Abstract
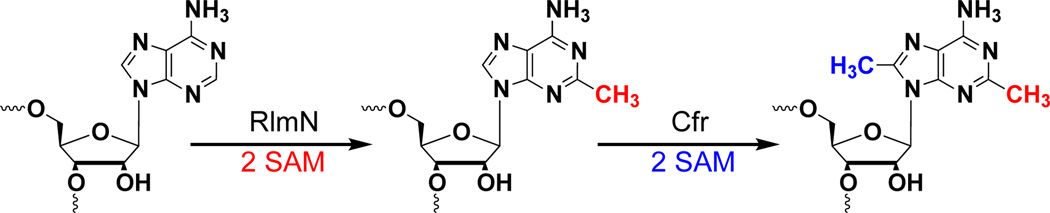
The radical SAM (RS) proteins RlmN and Cfr catalyze methylation of carbons 2 and 8, respectively, of adenosine 2503 in 23S rRNA. Both reactions are similar in scope, entailing the synthesis of a methyl group partially derived from S-adenosylmethionine (SAM) onto electrophilic sp2-hybridized carbon atoms via the intermediacy of a protein S-methylcysteinyl (mCys) residue. Both proteins contain five conserved Cys residues, each of which is required for turnover. Three cysteines lie in a canonical RS CxxxCxxC motif and coordinate a [4Fe–4S]-cluster cofactor. The remaining two cysteines are at opposite ends of the polypeptide. Herein we show that each protein contains only the one “radical SAM” [4Fe–4S] cluster, and that the two remaining conserved cysteines do not coordinate additional iron-containing species. In addition, we show that while wild-type RlmN bears the C355 mCys residue in its as-isolated state, RlmN that is either engineered to lack the [4Fe–4S] cluster by substitution of the coordinating cysteines, or isolated from Escherichia coli cultured under iron-limiting conditions, does not bear a C355 mCys residue. Reconstitution of the [4Fe–4S] cluster on wild-type apo RlmN followed by addition of SAM results in rapid production of S-adenosylhomocysteine (SAH) and the mCys residue, while treatment of apo RlmN with SAM affords no observable reaction. These results indicate that in Cfr and RlmN, SAM bound to the unique iron of the [4Fe–4S] cluster displays two reactivities. It serves to methylate C355 of RlmN (C338 of Cfr), or it serves to generate the 5’-deoxyadenosyl 5’-radical, required for substrate-dependent methyl synthase activity.
The addition of a methyl group to an acceptor molecule is among the most critical of cellular reactions. Although methyl groups can emanate from several complex cofactors (e.g. methylcobalamin and methylene- and methyltetrahydrofolate), the vast majority derive from the more understated molecule, S-adenosyl-l-methionine (SAM).1–3 Seminal model and enzymatic studies on SAM-dependent methyl-transfer reactions argue for a polar process, in which an appropriate nucleophile attacks the methyl group of SAM with concomitant elimination of S-adenosyl-l-homocysteine (SAH).4–6 Until recently, a direct SN2 displacement was the only means by which methyl groups derived from SAM were thought to be appended onto acceptor atoms.7, 8 However, RlmN and Cfr catalyze methyl transfer as well as methyl synthesis via radical-dependent methylene transfer to electrophilic carbon atoms.7, 8
Escherichia coli (Ec) RlmN and Staphylococcus aureus (Sa) Cfr share 33% sequence identity and modify the same nucleotide in 23S rRNA, adenosine 2503 (A2503). Although RlmN and Cfr act preferentially on naked rRNA,9 A2503 resides ultimately in the peptidyltransferase center of the 50S subunit of the bacterial ribosome.10–13 Methylation of C2 by RlmN is found throughout eubacteria (Scheme 1).14 Although this activity is nonessential, Ec mutants that lack it lose to wild-type (wt) Ec in co-growth competition experiments.15 Cfr, which is evolutionarily related to RlmN, also can catalyze methylation of C2 of A2503; however, C8 is its preferred target (Scheme 1).16 Methylation of C8 confers bacterial resistance to multiple classes of antibiotics, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A, as well as the macrolides josamycin and spiramycin.17
Scheme 1.
Reactions catalyzed by RlmN and Cfr in vivo.
Cfr and RlmN are members of the radical SAM (RS) superfamily, enzymes that cleave SAM reductively to a 5’- deoxyadenosyl 5’-radical (5’-dA•).18–21 The common feature of RS enzymes is the use of the 5’-dA• to abstract key substrate hydrogen atoms. Cleavage of SAM requires an electron, which is supplied by a reduced [4Fe–4S]+ cluster. The Cys residues that coordinate this essential [4Fe–4S] cluster typically reside in a CxxxCxxC motif,21 although exceptions have been reported.22–24
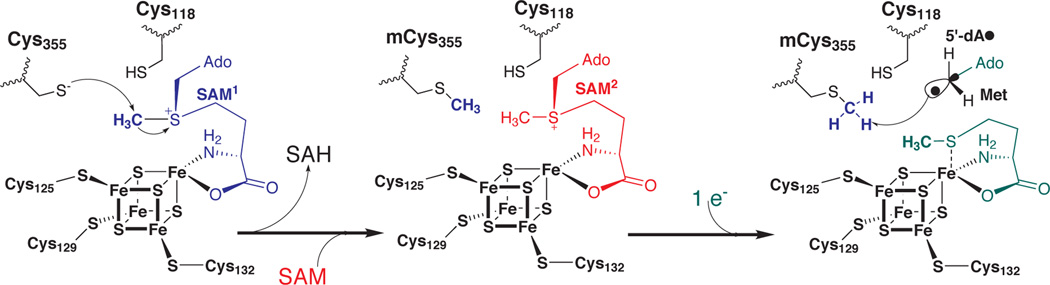
Our recent mechanistic studies of RlmN and Cfr have unraveled an unprecedented strategy for SAM-dependent methylation of an sp2-hybridized carbon atom.7 When RlmN or Cfr was incubated with S-adenosyl-l-[methyl-d3]-methionine (d3-SAM) or unlabeled SAM under single turnover conditions, the isotopic composition of the methyl group incorporated at C2 (RlmN) or C8 (Cfr) of the target adenosine nucleotide did not always reflect that of the methyl donor in the reaction, but rather the isotopic composition of l-methionine in the growth media of the Ec used for overproducing the protein. Consistent with this observation, analysis of RlmN by high-resolution mass spectrometry7 and X-ray crystallography25 showed the as-isolated (AI) protein to bear a methylcysteinyl (mCys) residue at C355. Moreover, methylation always involved incorporation of one hydrogen atom from the mCys residue into 5’-dA, indicating that the 5’-dA• activates the methyl group for radical addition rather than abstracts a hydrogen atom from the nucleotide to be modified. Cleavage of an ensuing methylene-bridged protein–nucleic acid crosslink was proposed to be catalyzed by disulfide-bond formation—with participation of a second absolutely conserved Cys residue (C118 in RlmN; C105 in Cfr), as shown in Scheme S1.7
A related study by Yan et al. also found that the 5’-dA• radical does not abstract a hydrogen atom from the substrate nucleotide. In that study, however, the authors were unaware of the initial transfer of a methyl group from SAM to C355 of RlmN (C338 of Cfr), and therefore proposed that the 5’-dA• abstracts a hydrogen atom from the methyl group of a second, simultaneously bound, SAM molecule, activating it for radical addition to the target nucleotide.8
The recent X-ray crystal structure of RlmN with SAM bound (2.05 Å)25 is consistent with the mechanism proposed by Grove et al.7 The structure shows C355 to reside in a flexible loop containing residues 350–358, which are strictly conserved. This loop is disordered in the absence of SAM, but is visible in the RlmN+SAM structure. Only one SAM-binding site was identified in the structure, wherein SAM is coordinated to the unique iron of the [4Fe–4S] cluster via its α-amino and α-carboxy groups, as is observed for other structurally characterized RS proteins.26 Although it was speculated that the related protein, Cfr, contains two Fe/S clusters,27 this stoichiometry is not supported by the structure of RlmN. The structure shows C355 to be S-methylated, and to be located ~6 Å from the sidechain of C118 and C5’ of SAM. This arrangement is consistent with disulfide-bond formation between C355 and C118 to resolve the protein–nucleic acid covalent adduct as well as abstraction of a hydrogen atom from the C355 mCys by the 5’-dA• as proposed by Grove et al.7 The absence of any other obvious SAM binding site led to the proposal that SAM coordinated to the [4Fe–4S] must exhibit dual functionality; it acts as both a methylating agent and as the source of the 5’-dA• intermediate.25 In this work we use Mössbauer spectroscopy in concert with quantitative analyses of iron and sulfide to show that both RlmN and Cfr contain only one [4Fe–4S] cluster, which is coordinated by cysteines in the canonical CxxxCxxC motif. This finding implies that the two remaining conserved Cys residues do not coordinate Fe/S species and are free to participate in other modes of catalysis. In addition, we overproduce RlmN in Ec under ironlimiting conditions and show that the AI protein is not S-methylated at C355 and does not catalyze methylation of C355 when incubated with SAM. Reconstitution of the protein with iron and sulfide followed by addition of SAM results in rapid methylation of C355, consistent with the proposal that SAM coordinated to the [4Fe–4S] cluster has a dual function.
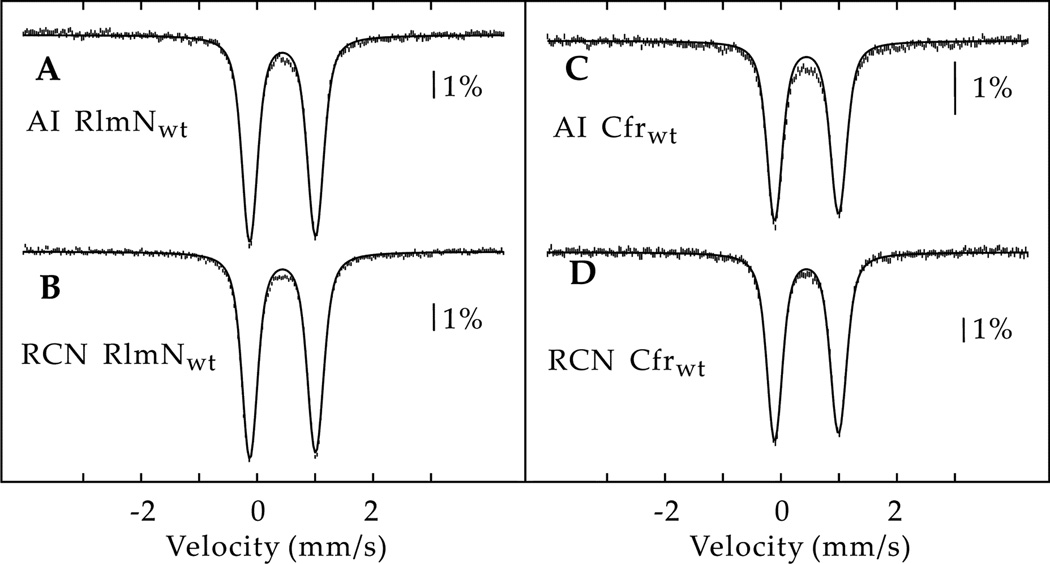
To determine the stoichiometry and configuration of iron-containing species associated with Cfr and RlmN, both proteins were purified from Ec cultured in minimal media supplemented with 57Fe, and then analyzed by Mössbauer spectroscopy. The UV–visible spectra of both AI proteins are consistent with the presence of [4Fe–4S] clusters (Figures S1 and S2), in agreement with previous studies.9, 27 The 4.2-K/53-mT spectrum of asisolated (AI) wild-type (wt) RlmN (RlmNwt) (Figure 1A) is dominated by a broad quadrupole doublet with parameters typical of [4Fe-4S]2+ clusters [isomer shift (δ) of 0.44 mm/s and quadrupole splitting parameter (ΔEQ) of 1.14 mm/s], and accounts for 93% of the total intensity. The weak absorption at −0.6 mm/s is most likely associated with a small amount (~3%) of [2Fe-2S]2+ clusters. Together with the ratio of 4.0 Fe per polypeptide, determined by quantitative analyses of acid-labile iron and sulfide (Table S1), the Mössbauer spectrum reveals a stoichiometry of 0.9 [4Fe-4S] clusters per AI RlmNwt. Reconstitution of AI RlmNwt with additional Fe and sulfide (RCN RlmNwt) does not result in an increase of Fe and sulfide associated with the protein (4.1 Fe per polypeptide). Consistent with this observation, the 4.2-K/53-mT Mössbauer spectrum of RCN RlmNwt (Figure 1B) is virtually identical to that of AI RlmNwt and reveals that RCN RlmNwt harbors 1.0 [4Fe-4S] cluster. This cluster is that coordinated by C125, C129, and C132, the indicated residues in the canonical CxxxCxxC RS motif.
Figure 1.
4.2-K/53-mT Mössbauer spectra of AI RlmNwt (A), RCN RlmNwt (B), AI Cfrwt (C), and RCN Cfrwt (D). Experimental spectra are shown as vertical bars. The solid lines are quadrupole doublet simulations with parameters quoted in the text, accounting for 93% (A), 95% (B), 86% (C), and 98% (D) of the total intensity.
The above cluster stoichiometry is corroborated by studies on an RlmN variant, in which C125, C129, and C132 have been changed to Ala (RlmNC125A-C129A-C132A), non-coordinating residues. AI RlmNC125A-C129A-C132A has only 0.8 Fe per polypeptide. Spectroscopic studies reveal an upper limit of 0.02 [4Fe–4S] clusters per protein (Figures S3 – S5), with the majority of Fe being unspecifically bound. Therefore, we conclude that RlmNC125AC129A-C132A does not harbor an additional Fe/S cluster.
In addition to C125, C129, and C132, RlmN has two additional, strictly conserved, cysteines that are absolutely required for complete turnover: C118 and C355.28 To show that they do not ligate Fe/S species, Cys→Ala variants of those residues (RlmNC118A and RlmNC355A) were engineered and studied with a combination of biochemical and spectroscopic methods. RlmNC118A and RlmNC355A contain 4.0–4.6 Fe per polypeptide in both their AI and RCN forms (Table S1), and display spectroscopic features and cluster stoichiometries that are virtually identical to those of RlmNwt, supporting the presence of one [4Fe–4S] cluster per polypeptide (Figures S6 and S7) ligated by C125, C129, and C132.
The 4.2-K/53-mT spectrum of AI Cfrwt (Figure 1C) is dominated by a broad quadrupole doublet indicative of [4Fe-4S]2+ clusters (δ = 0.44 mm/s and ΔEQ = 1.10 mm/s), which accounts for 86% of the total intensity. In addition, the spectrum of the sample exhibits a small amount (−15% of total Fe) of a broad, poorly resolved, feature that extends from −2 to 2 mm/s. Because an identical EPR sample does not show any significant signals associated with features of Fe/S clusters with S = 1/2 ground state (Figure S5), we assign the broad feature to unspecifically bound iron. Together with the ratio of 4.5 Fe per polypeptide (Table S1), the Mössbauer spectrum reveals a stoichiometry of 1.0 [4Fe-4S] cluster per AI Cfrwt. Reconstitution of AI Cfrwt with additional Fe and sulfide followed by purification (RCN Cfrwt) results in a slight decrease of Fe. The 4.2-K/53-mT Mössbauer spectrum of RCN Cfrwt (Figure 1D) can be simulated with a quadrupole doublet with identical parameters, and accounts for 98% of the total intensity. Thus, the results suggest that RCN Cfrwt harbors 1.0 [4Fe-4S] cluster, which is coordinated by C112, C116, and C119. We therefore conclude, that in contrast to radical SAM proteins that catalyze methylthiolation, which contain two distinct Fe/S clusters,18, 29, 30 RlmN and Cfr only contain the cluster housed in the canonical CxxxCxxC motif.
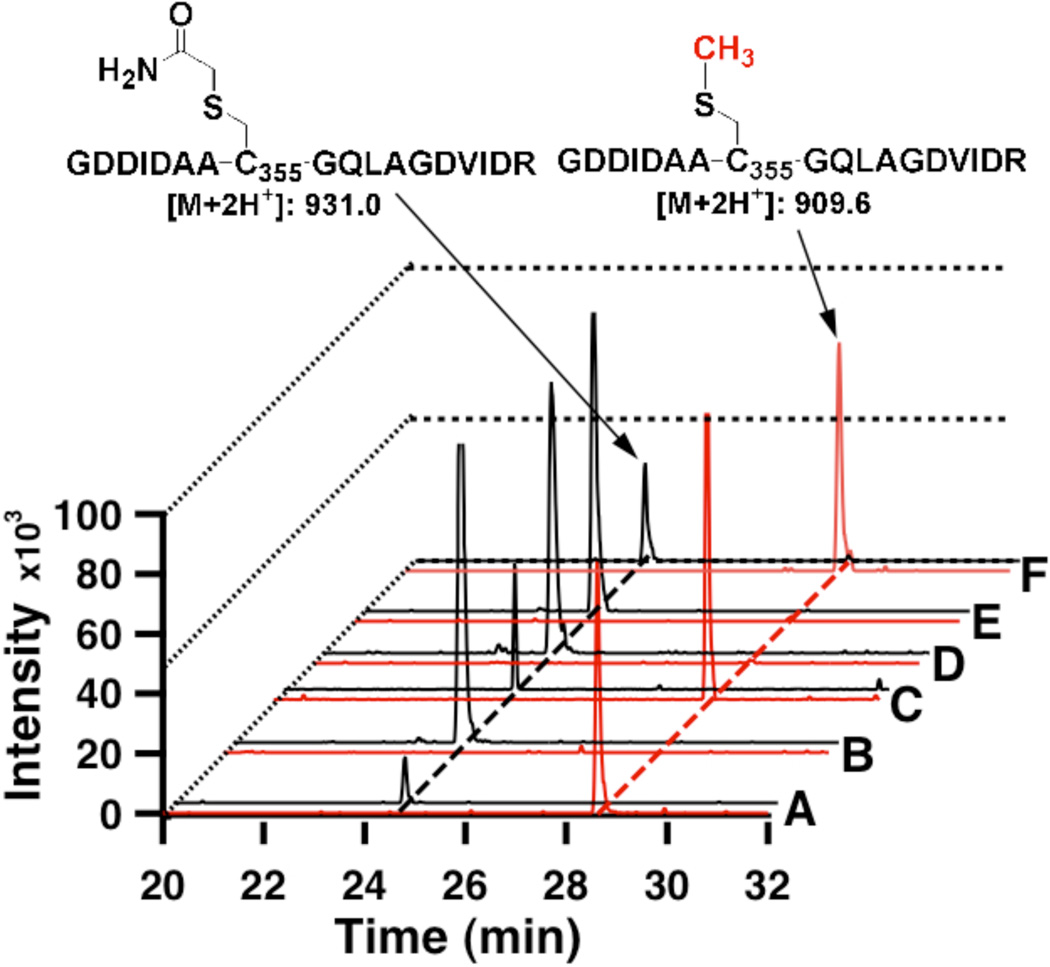
While previous studies have firmly established one role for the [4Fe–4S] cluster in the unique methylation reactions catalyzed by RlmN and Cfr,7–9, 25 — reductive cleavage of SAM to generate the 5’-dA• intermediate — the role of the [4Fe–4S] cluster, if any, in the first step of the reaction — methylation of a conserved cysteine (C355 in RlmN and C338 in Cfr) — has not been addressed. Interestingly, when the RlmNC125A-C129A-C132A triple variant was analyzed by ESI+-MS after alkylation with iodoacetamide and digestion with trypsin, the peptide containing C355 was found exclusively acetylated, indicating that it did not bear a mCys residue (Figure 2B). This observation contrasts with results obtained for RlmNwt, wherein almost 90% of the AI protein contained a mCys residue, and very little of it was alkylated by iodoacetamide during preparation for analysis by mass spectrometry (Figure 2A).7 To assess the effect of the iron–sulfur (Fe/S) cluster on generation of the mCys residue in RlmN, apo RlmNwt was produced by expression of the Ec yfgB gene — which encodes RlmN — in a modified M9 minimal medium containing 75 µM o-phenanthroline to chelate available iron (see supporting information for experimental details).31 A large fraction of RlmN was produced in inclusion bodies when expression was carried out in this manner. The soluble fraction was isolated anaerobically under conditions identical to those for isolation of holo RlmN, and a portion was set aside for reconstitution of its [4Fe–4S] cluster with Fe and sulfide by standard methods.32 Figure 3A shows UV–vis spectra of apo RlmNwt isolated from Ec cultured in o-phenanthroline-containing medium (solid black line), and RlmNwt reconstituted with iron and sulfide (apo RlmNwt→RCN) (solid red line). As can be observed, the telltale features of [4Fe–4S] clusters are significantly diminished in apo RlmNwt, consistent with the finding of 0.04 Fe and 0.07 sulfides per polypeptide (Table S1), but reappear in the apo RlmNwt→RCN sample. Analysis of trypsin-digested apo RlmNwt by LC-MS showed that it did not bear a C355 mCys modification, consistent with the premise that SAM bound to the [4Fe–4S] cluster acts as a methylating agent (Figure 2E). To ensure that in vivo methylation of apo RlmNwt was not the factor that distinguished soluble protein from insoluble protein, apo RlmNwt inclusion bodies were also analyzed by LC-MS; fragments obtained from trypsin digests of the inclusion body fraction did not bear a C355 mCys modification (Figure 2D).
Figure 2.
Extracted ion chromatograms (EIC) for the C355-containing peptide from trypsin digests of RlmN. Black traces (A, B, C, D, E, F) are EIC of m/z 931.0 [M+2H], indicative of carbamidomethyl modification to C355. Red traces (A, B, C, D, E, F) are EIC of m/z 909.6 [M+2H], indicative of a methyl modification of C355. Traces correspond to A) AI RlmNwt; B) AI RlmNC125A-C129A-C132A; C) AI RlmNC118A; D) apo RlmNwt, inclusion bodies; E) apo RlmNwt, soluble fraction; F) apo RlmNwt→RCN after addition of 1.5 mM SAM. Spectral pair intensities are normalized to the most abundant EIC.
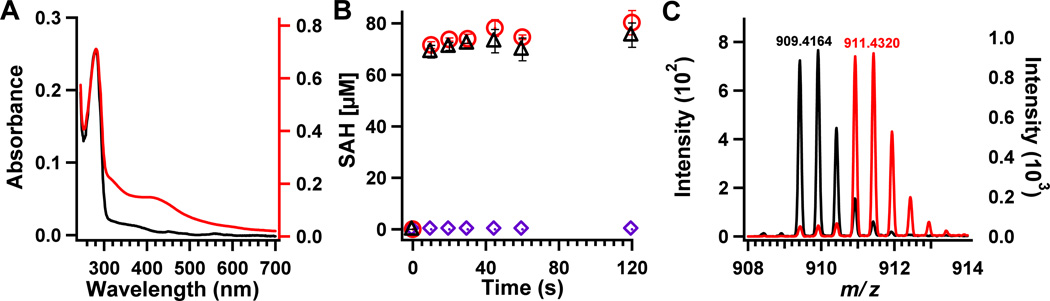
Figure 3.
A) UV–vis traces of apo RlmNwt (5 µM, solid black line) and apo RlmNwt→RCN (12 µM, solid red line). B) Methyl transfer catalyzed by apo RlmNwt (150 µM) or apo RlmNwt→RCN (150 µM). Production of SAH by apo RlmNwt (purple diamonds) or apo RlmNwt→RCN (red circles) in the presence of 1.5 mM SAM; or apo RlmNwt→RCN (150 µM) in the presence of 1.5 mM d3-SAM (black triangles). Error bars indicate one standard deviation from the average of three assays. C) Q-Tof MS analysis of tryptic peptides from apo RlmNwt→RCN after incubation in the presence of SAM (black trace) or d3- SAM (red trace). Indicated m/z values correspond to the +2 charge state.
Reconstitution of apo RlmNwt followed by further purification by size-exclusion chromatography afforded a protein containing 2.2 irons and 1.2 sulfides, respectively, per polypeptide (Table S1). This stoichiometry implies that 50% of the RCN protein contained a [4Fe–4S] cluster, given that its UV-vis spectrum does not contain telltale signatures of [2Fe–2S] clusters, and most, if not all, of the adventitiously bound iron was removed by molecular-sieve chromatography. When SAM was added to apo RlmNwt, methyl transfer to C355 did not occur, as evidenced by no significant production of SAH (Figure 3B, purple diamonds). By contrast, when SAM or d3-SAM was added to apo RlmNwt→RCN, ~0.5 equiv of SAH was produced within the first time point (15 s). Subsequent time points did not show increased amounts of SAH, suggesting that 50% of apo RlmNwt was reconstituted to its holo form (Figure 3B, red circles). Importantly, the quantity of SAH produced was directly proportional to the RlmN cluster stoichiometry, providing further evidence that the [4Fe–4S] cluster is required for methyl transfer. In addition, apo RlmNwt→RCN was capable of catalyzing time-dependent methylation of the target nucleotide in a 771-base synthetic RNA substrate (Figure S10). Analysis of apo RlmNwt→RCN by LC–MS after incubating it with SAM reveals a peak exhibiting a mass-to-charge ratio (m/z) of 909.6 (+2 charge state) for the C355-containing peptide fragment obtained after trypsin digestion, indicating that it bears a methyl group (Figures 2F and 4B, black trace). Moreover, the peak at m/z = 909.6 shifts to 911.2 (+2 charge state; thus the shift is ~1.5 units instead of the expected 3 for the +1 charge state) when apo RlmNwt→RCN is incubated with d3-SAM and similarly analyzed (Figure 4B, red trace). This modification is specific to C355 of RlmN, given that MS/MS analysis of the peptides shows that the y+ ion series is shifted by +15 for fragments that bear C355 (y11+1 and above) (Figure S8).7
The effect of C118 — another Cys residue that plays a key role in RlmN catalysis — on methylation of C355 was also assessed. As observed in Figure 2C, a large fraction of the AI C118A variant contains a mCys modification, consistent with this residue playing an insignificant role in SAM binding and methylation of C355. Last, the inclusion of the flavodoxin/flavodoxin reductase/NADPH reducing system had no effect on C355 methylation, indicating that methyl transfer to C355 does not require a reduced [4Fe–4S] cluster (Figure S9), and therefore most likely proceeds by a polar nucleophilic displacement, as do all characterized SAM-dependent methyltransferases.1
Herein we have shown that RlmN and Cfr catalyze two separate and distinct reactions, exploiting both polar and radical reactivities of SAM within a single polypeptide (Scheme 2). Indeed, these proteins both possess methyltransferase and methylsynthase activities. They contain only one [4Fe–4S] cluster — ligated by cysteines in CxxxCxxC motifs — to which SAM binds via its α-amino and carboxy groups.25 In this conformation, methyl transfer to unmodified C355 of RlmN (C338 of Cfr) takes place rapidly. Release of the product, SAH, permits binding of a second molecule of SAM to the exact same site; however, methyl transfer to the reactive cysteine is now blocked. Upon binding of the RNA substrate and the addition of an electron to the [4Fe–4S]2+ cluster, SAM is now induced to fragment into methionine and the 5’-dA•, the latter initiating catalysis by abstracting a hydrogen atom from the mCys residue. Our studies show that methyl transfer to the target Cys residue does not require the presence of substrate. Whether substrate binding accelerates this step is yet to be determined. However, our previous studies showed that substrate binding does indeed effect radical formation when the physiological reducing system is used to supply the requisite electron.7 Excitingly, this mode of catalysis is a clever twist on the “principle of economy in the evolution of binding sites,” wherein Nature evolves only a single substrate-binding site for reactions that involve two or more substrates with similar structural elements, and then mediates transfer of a group from one to the other by means of an intermediate enzyme–functional group covalent adduct.33 The Cfr and RlmN reactions are the first recognized instances in which a single substrate-binding site activates one molecule both for polar and radical-based chemistry.
Scheme 2.
SAM bound to the [4Fe–4S] cluster of RlmN or Cfr is activated toward two distinct reactivites.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (GM-63847 to S.J.B.). We thank Tatiana N. Laremore of the PSU MS facility for collecting high-resolution MS data.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: Materials and Methods; Table S1; and Figures S1–S10. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Frey PA, Hegeman AD. New York: Oxford University Press; 2007. [Google Scholar]
- 2.Markham GD. Encyclopedia of Life Sciences. John Wiley & Sons, Inc; 2010. S-adenosylmethionine. [Google Scholar]
- 3.Silverman RB. New York: Academic Press; 2002. [Google Scholar]
- 4.Hegazi MF, Borchardt RT, Schowen RL. J. Am. Chem. Soc. 1979;101:4359–4365. [Google Scholar]
- 5.Iwig DF, Grippe AT, McIntyre TA, Booker SJ. Biochemistry. 2004;43(42):13510–13524. doi: 10.1021/bi048692h. [DOI] [PubMed] [Google Scholar]
- 6.Woodard RW, Tsai M-D, Floss HG, Crooks PA, Coward JK. J. Biol. Chem. 1980;255:9124–9127. [PubMed] [Google Scholar]
- 7.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 8.Yan F, Fujimori DG. Proc. Natl. Acad. Sci. U S A. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan F, LaMarre JM, Röhrich R, Wiesner J, Jomaa H, Mankin AS, Galoníc Fujimori D. J. Am. Chem. Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ban N, Freborn B, Nissen P, Penczek P, Grassucci RA, Sweet R, Frank J, Moore PB, Steitz TA. Cell. 1998;93:1105–1115. doi: 10.1016/s0092-8674(00)81455-5. [DOI] [PubMed] [Google Scholar]
- 11.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Francheschi F, Yonath A. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 12.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Hoton JM, Cate JHD. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 13.Selmer M, Dunham CM, Murphy FV, IV, Weislbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 14.The_UniProt_Consortium. Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toh S-M, Xiong L, Bae T, Mankin AS. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giessing AMB, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long KS, Vester B, Kirpekar F. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith LK, Mankin AS. Antimicrob. Agents Chemother. 2008;52:1703–1712. doi: 10.1128/AAC.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Booker SJ. Curr. Opin. Chem. Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey PA, Booker SJ. Adv. Protein Chem. 2001;58:1–45. doi: 10.1016/s0065-3233(01)58001-8. [DOI] [PubMed] [Google Scholar]
- 20.Frey PA, Hegeman AD, Ruzicka FJ. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 21.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat. Chem. Biol. 2008;4:758–765. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez–Gomez NC, Downs DM. Biochemistry. 2008;47:9054–9056. doi: 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGlynn SE, Boyd ES, Shepard EM, Lange RK, Gerlach R, Broderick JB, Peters JW. J. Bacteriol. 2010;192:595–598. doi: 10.1128/JB.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vey JL, Drennan CL. Chem. Rev. 2011;111:2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Booth MPS, Challand MR, Emery DC, Roach PL, Spencer J. Protein Expr. Purif. 2010;74:204–210. doi: 10.1016/j.pep.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. Nuc. Acids. Res. 2010 doi: 10.1093/nar/gkp1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atta M, Mulliez E, Arragain S, Forouhar F, Hunt JF, Fontecave M. Curr. Opin. Struct. Biol. 2010;20:1–9. doi: 10.1016/j.sbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Booker SJ, Cicchillo RM, Grove TL. Curr. Opin. Chem. Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkin SE, Chen S, Ley BA, Mangravite L, Edmondson DE, Huynh BH, Bollinger JM., Jr Biochemistry. 1998;37:1124–1130. doi: 10.1021/bi9723717. [DOI] [PubMed] [Google Scholar]
- 32.Cicchillo RM, Lee K-H, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ. Biochemistry. 2004;43:11770–11781. doi: 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 33.Frey PA. Nucleotidyltransferases and phosphotransferases: Stereochemistry and covalent intermediates. In: Sigman DS, editor. The Enzymes. 3rd Ed. Vol. 20. San Diego: Academic Press, Inc; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.