Abstract
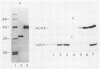
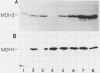
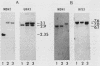
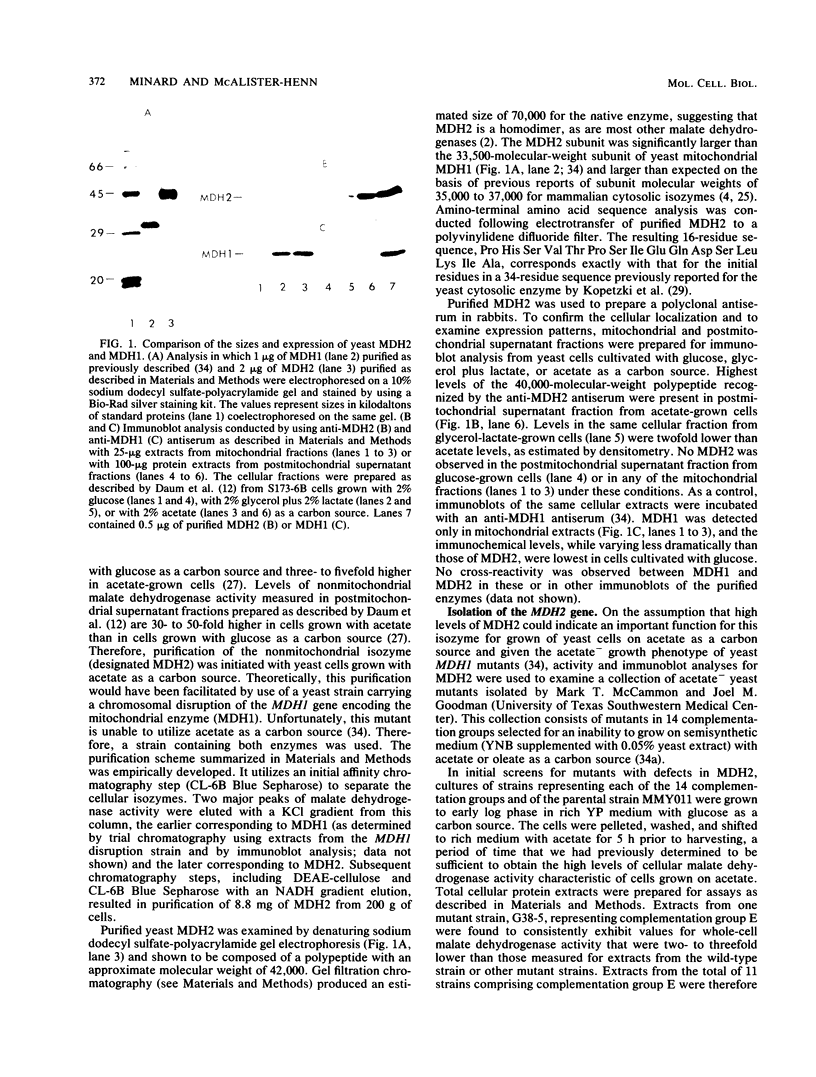
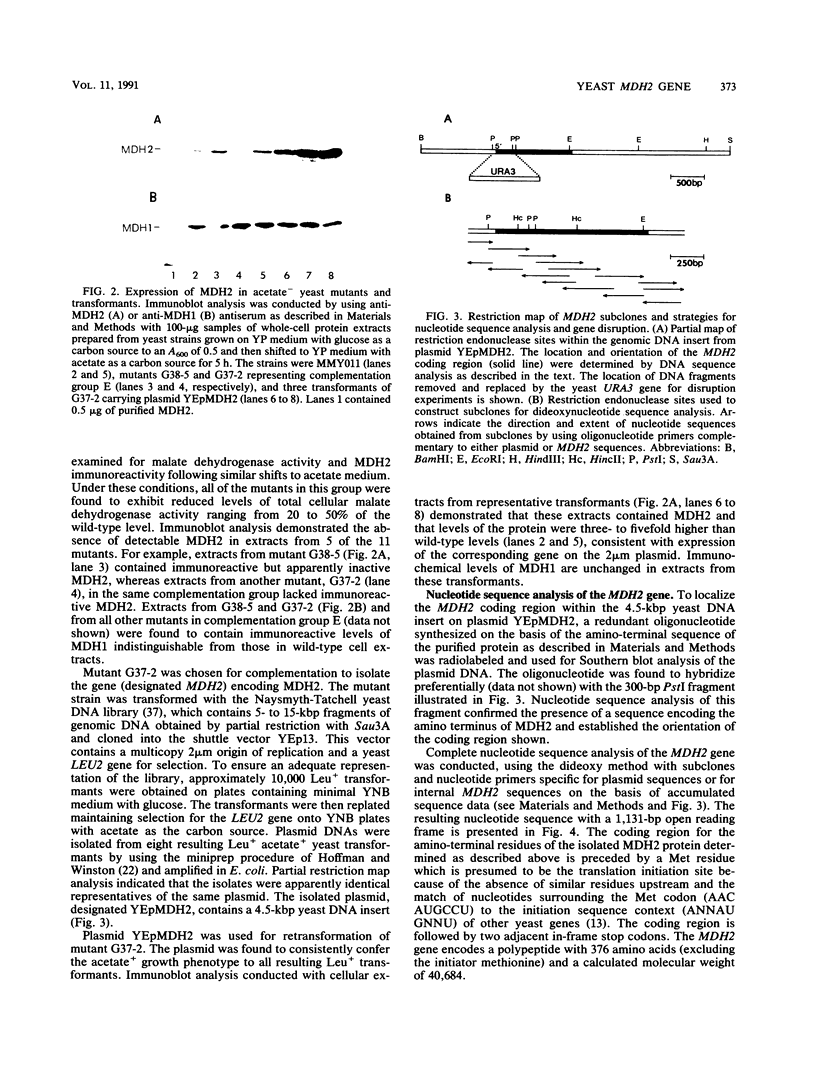
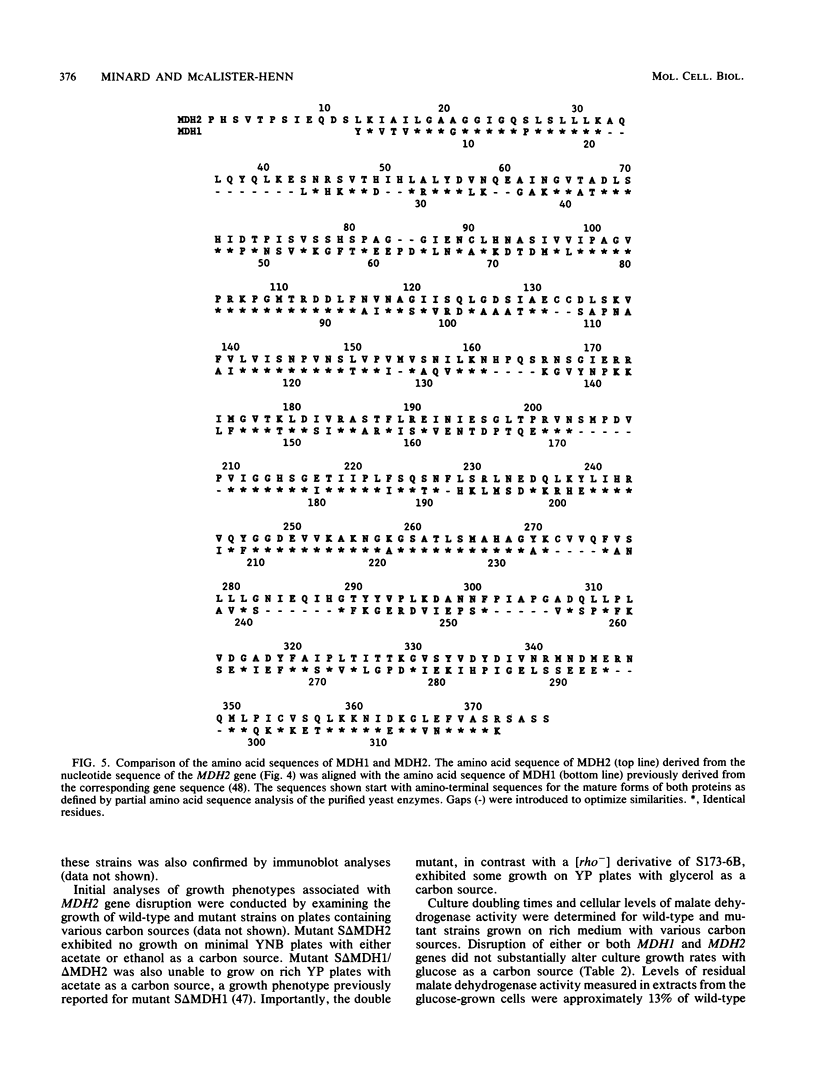
The major nonmitochondrial isozyme of malate dehydrogenase (MDH2) in Saccharomyces cerevisiae cells grown with acetate as a carbon source was purified and shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to have a subunit molecular weight of approximately 42,000. Enzyme assays and an antiserum prepared against the purified protein were used to screen a collection of acetate-nonutilizing (acetate-) yeast mutants, resulting in identification of mutants in one complementation group that lack active or immunoreactive MDH2. Transformation and complementation of the acetate- growth phenotype was used to isolate a plasmid carrying the MDH2 gene from a yeast genomic DNA library. The amino acid sequence derived from complete nucleotide sequence analysis of the isolated gene was found to be extremely similar (49% residue identity) to that of yeast mitochondrial malate dehydrogenase (molecular weight, 33,500) despite the difference in sizes of the two proteins. Disruption of the MDH2 gene in a haploid yeast strain produced a mutant unable to grow on minimal medium with acetate or ethanol as a carbon source. Disruption of the MDH2 gene in a haploid strain also containing a disruption in the chromosomal MDH1 gene encoding the mitochondrial isozyme produced a strain unable to grow with acetate but capable of growth on rich medium with glycerol as a carbon source. The detection of residual malate dehydrogenase activity in the latter strain confirmed the existence of at least three isozymes in yeast cells.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atzpodien W., Gancedo J. M., Duntze W., Holzer H. Isoenzymes of malate dehydrogenase in Saccharomyces cerevisiae. Eur J Biochem. 1968 Dec;7(1):58–62. doi: 10.1111/j.1432-1033.1968.tb19573.x. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Banaszak L. J. The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J Biol Chem. 1983 Jan 10;258(1):472–482. doi: 10.2210/pdb2mdh/pdb. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Bradshaw R. A., Banaszak L. J. Structure of porcine heart cytoplasmic malate dehydrogenase: combining X-ray diffraction and chemical sequence data in structural studies. Biochemistry. 1987 May 19;26(10):2722–2734. doi: 10.1021/bi00384a011. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Fernley R. T., Bradshaw R. A., Banaszak L. J. Amino acid sequence homology among the 2-hydroxy acid dehydrogenases: mitochondrial and cytoplasmic malate dehydrogenases form a homologous system with lactate dehydrogenase. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6166–6170. doi: 10.1073/pnas.79.20.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birktoft J. J., Fu Z., Carnahan G. E., Rhodes G., Roderick S. L., Banaszak L. J. Comparison of the molecular structures of cytoplasmic and mitochondrial malate dehydrogenase. Biochem Soc Trans. 1989 Apr;17(2):301–304. doi: 10.1042/bst0170301. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breidenbach R. W. Characterization of some glyoxysomal proteins. Ann N Y Acad Sci. 1969 Dec 19;168(2):342–347. doi: 10.1111/j.1749-6632.1969.tb43120.x. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chapman C., Bartley W. The kinetics of enzyme changes in yeast under conditions that cause the loss of mitochondria. Biochem J. 1968 Apr;107(4):455–465. doi: 10.1042/bj1070455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13028–13033. [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duntze W., Neumann D., Gancedo J. M., Atzpodien W., Holzer H. Studies on the regulation and localization of the glyoxylate cycle enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1969 Aug;10(1):83–89. doi: 10.1111/j.1432-1033.1969.tb00658.x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gangloff S. P., Marguet D., Lauquin G. J. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol. 1990 Jul;10(7):3551–3561. doi: 10.1128/mcb.10.7.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C. Glyoxysomal malate dehydrogenase from watermelon is synthesized with an amino-terminal transit peptide. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5773–5777. doi: 10.1073/pnas.87.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C., Lehnerer M., Olsen O. Mitochondrial malate dehydrogenase from watermelon: sequence of cDNA clones and primary structure of the higher-plant precursor protein. Plant Mol Biol. 1990 Jun;14(6):1019–1030. doi: 10.1007/BF00019398. [DOI] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989 May;108(5):1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. M., Tellam J., May V. L., Strauss A. W. Isolation and nucleotide sequence of a cDNA clone encoding rat mitochondrial malate dehydrogenase. Nucleic Acids Res. 1986 Aug 11;14(15):6053–6066. doi: 10.1093/nar/14.15.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hägele E., Neeff J., Mecke D. The malate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, characterisation and studies on their regulation. Eur J Biochem. 1978 Feb 1;83(1):67–76. doi: 10.1111/j.1432-1033.1978.tb12069.x. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh T., Takeshima H., Tsuzuki T., Setoyama C., Shimada K., Tanase S., Kuramitsu S., Kagamiyama H., Morino Y. Cloning and sequence analysis of cDNAs encoding mammalian cytosolic malate dehydrogenase. Comparison of the amino acid sequences of mammalian and bacterial malate dehydrogenase. J Biol Chem. 1987 Nov 5;262(31):15127–15131. [PubMed] [Google Scholar]
- Joh T., Takeshima H., Tsuzuki T., Shimada K., Tanase S., Morino Y. Cloning and sequence analysis of cDNAs encoding mammalian mitochondrial malate dehydrogenase. Biochemistry. 1987 May 5;26(9):2515–2520. doi: 10.1021/bi00383a017. [DOI] [PubMed] [Google Scholar]
- Keys D. A., McAlister-Henn L. Subunit structure, expression, and function of NAD(H)-specific isocitrate dehydrogenase in Saccharomyces cerevisiae. J Bacteriol. 1990 Aug;172(8):4280–4287. doi: 10.1128/jb.172.8.4280-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Rosenkrantz M., Guarente L., Srere P. A. Metabolic changes in Saccharomyces cerevisiae strains lacking citrate synthases. J Biol Chem. 1988 Aug 15;263(23):11145–11149. [PubMed] [Google Scholar]
- Kopetzki E., Entian K. D., Lottspeich F., Mecke D. Purification procedure and N-terminal amino acid sequence of yeast malate dehydrogenase isoenzymes. Biochim Biophys Acta. 1987 Apr 30;912(3):398–403. doi: 10.1016/0167-4838(87)90044-6. [DOI] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- McAlister-Henn L., Blaber M., Bradshaw R. A., Nisco S. J. Complete nucleotide sequence of the Escherichia coli gene encoding malate dehydrogenase. Nucleic Acids Res. 1987 Jun 25;15(12):4993–4993. doi: 10.1093/nar/15.12.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister-Henn L., Thompson L. M. Isolation and expression of the gene encoding yeast mitochondrial malate dehydrogenase. J Bacteriol. 1987 Nov;169(11):5157–5166. doi: 10.1128/jb.169.11.5157-5166.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982 Jun 25;257(12):7181–7188. [PubMed] [Google Scholar]
- McCammon M. T., Veenhuis M., Trapp S. B., Goodman J. M. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J Bacteriol. 1990 Oct;172(10):5816–5827. doi: 10.1128/jb.172.10.5816-5827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Holzer H. Regulation of fructose-1,6-bisphosphatase in yeast by phosphorylation/dephosphorylation. Biochem Biophys Res Commun. 1981 Dec 15;103(3):926–933. doi: 10.1016/0006-291x(81)90899-8. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Polakis E. S., Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965 Oct;97(1):284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenhouse J., Moberly L., Marcus F. Phosphorylation in vivo of yeast (Saccharomyces cerevisiae) fructose-1,6-bisphosphatase at the cyclic AMP-dependent site. J Biol Chem. 1987 Jul 25;262(21):10114–10119. [PubMed] [Google Scholar]
- Roderick S. L., Banaszak L. J. The three-dimensional structure of porcine heart mitochondrial malate dehydrogenase at 3.0-A resolution. J Biol Chem. 1986 Jul 15;261(20):9461–9464. [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small G. M., Lewin A. S. Protein targeting to peroxisomes. Biochem Soc Trans. 1990 Feb;18(1):85–87. doi: 10.1042/bst0180085. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thompson L. M., Sutherland P., Steffan J. S., McAlister-Henn L. Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 1988 Nov 1;27(22):8393–8400. doi: 10.1021/bi00422a015. [DOI] [PubMed] [Google Scholar]
- Witt I., Kronau R., Holzer H. Isoenzyme der malatdehydrogenase und ihre regulation in Saccharomyces cerevisiae. Biochim Biophys Acta. 1966 Oct 17;128(1):63–73. [PubMed] [Google Scholar]
- Witt I., Kronau R., Holzer H. Repression von Alkoholdehydrogenase, Malatdehydrogenase, Isocitratlyase und Malatsynthase in Hefe durch Glucose. Biochim Biophys Acta. 1966 Jun 15;118(3):522–537. [PubMed] [Google Scholar]