Abstract
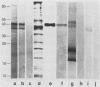
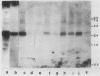
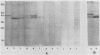
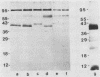
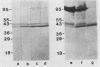
Variants of the Cx43 gap junction protein have been detected on Western immunoblots by using an antipeptide antibody to the N-terminus of the protein. In heart ventricle, atrium, brain, retina, and uterus, different yet characteristic ratios of a broad 43-kDa band and a 39- to 40-kDa doublet were observed. These proteins (in lens epithelium, testes, and spleen) or their messages (in stomach, duodenum, kidney, and lung) were also detected in several nonexcitable systems but at consistently lower levels than found in electrically excitable tissues. The reproducible heterogeneity in electrophoretic mobility of Cx43 seen in different tissues does not appear to be due to proteolysis, since both the 43-kDa band and the 39- to 40-kDa doublet were recognized by an N-terminal as well as a C-terminal antibody. Furthermore, Northern (RNA) blots from different tissues show that both polypeptide profiles arise from indistinguishable transcripts. The conversion by alkaline phosphatase treatment of a predominantly 43-kDa profile (in heart) to a 39- to 40-kDa profile (characteristic of brain and protein translated in vitro from the RNA) suggests that the observed electrophoretic heterogeneity arises from tissue-wide differences in the phosphorylation state of Cx43.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett M. V., Goodenough D. A. Gap junctions, electrotonic coupling, and intercellular communication. Neurosci Res Program Bull. 1978 Sep;16(3):1–486. [PubMed] [Google Scholar]
- Beyer E. C., Kistler J., Paul D. L., Goodenough D. A. Antisera directed against connexin43 peptides react with a 43-kD protein localized to gap junctions in myocardium and other tissues. J Cell Biol. 1989 Feb;108(2):595–605. doi: 10.1083/jcb.108.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt J. M., Spray D. C. Inotropic agents modulate gap junctional conductance between cardiac myocytes. Am J Physiol. 1988 Jun;254(6 Pt 2):H1206–H1210. doi: 10.1152/ajpheart.1988.254.6.H1206. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- De Mello W. C. Increase in junctional conductance caused by isoproterenol in heart cell pairs is suppressed by cAMP-dependent protein-kinase inhibitor. Biochem Biophys Res Commun. 1988 Jul 29;154(2):509–514. doi: 10.1016/0006-291x(88)90169-6. [DOI] [PubMed] [Google Scholar]
- Dermietzel R., Leibstein A., Frixen U., Janssen-Timmen U., Traub O., Willecke K. Gap junctions in several tissues share antigenic determinants with liver gap junctions. EMBO J. 1984 Oct;3(10):2261–2270. doi: 10.1002/j.1460-2075.1984.tb02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermietzel R., Traub O., Hwang T. K., Beyer E., Bennett M. V., Spray D. C., Willecke K. Differential expression of three gap junction proteins in developing and mature brain tissues. Proc Natl Acad Sci U S A. 1989 Dec;86(24):10148–10152. doi: 10.1073/pnas.86.24.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara L., Beyer E. C., Swenson K. I., Paul D. L., Goodenough D. A. Cloning and expression of a Xenopus embryonic gap junction protein. Science. 1989 Mar 3;243(4895):1194–1195. doi: 10.1126/science.2466337. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Marchiori F., Borin G., Pinna L. A. Distinct structural requirements of Ca2+/phospholipid-dependent protein kinase (protein kinase C) and cAMP-dependent protein kinase as evidenced by synthetic peptide substrates. FEBS Lett. 1985 May 6;184(1):72–77. doi: 10.1016/0014-5793(85)80656-6. [DOI] [PubMed] [Google Scholar]
- Fraser S. E., Green C. R., Bode H. R., Gilula N. B. Selective disruption of gap junctional communication interferes with a patterning process in hydra. Science. 1987 Jul 3;237(4810):49–55. doi: 10.1126/science.3037697. [DOI] [PubMed] [Google Scholar]
- Fyrberg E. A., Kindle K. L., Davidson N., Kindle K. L. The actin genes of Drosophila: a dispersed multigene family. Cell. 1980 Feb;19(2):365–378. doi: 10.1016/0092-8674(80)90511-5. [DOI] [PubMed] [Google Scholar]
- Garfield R. E., Kannan M. S., Daniel E. E. Gap junction formation in myometrium: control by estrogens, progesterone, and prostaglandins. Am J Physiol. 1980 Mar;238(3):C81–C89. doi: 10.1152/ajpcell.1980.238.3.C81. [DOI] [PubMed] [Google Scholar]
- Gilula N. B., Reeves O. R., Steinbach A. Metabolic coupling, ionic coupling and cell contacts. Nature. 1972 Feb 4;235(5336):262–265. doi: 10.1038/235262a0. [DOI] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Differential regulation of the levels of three gap junction mRNAs in Xenopus embryos. J Cell Biol. 1990 Mar;110(3):597–605. doi: 10.1083/jcb.110.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Sequence and developmental expression of mRNA coding for a gap junction protein in Xenopus. J Cell Biol. 1988 Sep;107(3):1065–1073. doi: 10.1083/jcb.107.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros D. B., Nicholson B. J., Revel J. P. Comparative analysis of the gap junction protein from rat heart and liver: is there a tissue specificity of gap junctions? Cell. 1983 Dec;35(2 Pt 1):539–549. doi: 10.1016/0092-8674(83)90188-5. [DOI] [PubMed] [Google Scholar]
- Henderson D., Eibl H., Weber K. Structure and biochemistry of mouse hepatic gap junctions. J Mol Biol. 1979 Aug 5;132(2):193–218. doi: 10.1016/0022-2836(79)90391-7. [DOI] [PubMed] [Google Scholar]
- Hertzberg E. L., Skibbens R. V. A protein homologous to the 27,000 dalton liver gap junction protein is present in a wide variety of species and tissues. Cell. 1984 Nov;39(1):61–69. doi: 10.1016/0092-8674(84)90191-0. [DOI] [PubMed] [Google Scholar]
- Kistler J., Christie D., Bullivant S. Homologies between gap junction proteins in lens, heart and liver. Nature. 1988 Feb 25;331(6158):721–723. doi: 10.1038/331721a0. [DOI] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986 Sep;103(3):767–776. doi: 10.1083/jcb.103.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird D. W., Revel J. P. Biochemical and immunochemical analysis of the arrangement of connexin43 in rat heart gap junction membranes. J Cell Sci. 1990 Sep;97(Pt 1):109–117. doi: 10.1242/jcs.97.1.109. [DOI] [PubMed] [Google Scholar]
- Lee S., Gilula N. B., Warner A. E. Gap junctional communication and compaction during preimplantation stages of mouse development. Cell. 1987 Dec 4;51(5):851–860. doi: 10.1016/0092-8674(87)90108-5. [DOI] [PubMed] [Google Scholar]
- Loewenstein W. R. Junctional intercellular communication and the control of growth. Biochim Biophys Acta. 1979 Feb 4;560(1):1–65. doi: 10.1016/0304-419x(79)90002-7. [DOI] [PubMed] [Google Scholar]
- Loewenstein W. R. Junctional intercellular communication: the cell-to-cell membrane channel. Physiol Rev. 1981 Oct;61(4):829–913. doi: 10.1152/physrev.1981.61.4.829. [DOI] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Goodenough D. A. Gap junction structures. II. Analysis of the x-ray diffraction data. J Cell Biol. 1977 Aug;74(2):629–645. doi: 10.1083/jcb.74.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath C. K., Goings G. E., Page E. Proteolysis of cardiac gap junctions during their isolation from rat hearts. J Membr Biol. 1985;85(2):159–168. doi: 10.1007/BF01871268. [DOI] [PubMed] [Google Scholar]
- Manjunath C. K., Nicholson B. J., Teplow D., Hood L., Page E., Revel J. P. The cardiac gap junction protein (Mr 47,000) has a tissue-specific cytoplasmic domain of Mr 17,000 at its carboxy-terminus. Biochem Biophys Res Commun. 1987 Jan 15;142(1):228–234. doi: 10.1016/0006-291x(87)90475-x. [DOI] [PubMed] [Google Scholar]
- Manjunath C. K., Page E. Rat heart gap junctions as disulfide-bonded connexon multimers: their depolymerization and solubilization in deoxycholate. J Membr Biol. 1986;90(1):43–57. doi: 10.1007/BF01869685. [DOI] [PubMed] [Google Scholar]
- Miller T., Dahl G., Werner R. Structure of a gap junction gene: rat connexin-32. Biosci Rep. 1988 Oct;8(5):455–464. doi: 10.1007/BF01121644. [DOI] [PubMed] [Google Scholar]
- Musil L. S., Beyer E. C., Goodenough D. A. Expression of the gap junction protein connexin43 in embryonic chick lens: molecular cloning, ultrastructural localization, and post-translational phosphorylation. J Membr Biol. 1990 Jun;116(2):163–175. doi: 10.1007/BF01868674. [DOI] [PubMed] [Google Scholar]
- Nicholson B. J., Gros D. B., Kent S. B., Hood L. E., Revel J. P. The Mr 28,000 gap junction proteins from rat heart and liver are different but related. J Biol Chem. 1985 Jun 10;260(11):6514–6517. [PubMed] [Google Scholar]
- Nicholson B., Dermietzel R., Teplow D., Traub O., Willecke K., Revel J. P. Two homologous protein components of hepatic gap junctions. Nature. 1987 Oct 22;329(6141):732–734. doi: 10.1038/329732a0. [DOI] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez J. C., Spray D. C., Nairn A. C., Hertzberg E., Greengard P., Bennett M. V. cAMP increases junctional conductance and stimulates phosphorylation of the 27-kDa principal gap junction polypeptide. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2473–2477. doi: 10.1073/pnas.83.8.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub O., Look J., Paul D., Willecke K. Cyclic adenosine monophosphate stimulates biosynthesis and phosphorylation of the 26 kDa gap junction protein in cultured mouse hepatocytes. Eur J Cell Biol. 1987 Feb;43(1):48–54. [PubMed] [Google Scholar]
- Woodgett J. R., Gould K. L., Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986 Nov 17;161(1):177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- Yancey S. B., John S. A., Lal R., Austin B. J., Revel J. P. The 43-kD polypeptide of heart gap junctions: immunolocalization, topology, and functional domains. J Cell Biol. 1989 Jun;108(6):2241–2254. doi: 10.1083/jcb.108.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. T., Nicholson B. J. Sequence and tissue distribution of a second protein of hepatic gap junctions, Cx26, as deduced from its cDNA. J Cell Biol. 1989 Dec;109(6 Pt 2):3391–3401. doi: 10.1083/jcb.109.6.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]