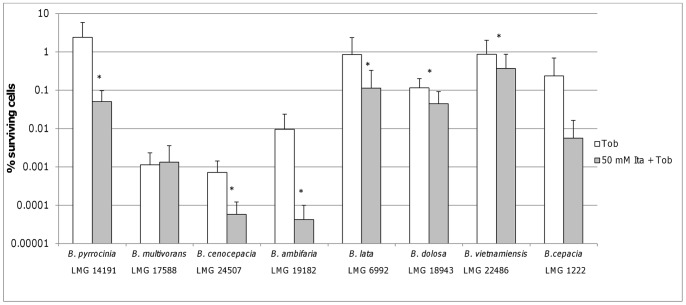
Persister cells are typically considered as “dormant” (i.e. metabolically inactive) cells [11] but our data challenge that paradigm. While it is true that some pathways (i.e. TCA cycle) were found to be downregulated, other pathways were upregulated, and these may be a novel therapeutic target for combination therapy. The importance of the upregulation of the glyoxylate shunt for the survival of persister cells was confirmed by inhibition of ICL, the first enzyme in the shunt. Because persisters are thought to be pre-existent, biofilms were grown in the presence of these inhibitors. During treatment the inhibitors were removed because of pH incompatibility with the antibiotics used. Pre-treatment of B. cenocepacia biofilms with the ICL inhibitor itaconate in a concentration of 50 mM reduced the persister fraction approximately 10 fold when the biofilms were subsequently treated with tobramycin. Pre-treatment with 10 mM 3-NP, a more potent ICL inhibitor resulted in similar reductions (Fig. 5). The concentrations of the ICL inhibitor used were below the MIC and they did not affect growth in the untreated biofilms. In addition, we did not observe an effect of either inhibitor on ROS production (data not shown). The additional killing observed after combined treatment with tobramycin and an ICL inhibitor was not observed with ciprofloxacin (Fig. 5). This may suggest that this effect is related to the type of antibiotic and/or the magnitude of its effect on biofilms. However, this remains to be investigated. Similar results were obtained in other Bcc bacteria (Fig. 6).
Figure 5. Effect of isocitrate lyase inhibition on B. cenocepacia J2315.
The number of surviving cells in B. cenocepacia J2315 biofilms after treatment with tobramycin (4×MIC, 24 h) or ciprofloxacin (4×MIC, 24 h) for biofilms grown in LB or LB supplemented with 50 mM itaconate (ita) or 10 mM 3-nitropropionate (3-NP). Error bars represent standard deviation. Statistically significant differences are indicated by an asterix (p<0.05, n ≥3).
Figure 6. Effect of iocitrate lyase inhibition on different Bcc strains.
The number of surviving cells in different Bcc biofilms after treatment with tobramycin (4×MIC, 24 h) for biofilms grown in LB or LB supplemented with 50 mM itaconate. Statistically significant differences are indicated by an asterix (p<0.05) (n≥3).
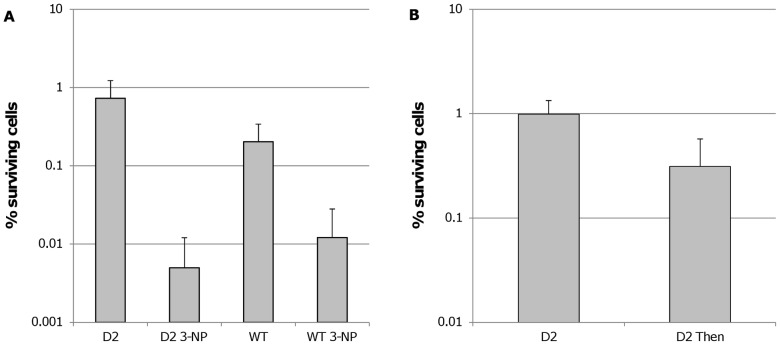
To confirm the results obtained by chemically inhibiting ICL, we constructed a B. cenocepacia J2315 mutant in which both ICL genes were inactivated. Surprisingly, there was no significant decrease in the number of surviving cells after treatment with tobramycin in the D2 mutant compared to wild type (Fig. 7A), but we did find an additional effect after adding 3-NP to the mutant. Itaconate and 3-NP are succinate analogues and besides inhibiting ICL, they also inhibit succinate dehydrogenase (SDH) [33].
Figure 7. A. The number of surviving cells in a double ICL mutant (D2) compared to wild type (WT).
The number of surviving cells in a double ICL mutant (D2) and in a wild type B. cenocepacia J2315 (WT) biofilm after treatment with tobramycin (4 MIC, 24h) for biofilms grown in LB or LB supplemented with 10 mM 3-NP. Error bars represent standard deviation (n ≥3). For both strains, treatment with 3-NP resulted in significantly (p<0.05) less surviving cells compared to the untreated control. B. Effect of succinate dehydrogenase inhibition on B. cenocepacia D2. The number of surviving cells in B. cenocepacia D2 biofilms after treatment with tobramycin (4×MIC, 24 h) for biofilms grown in LB or LB supplemented with 250 nM 2-thenoyltrifluoroacetone (Then). Error bars represent standard deviation (n = 4). Statistically significant differences are indicated by an asterix (p<0.05).
We hypothesised that an upregulation of ICL in persisters not only limits the production of NADH (and thus limits ROS production) but also leads to an increased intracellular succinate concentration. Succinate can be oxidised by SDH, thereby generating FADH2 which can lead to basal production of ATP in persister cells in which other energy-generating systems are downregulated. Itaconate and 3-NP likely kill persisters by shutting down the remaining ATP production by inhibiting both ICL and SDH. To test this hypothesis we constructed a SDH antisense overexpression mutant in the D2 background, in which SDH is inactivated if rhamnose is present. However, we noticed that Tp (required to maintain the plasmid) and/or rhamnose (required to induce antisense RNA expression) had a mild influence of biofilm formation as such, and on the number of persisters in biofilms (data not shown), which made interpretation of these data impossible. However, addition of an ubiquinone type SDH inhibitor, 2-thenoyltrifluoroacetone, to B. cenocepacia D2 biofilms significantly (p<0.05) reduced the number of surviving persisters (Fig. 7B), while not affecting the MIC for tobramycin (data not shown).
The reaction catalysed by ICL also leads to the production of glyoxylate. In B. cenocepacia glyoxylate can be used to form malate (through the activity of malate synthase) or tartronate semialdehyde (through the activity of glyoxylate carboligase). There are two malate synthases of which only one (BCAL2122) was slightly upregulated and glyoxylate carboligase (BCAL1949) was slightly downregulated (Table 5).
Conclusion
Our results contribute to a better understanding of the molecular mechanisms responsible for the antimicrobial tolerance of Bcc biofilms by demonstrating that these biofilms contain tolerant persister cells. In these surviving persister cells, the TCA cycle was downregulated and the expression of genes involved in the electron transport chain was also downregulated. This way the cells avoid the production of ROS. At the same time, persister cells activate an alternative pathway, i.e. the glyoxylate shunt. When biofilms were grown in the presence of an inhibitor of ICL and SDH, less persisters survived. Similar results were obtained in different species, indicating that this mechanism is widespread within the Bcc. So far, most anti-persister strategies focussed on “reawaking” persisters for efficient killing [34], but we found that inhibiting a cellular target can also reduce the number of surviving persisters. This finding may provide novel approaches for treatment of persister-related infections.
Supporting Information
Average expression stability values (M) of remaining control genes during stepwise exclusion of the least stable control gene (between brackets). Genes are ranked from left to right in order of increasing expression stability (decreasing M value). Genes labeled with an asterix were used for normalization.
(TIF)
Differentially expressed protein-coding genes ordered by functional category.
(DOCX)
Acknowledgments
We thank Miguel Valvano for providing us with B. cenocepacia C524 and the B. cenocepacia MDL1 and MDL2 mutant strains and Pam Sokol, James Zlosnik and John LiPuma for the early and late clonal pairs. TC acknowledges the support of the Special Research Fund of Ghent University and the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office. EM and TC acknowledge the European Cooperation in Science and Technology (COST) action BM1003 for facilitating collaborative networking that assisted with these studies.
Funding Statement
This work was financially supported by The Research Foundation - Flanders FWO-Vlaanderen, Special Research Fund of Ghent University, Cystic Fibrosis Foundation Therapeutics Inc and the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office. EM and TC acknowledge the European Cooperation in Science and Technology (COST) action BM1003 for facilitating collaborative networking that assisted with these studies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, et al. (2009) Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp.nov. Int J Syst Evol Microbiol 59: 102–111. [DOI] [PubMed] [Google Scholar]
- 2. LiPuma JJ (2005) Update on the Burkholderia cepacia complex. Curr Opin Pulm Med 11: 528–533. [DOI] [PubMed] [Google Scholar]
- 3. Jones AM, Dodd ME, Govan JRW, Barcus V, Doherty CJ, et al. (2004) Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59: 948–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Soyza A, Morris K, McDowell A, Doherty C, Archer L, et al. (2004) Prevalence and clonality of Burkholderia cepacia complex genomovars in UK patients with cystic fibrosis referred for lung transplantation. Thorax 59: 526–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mahenthiralingam E, Urban TA, Goldberg JB (2005) The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3: 144–156. [DOI] [PubMed] [Google Scholar]
- 6. Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15: 167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caraher E, Reynolds G, Murphy P, McClean S, Callaghan M (2006) Comparison of antibiotic susceptibility of Burkholderia cepacia complex organisms when grown planktonically or as biofilm in vitro. Eur J Clin Microbiol Infect Dis 26: 213–216. [DOI] [PubMed] [Google Scholar]
- 8. Lewis K (2001) Riddle of biofilm resistance. Antimicrob Agents Chemother 45: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mah TFC, O’Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9: 34–39. [DOI] [PubMed] [Google Scholar]
- 10. George AM, Jones PM, Middleton PG (2009) Cystic fibrosis infections: treatment strategies and prospects. FEMS Microbiol Lett 300: 153–164. [DOI] [PubMed] [Google Scholar]
- 11. Lewis K (2010) Persister cells. Annu. Rev. Microbiol 64: 357–372. [DOI] [PubMed] [Google Scholar]
- 12. Keren I, Shah D, Spoering A, Kaldalu N, Lewis K (2004) Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. FEMS Microbiol. Lett. 230: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810. [DOI] [PubMed] [Google Scholar]
- 14.Berg JM, Tymoczko JL, Stryer L (2002) Biochemistry. 5th edition. New York: WH Freeman. Section 17.4. [Google Scholar]
- 15. Van Schaik EJ, Tom M, Woods DE (2009) Burkholderia pseudomallei isocitrate lyase is a persistence factor in pulmonary melioidosis: implications for the development of isocitrate lyase inhibitors as novel antimicrobials. Infection and immunity 77: 4275–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lefebre MD, Flannagan RS, Valvano MA (2005) A minor catalase/peroxidase from Burkholderia cenocepacia is required for normal aconitase activity. Microbiol 151: 1975–1985. [DOI] [PubMed] [Google Scholar]
- 17. LiPuma JJ, Spiker T, Coenye T, Gonzalez CF (2002) An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359: 2002–2003. [DOI] [PubMed] [Google Scholar]
- 18. Peeters E, Nelis HJ, Coenye T (2009) In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J Antimicrob Chemother 64: 801–809. [DOI] [PubMed] [Google Scholar]
- 19. Van Acker H, Van Snick E, Nelis HJ, Coenye T (2010) In vitro activity of temocillin against planktonic and sessile Burkholderia cepacia complex bacteria. J Cyst Fibros 9: 450–454. [DOI] [PubMed] [Google Scholar]
- 20. Peeters E, Sass A, Mahenthiralingam E, Nelis H, Coenye T (2010) Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of H2O2 and NaOCl. BMC Genomics 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamad MA, Skeldon AM, Valvano MA (2010) Construction of aminoglycoside -sensitive Burkholderia cenocepacia strains for use in studies of intracellular bacteria with the gentamycin protection assay. Appl. Environ. Microbiol 76: 3170–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cardona ST, Mueller CL, Valvano MA (2006) Identification of essential operons with a rhamnose-inducible promotor in Burkholderia cenocepacia App Environ Microbiol. 72: 2547–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T (1990) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory. [Google Scholar]
- 25. Cohen SN, Chang AC, Hsu L (1972) Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69: 2110–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ (1989) A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135: 2885–2890. [DOI] [PubMed] [Google Scholar]
- 27. Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans Proc. Natl. Acad. Sci. USA 76: 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mulcahy LR, Burns JL, Lory S, Lewis K (2010) Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192: 6191–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Young JP, Crossman LC, Johnston AW, Thomson NR, Ghazoui ZF, et al. (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riley M (1993) Functions of the gene products of Escherichia coli . Microbiol Rev 57: 862–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sing R, Lemire J, Mailloux RJ, Chénier D, Hamel R, Appanna VD (2009) An ATP and oxalate generating variant tricarboxylic acid cycle counters aluminium toxicity in Pseudomonas fluorescens . Plos One 4: e7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hagins JM, Scoffield JA, Suh SJ, Silo-Suh L (2010) Influence of RpoN on isocitrate lyase activity in Pseudomonas aeruginosa . Microbiology 156: 1201–1210. [DOI] [PubMed] [Google Scholar]
- 33. Alston TA, Mela L, Bright HJ (1977) 3-nitropropionate, the toxic substance of Indigofera, is a suicide inactivator of succinate dehydrogenase Proc Natl Acad Sci USA. 74: 3767–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Allison K, Brynildsen M, Collins J (2011) Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average expression stability values (M) of remaining control genes during stepwise exclusion of the least stable control gene (between brackets). Genes are ranked from left to right in order of increasing expression stability (decreasing M value). Genes labeled with an asterix were used for normalization.
(TIF)
Differentially expressed protein-coding genes ordered by functional category.
(DOCX)