Abstract
During development, Myxococcus xanthus cells undergo programmed cell death (PCD) whereby 80% of vegetative cells die. Previously, the MazF RNA interferase has been implicated in this role. Recently, it was shown that deletion of the mazF gene does not eliminate PCD in wild-type strain DK1622 as originally seen in DZF1. To clarify the role of MazF, recombinant enzyme was characterized using a highly sensitive assay in the presence and absence of the proposed antitoxin MrpC. In contrast to previous reports that MrpC inhibits MazF activity, the hydrolysis rate was enhanced in a concentration-dependent manner with MrpC or MrpC2, an N-terminally truncated form of MrpC. Furthermore, MazF transcripts were not detected until 6–8 hours post-induction, suggesting an antitoxin is unnecessary earlier. Potential MazF targets were identified and their transcript levels were shown to decline in DK1622 while remaining steady in a mazF deletion strain. Elimination of the mazF hydrolysis site in the nla6 transcript resulted in overproduction of the mRNA. Thus, MazF negatively regulates specific transcripts. Additionally, we show that discrepancies in the developmental phenotypes caused by removal of mazF in DK1622 and DZF1 are due to the presence of the pilQ1 allele in the latter strain.
Keywords: MazF, Myxococcus xanthus, mRNA interferase, development
Introduction
Microbes regulate many complex processes with networks of sensing and signaling pathways. Programmed cell death (PCD) is perhaps the most intriguing because it results in altruistic suicide for the benefit of neighboring organisms (Perry et al., 2009, Blower et al., 2009). The δ-Proteobacterium Myxococcus xanthus is an excellent model for the study of cell-to-cell communication and PCD. In response to starvation, M. xanthus forms multicellular fruiting bodies containing spores. Development is directed by a series of extracellular signals that occur in a precise temporal order, ultimately leading to PCD for roughly 80 percent of cells (Shimkets, 1999, Wireman & Dworkin, 1977).
Recently, the M. xanthus genome was found to encode an orphan mazF toxin gene (Nariya & Inouye, 2008). Removal of mazF resulted in greatly reduced PCD and sporulation in strain DZF1. MazF belongs to the family of enzymes known as mRNA interferases, which function as endoribonucleases to hydrolyze RNA molecules at specific sequences (Yamaguchi & Inouye, 2009). In other organisms, mazF is normally found cotranscribed with mazE. MazF activity is inhibited by MazE binding and initiated when MazE is degraded in response to stress conditions (Gerdes et al., 2005). Nariya et al. searched for potential M. xanthus MazF binding partners and identified MrpC, which binds MazF in vitro and in vivo and was reported to inhibit endoribonuclease activity (Nariya & Inouye, 2008).
MrpC is a transcription factor that activates expression of many development-specific genes (Ueki & Inouye, 2003, Sun & Shi, 2001). Strains lacking mrpC fail to develop and sporulate (Nariya & Inouye, 2008). Pre-starvation, MrpC is negatively regulated by phosphorylation through the Pkn8/Pkn14 kinase cascade (Nariya & Inouye, 2005). Upon starvation, a shortened form of MrpC is observed that lacks the first 33 N-terminal residues, eliminating the predicted site of phosphorylation (Ueki & Inouye, 2003). Subsequent work using a recombinant form lacking only 25 N-terminal residues, named MrpC2, showed it is not present in cells lacking the protease LonD (Nariya & Inouye, 2006). Direct hydrolysis has not been demonstrated. MrpC2 binds DNA with greater affinity and positively regulates expression of mrpC (Nariya & Inouye, 2006). MrpC2 also binds to the promoter region upstream of the mazF gene to activate transcription of the mazF gene (Nariya & Inouye, 2008).
Together, these results point to an elegant system of PCD that is fundamentally tied to development through the use of an essential transcription factor. Unfortunately, conflicting data have recently been presented with regard to MazF function. Lee et al. showed that removal of mazF from wild-type strains DK1622 and DZ2 had little to no effect on development or PCD (Lee et al., 2012). The earlier experiments had been done in the DZF1 background, which contains a pilQ1 allele that compromises an outer membrane secretin required for S-motility (Wall et al., 1999). This allele bears two missense mutations in pilQ that cause amino acid substitutions G741S and N762G. PilQ1 greatly sensitizes cells to antibiotics and could conceivably render cells more susceptible to PCD. Thus the role of MazF in Myxococcus PCD remains in question.
A larger question is whether MazF mediates PCD in any organism. MazF was initially suggested to mediate PCD in Escherichia coli (Aizenman et al., 1996). E. coli MazF has a small target sequence, ACA (Zhang et al., 2003). Consequently, E. coli MazF degrades most mRNAs, arresting protein synthesis and leading to growth arrest (Zhang et al., 2003). However, these cells can be fully revived upon induction of mazE and seem not undergo PCD (Pedersen et al., 2002). Furthermore, MazEF was shown to contribute to cell survival and persistence in E. coli (Gerdes & Maisonneuve, 2012). In other organisms, specific MazF regulatory roles have been determined. Mycobacterium tuberculosis contains several MazF homologues, each recognizing a different pentameric sequence. Each MazF eliminates a subset of mRNAs to alter protein expression through a regulatory mechanism of specific mRNA degradation (Zhu et al., 2008). In Staphylococcus aureus, the MazF recognition sequence is unusually abundant in virulence factor mRNA allowing induction of MazF to diminish pathogenesis (Zhu et al., 2009). Similar trends of regulatory cleavage can be seen in Clostridium difficile and Bacillus subtilis (Rothenbacher et al., 2012, Park et al., 2011). Thus it has become apparent that MazF homologues display a wide variety of functions, none directly related to PCD. The predominant recognition sequence of M. xanthus MazF was determined to be an octamer, GAGUUGCA (Nariya & Inouye, 2008). Such a long recognition sequence is unique to M. xanthus, suggesting a specific regulatory role for the enzyme.
We employed a highly sensitive assay for MazF activity and show that recombinant MrpC and MrpC2 increase rather than inhibit recombinant MazF RNA interferase activity in vitro. In addition, we identify target molecules for MazF hydrolysis and show that transcript levels are greatly diminished upon expression of MazF during development unless the MazF hydrolysis site is removed. Finally, we show that the pilQ1 allele is responsible for stimulating PCD in DZF1 cells containing a mazF mutation.
Results
Expression and Purification of MazF, MrpC, and MrpC2
The M. xanthus mazF gene (MXAN_1659) was cloned with a N-terminal six-histidine tag. The protein was expressed and purified in E. coli with a yield of approximately 15 mg protein L−1 culture. A band corresponding to approximately 15 kDa was observed with SDS-PAGE in agreement with the theoretical molecular weight of the recombinant protein (Figure S1). No MazF proteins are known to utilize cofactors, so no distinct features were present in the UV-VIS spectrum (not shown).
The M. xanthus mrpC gene (MXAN_5125) was cloned and the protein expressed as a C-terminal fusion to a 55 kDa chitin binding domain with the Saccharomyces cerevisiae VMA intein. Cleavage of the intein with DTT yielded approximately 3 mg unmodified protein L−1 culture. For MrpC2, we took advantage of a natural methionine codon 75 bp downstream from the start codon of mrpC, which has previously been used in recombinant MrpC2 studies (Nariya & Inouye, 2006, Nariya & Inouye, 2008). MrpC2 was expressed and purified identically to MrpC with similar yields. MrpC and MrpC2 showed approximately 27 and 25 kDa bands respectively, in line with the theoretical values calculated from amino acid sequence (Figure S1). As expected, no distinct features were present in the UV-VIS spectrum.
MazF acts as a mRNA interferase
A continuous fluorometric assay was developed using an RNA oligonucleotide bearing a fluorophore at the 5’- position and a quencher at the 3’-position (Zhang et al., 2005). Fluorescence is detected when this synthetic oligonucleotide is cleaved by MazF. Since the cleavage site of M. xanthus MazF is known (Nariya & Inouye, 2008), the substrate was designed to be 13 nucleotides long with the octamer recognition sequence (rGrArGrUrUrGrCrA) at the center.
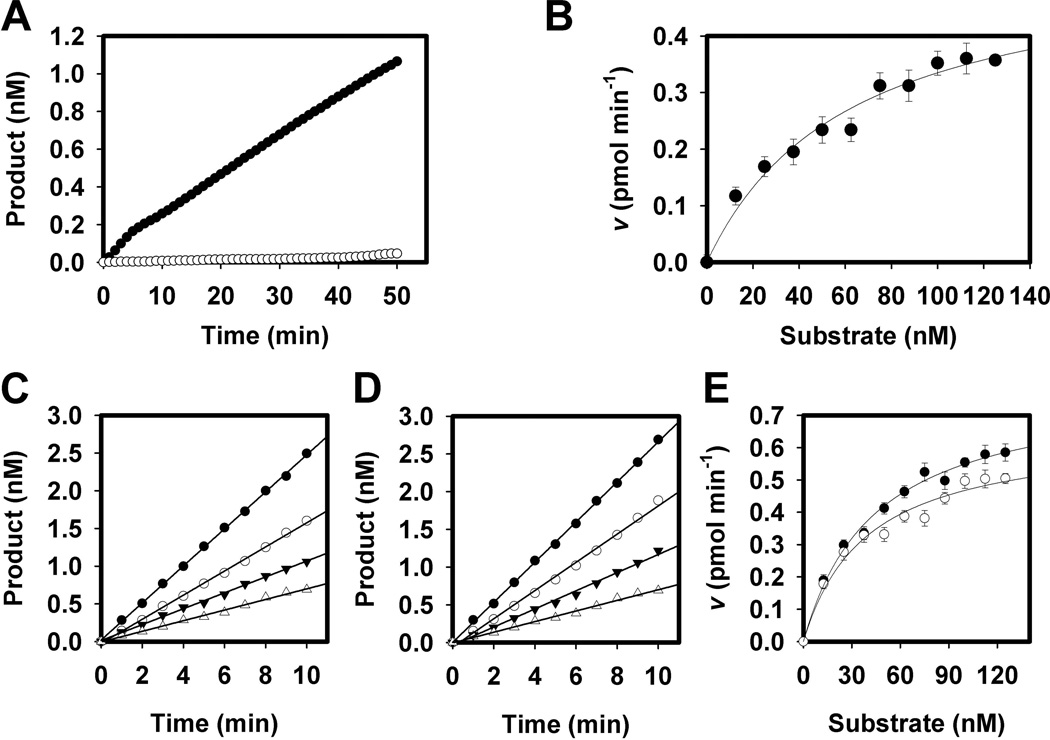
Fluorescence was only detectable after cleavage by MazF (Figure 1A). The reaction follows standard first-order Michaelis-Menten kinetics when fitted to the Michaelis equation (Figure 1B). The Km and Vmax for the substrate were 62 nM and 0.4 pmol min−1 respectively. The maximum rate is comparable with the E. coli homologue having a Vmax of 0.37 pmol min−1. M. xanthus MazF displays a very high substrate affinity when compared to E. coli MazF however, which has a Km over two orders of magnitude higher at 6.9 µM (Wang & Hergenrother, 2007).
Figure 1.
Enzymatic hydrolysis of synthetic substrate by MazF. A. MazF mRNA interferase assays are shown for 250 nM MazF + 25 nM substrate (6-carboxyfluorescein 5’-rArArGrArGrUrUrGrCrArCrArG-3’-Black Hole Quencher 1) (●) versus substrate only (○). Activity is only seen in the presence of enzyme. B. Determination of apparent Michaelis constants for MazF. 250 nM MazF was assayed in the presence of varying concentrations of substrate. Data were fitted to equation 1. The Km and Vmax were 62 nM and 0.4 pmol min−1 respectively. Error bars represent the standard deviation of triplicate experiments. C–E. Enzymatic activity of MazF is enhanced in the presence of MrpC or MrpC2. C. Sample assay of 250 nM MazF alone (△) or in the presence of 250 nM (▼), 2.5 µM (○), or 25 µM (●) MrpC. D. Sample assay of 250 nM MazF alone (△) or in the presence of 250 nM (▼), 2.5 µM (○), or 25 µM (●) MrpC2. E. Effect of MrpC and MrpC2 on Michaelis constants for the MazF reaction. Equimolar amounts of MrpC (○) or MrpC2 (●) were used. Vmax and Km for 250 nM MazF + 250 nM MrpC were 45 nM and 0.8 pmol min−1 respectively. Vmax and Km for 250 nM MazF + 250 nM MrpC2 were 42 nM and 0.7 pmol min−1 respectively. Error bars represent the standard deviation of triplicate experiments.
MazF activity is enhanced, not inhibited, by MrpC/MrpC2
MazF was assayed in the presence of varying concentrations of MrpC and MrpC2 to examine potential interactions. Unexpectedly, the reaction rates were doubled in the presence of MrpC or MrpC2 (Figure 1C and 1D). With equimolar amounts of MrpC, the Km and Vmax of MazF became 45 nM and 0.8 pmol min−1, respectively (Figure 1E). With equimolar amounts of MrpC2, the Km and Vmax were 42 nM and 0.7 pmol min−1, respectively. Greater concentrations of MrpC or MrpC2 also enhanced the rate of hydrolysis, but with diminishing returns. For example, a ten-fold increase in MrpC:MazF ratio yielded only a four-fold increase in activity. MazF was also assayed identically in the presence of BSA to rule out the possibility that the enzymatic enhancement is nonspecific (data not shown). No stimulation was observed even at the highest BSA concentration.
mazF transcripts appear during the peak of mrpC levels
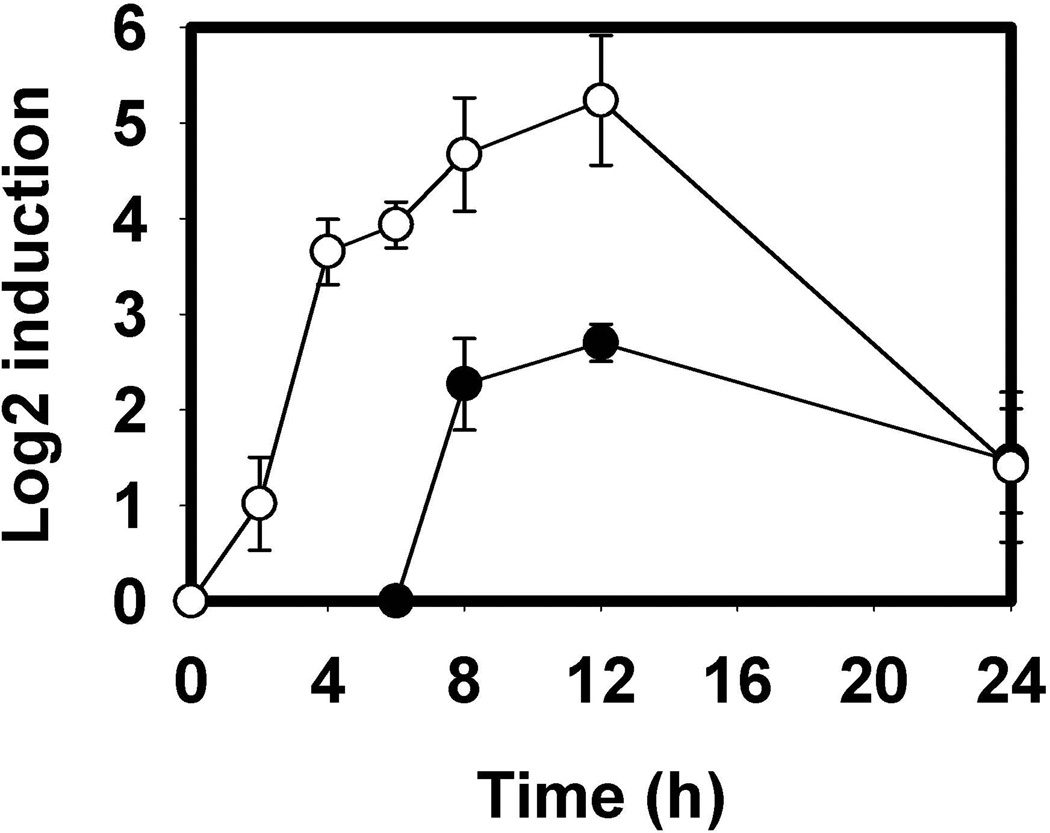
Expression of MrpC increases 6–12 hours post-starvation, when MrpC2 first appears (Nariya & Inouye, 2005). MrpC2 increases mrpC expression during this time by binding to its own promoter region with a 4-fold higher affinity than MrpC (Nariya & Inouye, 2006). Two putative MrpC binding sequences are also present in the mazF promoter region leading one to suspect that MazF is produced in the same time frame (Nariya & Inouye, 2008). The relative transcript levels of mazF and mrpC were determined via quantitative RT-PCR in the wild-type strain DK1622 at 2, 4, 6, 8, 12, and 24 hours post-starvation, with each timepoint normalized against the housekeeping gene rpoC (MXAN_3078). The 0 hour samples represent transcript levels present during vegetative growth. mrpC transcript levels are in agreement with Western blots that show MrpC levels peaking around 12 hours followed by a sharp decline (Konovalova et al., 2012) (Figure 2). In DK1622, mazF transcripts could not be detected until 6–8 hours, peaking around 12 hours. These data indicate that MazF is a development-specific protein.
Figure 2.
Relative transcript levels of mrpC and mazF during development. Transcript levels of mrpC (○) and mazF (●) were determined using quantitative RT-PCR during 2, 4, 6, 8, 12, and 24 hour time points post-starvation compared with vegetative cells. mazF transcripts were not detected until 6–8 hours, during the peak of mrpC expression. Relative levels were calculated using the ΔΔCT method with error bars representing the standard deviation of triplicate samples.
MazF regulates transcript levels of genes required for development and cell viability
MazF cannot compromise global transcript levels due to the specificity of the M. xanthus MazF recognition sequence. To address what portions of the transcriptome are altered in vivo, Pattern Locator (http://www.cmbl.uga.edu/software/patloc.html) was used to generate a list of coding sequences containing the cleavage site 5’-GAGUUGCA-3’ (Mrazek & Xie, 2006) (Table S1). Five potential candidates were selected from the list of 72 due to their ability to alter cell fate or mediate development (highlighted on table S1). nuoM encodes the M subunit of NADH dehydrogenase, a complex required for stationary phase survival in E. coli and many other organisms (Zambrano & Kolter, 1993). ClpA is part of the multimeric ClpAP protease, which mediates protein degradation and is also required for E. coli stationary phase survival (Weichart et al., 2003). The remaining three genes are required for M. xanthus development. FrzZ is the output of the Frz chemosensory pathway and is required for fruiting body formation (Inclan et al., 2007). EpsE is a glycosyl transferase involved in exopolysaccharide biosynthesis and is required for S-motility and fruiting body formation (Lu et al., 2005). Nla6 is an enhancer protein that is required for sporulation (Caberoy et al., 2003). It should be noted that the list of genes presented in Table S1 may not contain all MazF targets, as cleavage of any of these transcripts may also affect translation of other genes within the same transcript. Intergenic regions were not included but could produce as yet unidentified small RNAs.
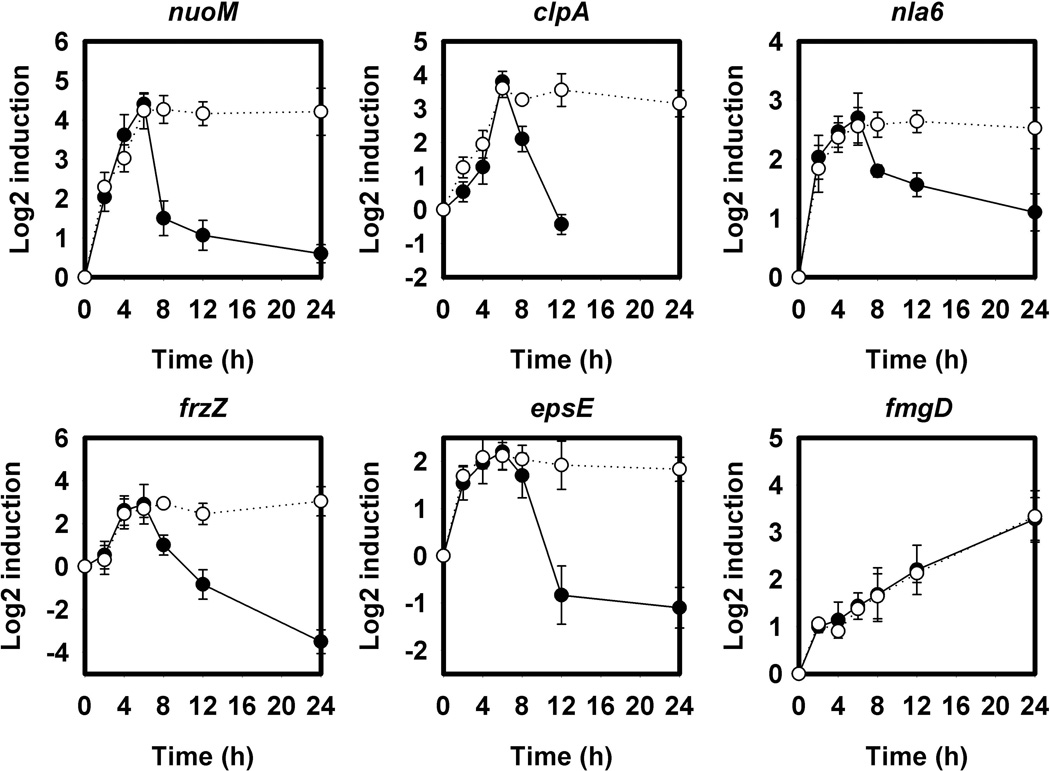
RT-PCR was used to quantify relative transcript levels 2, 4, 6, 8, 12, and 24 hours post-starvation in DK1622 and LS3118 (ΔmazF) (Figure 3). All five MazF target transcripts peaked 4–6 hours after development. They dropped off in DK1622 but remained high in cells lacking MazF. A gene lacking the MazF hydrolysis site, fmgD (MXAN_1501), was also examined and had identical profiles in both wild type and LS3118 (Figure 3).
Figure 3.
Relative transcript levels of potential MazF targets during development. Transcript levels were determined as in Figure 2 for each of the 5 candidate genes in wild type DK1622 (●) and the ΔmazF strain LS3118 (○). In cells containing MazF, transcripts of each of these genes began to drop significantly around 8 hours at the time MazF is expressed, unlike LS3118. fmgD lacks the MazF recognition sequence and was used as a control. Relative levels were calculated using the ΔΔCT method with error bars representing the standard deviation of triplicate samples.
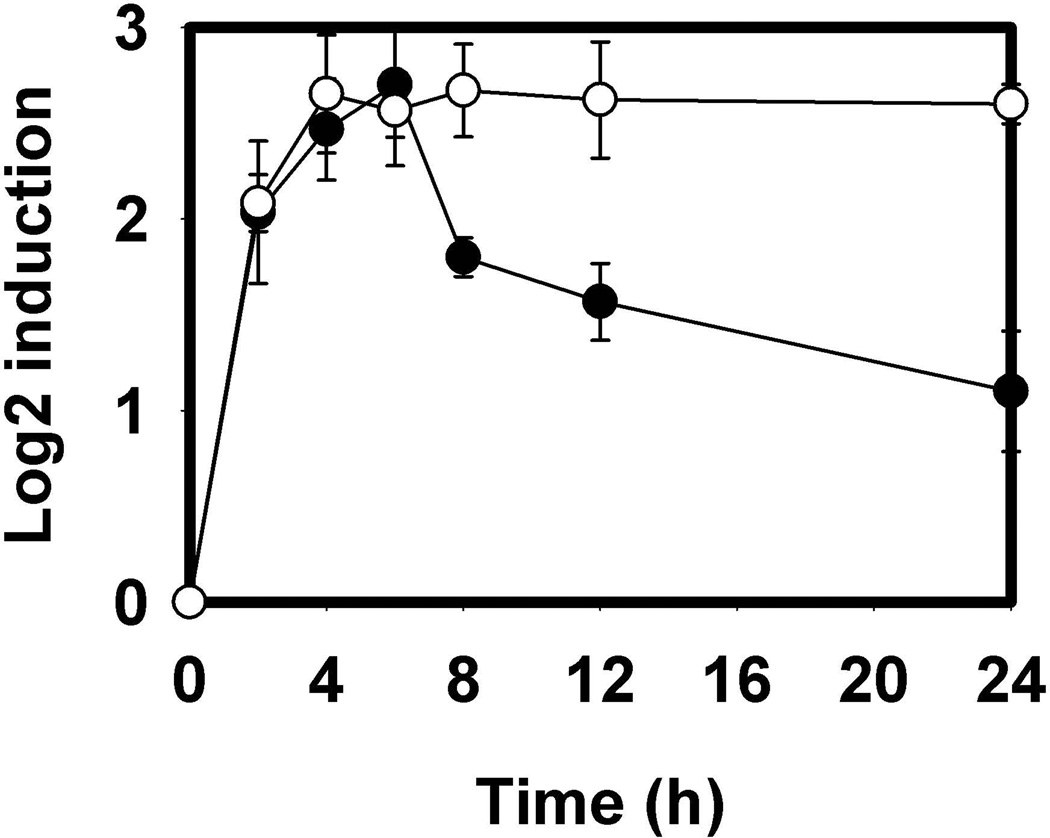
To prove that MazF is directly responsible for the changes in target mRNA levels, LS3160 was created in which nla6 was desensitized to MazF. Two silent mutations were introduced, T751C and G753C, abolishing the MazF hydrolysis site but maintaining the amino acid sequence. Transcript levels were found to remain high despite the presence MazF, confirming the biological relevance of MazF in reducing these transcripts (Figure 4).
Figure 4.
Decreased levels of MazF target mRNA is enzyme-specific. Relative transcript levels of wild type nla6 mRNA (●) decrease during development while LS3160 (○) do not. LS3160 contains two silent mutations in nla6 that abolish the MazF hydrolysis site. Relative levels were calculated using the ΔΔCT method with error bars representing the standard deviation of triplicate samples.
The pilQ1 allele blocks development in mazF mutants
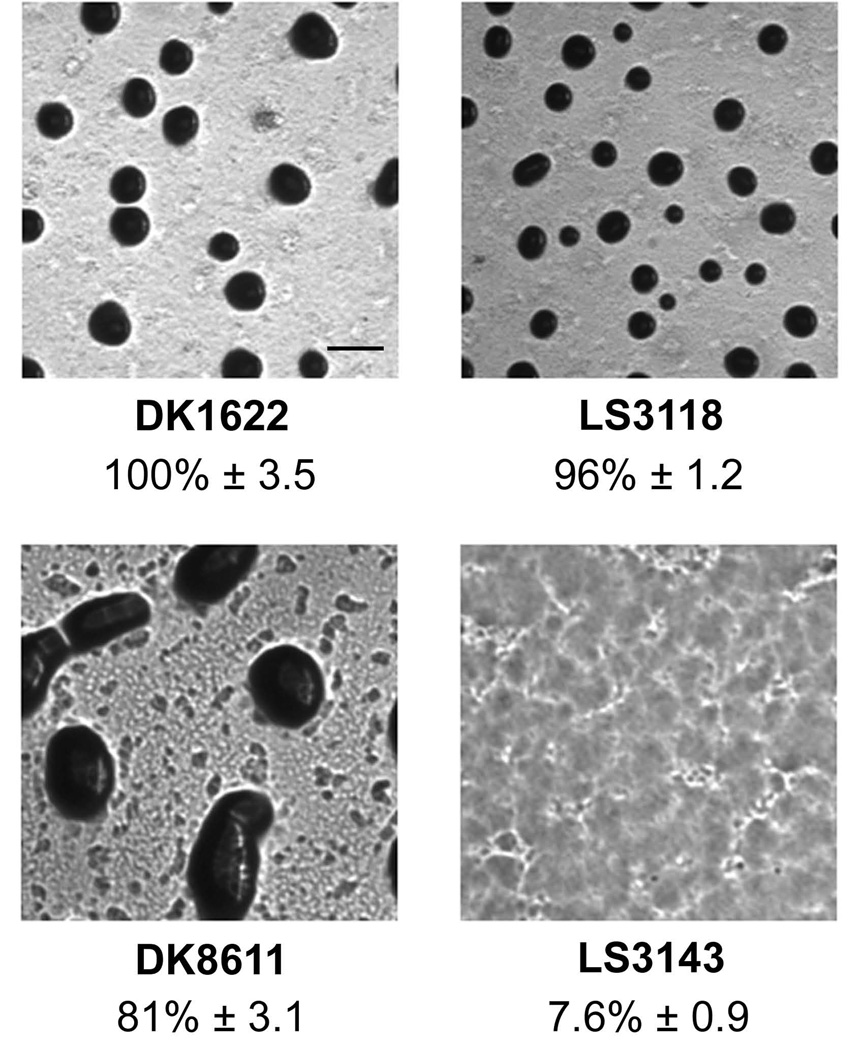
Removal of mazF from the M. xanthus genome has a deleterious effect on development of DZF1, but not in the wild type strains DK1622 and DZ2 (Lee et al., 2012). Of the many differences in DZF1, one key mutation is the pilQ1 allele, which renders M. xanthus incapable of social motility and nearly 1000-fold more susceptible to certain lethal substances such as vancomycin (Wall et al., 1999). A DK1622 derived strain called LS3143 bearing pilQ1 and ΔmazF was created to examine whether it would recreate the developmental defect observed with DZF1. Fruiting body formation and sporulation were compared with DK1622, DK8611 (DK1622 bearing the pilQ1 allele of DZF1), and LS3118 (ΔmazF) (Figure 5). DK1622, DK8611, and LS3118 were all competent in fruiting body formation and sporulation. In contrast, LS3143 was unable to form fruiting bodies or spores, mimicking the characteristics reported for a mazF deletion in DZF1.
Figure 5.
Effect of mazF removal on fruiting body formation. Deletion of mazF (LS3118) has no effect on fruiting body formation compared to wild type DK1622. The pilQ1 mutant (DK8611) is also able to form fruiting bodies. Only when both mutations are present (LS3143) is fruiting body development impaired. All images are taken approximately 48 hours post-initiation under equal magnification. % sporulation relative to DK1622 is given beneath each photograph ± standard deviation. Black bar = 200 µm.
Discussion
During M. xanthus development roughly 80% of the population is sacrificed by PCD. Neither the mechanism of PCD, nor the method by which cells acquire a specific fate are understood. The recent discovery of MazF in M. xanthus suggested a model whereby the mRNA interferase remained inactive via a complex with the transcription factor MrpC. In this model, MrpC is degraded by proteases, allowing MazF to stimulate lysis. However, while MazF is necessary for PCD and sporulation in strain DZF1, it is not essential for PCD or sporulation in wild type strains DK1622 and DZ2. Thus, we reexamined the function of MazF.
Our data shows enhancement of MazF endoribonuclease activity by MrpC and MrpC2 in direct opposition to the inhibition reported previously by Nariya and Inouye. One possible explanation for this is the type of assay used in the respective experiments. Whereas the previous observations used a qualitative gel electrophoresis assay that relied on the cleavage of MS2 ssRNA (Nariya & Inouye, 2008), we used a sensitive, continuous fluorometric assay with an artificial target with the M. xanthus MazF hydrolysis site. Another source of variation is the recombinant proteins. In the Nariya and Inouye study, MazF, MrpC, and MrpC2 are expressed with 22 amino acid, N-terminal additions containing 10× histidine tags. While our MazF has a 12 amino acid N-terminal addition containing a 6× histidine tag, our MrpC and MrpC2 proteins do not contain additional sequences and should more closely mimic the native proteins.
The hydrolysis site of M. xanthus MazF is an octamer found in only about 1% of M. xanthus genes. In organisms where the recognition sequence is 4–5 nucleotides, such as S. aureus and M. tuberculosis, MazF negatively regulates the expression of a subset of genes (Zhu et al., 2008, Zhu et al., 2009). Thus it is likely that M. xanthus MazF functions similarly, with a much greater specificity. Our data show that MazF degrades a subset of mRNAs, a few of which are tied to cell viability and development. Of the five genes selected from the list of potential targets, all were reduced in vivo, some below levels found in vegetative cells. Through these genes and other potential targets, MazF regulates synthesis of proteins at the posttranscriptional level.
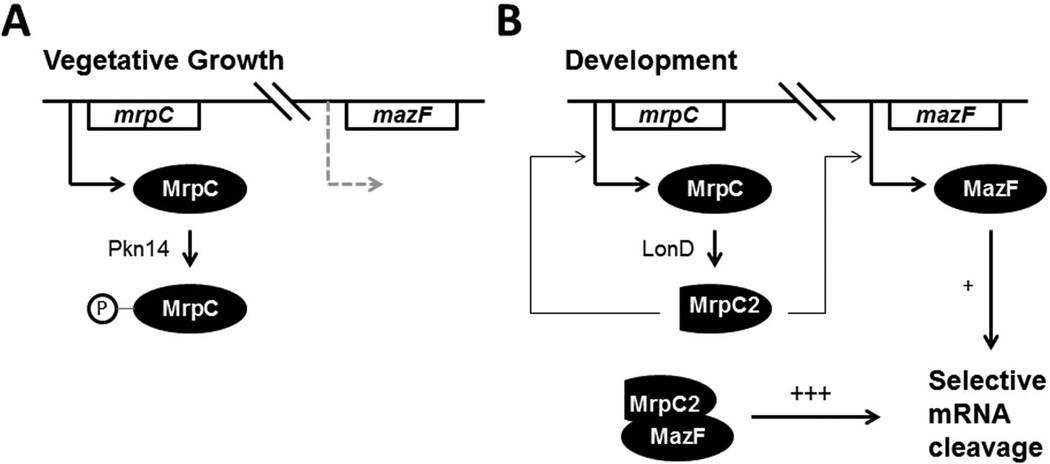
Unlike the previous model of MazF action (Nariya & Inouye, 2008), our data point to a model in which MazF functions without an antitoxin (Figure 6). In vegetative cells, MrpC is sequestered into an inactive state through a previously established protein kinase cascade whereby Pkn14 phosphorylates MrpC. With MrpC phosphorylated and unable to bind the mazF promoter, MazF is not expressed at appreciable levels. Once development is initiated, MrpC is hydrolyzed to MrpC2 by a process requiring LonD protease and is no longer able to be phosphorylated (Nariya & Inouye, 2006). Cleavage through LonD may constitute a bistable switch, as conversion to MrpC2 positively regulates production of MrpC. Now active, MrpC2 upregulates its own transcription and initiates transcription of mazF, which then negatively regulates mRNA transcript levels. Interestingly, Myxococcus has recruited its own transcriptional activator as an enhancer of enzymatic activity. This novel mode of action for MazF means that MrpC regulates MazF at both the transcriptional and post-translational levels.
Figure 6.
Model of MazF function. A. During vegetative growth, MrpC phosphorylation by Pkn14 renders it unable to activate transcription of MazF. B. Upon starvation, MrpC is cleaved to MrpC2 directly or indirectly by protease LonD and can no longer be phosphorylated. MrpC and MazF expression are upregulated and the interaction of MazF and MrpC leads to mRNA interferase activity.
Deletion of mazF causes different phenotypes in different genetic backgrounds. From the DZF1 data, it would appear that MazF is required for PCD (Nariya & Inouye, 2008). In DK1622 and DZ2, however, mazF mutant cells develop and sporulate virtually identical to their parental strains (Lee et al., 2012). The phenotypic effects of mazF removal in DZF1 were recreated in DK1622 by introducing the pilQ1 mutation into a ΔmazF mutant. Two possible explanations are apparent. As proposed by Lee et al., two parallel, redundant pathways of PCD could exist in DK1622 and DZ2, one controlled by MazF, and the other by an as yet unknown mechanism disrupted in DZF1. Removal of mazF in the strain would then be deleterious to development. Alternatively, the observed phenotypic differences may be more artifactual than revealing, and result from the increased sensitivity of the pilQ1 allele to toxic molecules. pilQ1 results in a 1000-fold increased sensitivity to vancomycin suggesting that the porin is held in an open state. The increased membrane permeability coupled with removal of MazF activity could then indirectly affect cell fate independent of PCD. Further experimentation is required to distinguish between these possibilities.
In conclusion, M. xanthus MazF RNA interferase controls expression of roughly 1% of the genome by a novel mechanism involving stimulation by its transcription factor MrpC. This represents the first description of a so-called toxin devoid of its corresponding antitoxin. These results further support the idea that MazF proteins are not global toxins, but instead play regulatory roles distinct to each organism.
Experimental Procedures
Bacterial Strains and Plasmids and Development of Myxococcus xanthus
All strain and plasmids used in this study are listed in Table 1. Sequences of all PCR primers can be found in table S2. Unless noted, M. xanthus and E. coli strains were grown in CYE (1.0% Bacto Casitone, 0.5% Difco yeast extract, 10 mM 3-[N-morpholino] propanesulfonic acid, pH 7.6, and 0.1% MgSO4) and LB respectively, and supplemented with 50 µg ml−1 kanamycin and 50 µg ml−1 ampicillin where appropriate. M. xanthus and E. coli were grown at 32°C and 37°C respectively. Development of M. xanthus was induced by first growing cells to a concentration of 5×108 cells ml−1 in CYE with shaking. Cells were pelleted and then resuspended in sterile water at 5×109 cells ml−1. 20 µl of this suspension was then spotted onto TPM agar (10 mM Tris-HCl, 8 mM MgSO4, 1 mM K2HPO4-KH2PO4, 1.5% Difco agar, pH 7.6) and incubated at 32°C. For quantification of spores, spots were resuspended 5 days post-starvation in 1 ml dH20 and incubated 55 °C for one hour followed by brief sonication to kill vegetative cells. Spores were then counted directly using a Petroff-Hauser chamber.
Table 1.
Bacterial strains and plasmids used in the course of this study.
| Strain or Plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH10B | Used for cloning of plasmids and protein expression | (Durfee et al., 2008) |
| M. xanthus | ||
| DK1622 | Wild-type strain | (Chen et al., 1990) |
| LS3118 | DK1622 bearing a clean deletion of mazF | This study |
| DK8611 | DK1622 bearing the pilQ1 allele | (Wall et al., 1999) |
| LS3143 | DK1622 bearing both LS3118 and DK8611 mutations | This study |
| LS3160 | DK1622 bearing two silent mutations, T751C and G753C in the nla6 (MXAN4042) gene | This study |
| Plasmids | ||
| pUC57 | Derivative of pUC19, E. coli vector | (Yanisch-Perron et al., 1985) |
| pTOB12 | Synthetic clone of M. xanthus mazF (MXAN1659) flanking regions inserted into pUC57 | This study |
| pBJ113 | M. xanthus recombination backbone plasmid | (Ueki et al., 1996) |
| pTOB13 | pBJ113 containing inserted sequence of pTOB12 | This study |
| pTOB1 | Synthetic clone of M. xanthus mazF codon optimized for E. coli inserted into pUC57 | This study |
| pTrcHisB | E. coli Expression vector | (Amann et al., 1983) |
| pTOB2 | Codon optimized mazF sequence inserted into pTrcHisB | This study |
| pTOB7 | Synthetic clone of M. xanthus mrpC (MXAN5125) codon optimized for E. coli inserted into pUC57 | |
| pTYB2 | E. coli Expression vector | (Chong et al., 1997) |
| pTOB14 | Codon optimized mrpC sequence inserted in pTYB2 | This study |
| pTOB15 | Codon optimized mrpC2 sequence inserted in pTYB2 | This study |
| pTOB30 | pBJ113 containing nla6 (MXAN4042) bearing T751C and G753C mutations |
Strain LS3118 (ΔmazF) was generated through the double recombination method as previously described (Ueki et al., 1996) using the vector pBJ113 (Julien et al., 2000). Briefly, plasmid pTOB12 was synthesized by Genscript (Piscataway, NJ, USA) to contain the 700 bp immediately upstream of the M. xanthus mazF (MXAN_1659) coding sequence fused to the 700 bp immediately downstream. This 1400 bp fragment was flanked by 5’ KpnI and 3’ HindIII restriction sequences and inserted into the EcoRV site of pUC57. pTOB12 was then digested with KpnI and HindIII and the 1400 bp insert was ligated into the respective sites of the plasmid vector pBJ113 to create pTOB13. This plasmid was then transformed into electrocompetent DK1622 cells with selection for kanamycin resistance where it integrated by a single homologous crossover. A second recombination event was counterselected with CYE containing 1% galactose and resulted in an in-frame deletion that was identified by PCR.
Strain LS3160 was created using the double recombination method above with pTOB30. pTOB30 was created by first amplifying nucleotides 1–747 of nla6, adding a 5’ KpnI site and 3’ SacI site. Nucleotides 754–1455 were also amplified adding a 5’ SacI site and 3’ HindIII site. Ligation of these two products into the KpnI site and HindIII site of pBJ113 yielded a plasmid in which the SacI site changed nucleotides 748–753 of nla6 from 5’-GAGTTG-3’ to 5’-GAGCTC-3’, introducing the silent mutations T751C and G753C.
Strain LS3143 was generated by transduction of the pilQ1 allele from DK8611 into LS3118 via bacteriophage Mx4 (Campos et al., 1978).
For the mazF expression vector pTOB2, pTOB1 was first synthesized by Genscript (Piscataway, NJ, USA) to contain M. xanthus mazF codon-optimized for E. coli flanked by 5’ NheI and 3’ HindIII restriction sequences. The insert was removed and ligated into the respective sites of pTrcHisB (Invitrogen, Grand Island, NY, USA). The codon optimized sequenced of pTOB1 can be seen in Figure S2 and lacks the natural start codon. The protein product contains a 12 N-terminal amino acid addition including a 6× His tag.
For the mrpC expression vector pTOB14, the M. xanthus mrpC sequence was codon-optimized (Figure S3) and synthesized as before to obtain plasmid pTOB7. Optimized mrpC was amplified by PCR from pTOB7 to contain no stop codon and flanking 5’ NdeI and 3’ XhoI restriction sites. The amplicon was ligated into the respective sites of the commercial vector pTYB2 (New England Biolabs, Ipswich, MA, USA). pTOB15 was generated in a similar fashion, except that the amplified PCR product lacked the first 75 bp of the optimized mrpC gene using a natural ATG at this position as a start codon (Nariya & Inouye, 2006). The protein products of pTOB14 and pTOB15 were designed so that intein self-splicing generates products with the native amino acid sequences.
Expression and Purification of Recombinant Proteins
Expression and purification of MazF was performed as previously described (Boynton et al., 2009). Briefly, E. coli bearing pTOB2 were grown for 6 hours at 30°C in 100 mL Circlegrow media (MP Biomedicals, Solon, OH, USA) containing 50 µg ml−1 ampicillin and then transferred to 1 L of Circlegrow media and grown for an additional 22 hours at 30°C. Cells collected by centrifugation were lysed by sonication in a solubilization buffer containing 50 mM Tris-buffer 3-(N-Morpholino) propane-sulfonic acid (MOPS) pH 8.0, 100 mM KCl, 1% sodium cholate, and 10 µg ml−1 phenylmethylsulfonyl fluoride (PMSF). The lysate was then centrifuged for 30 minutes at 100,000 × g and the resulting supernatant applied to a column containing 1.25 mL bed volume HisPur Cobalt Affinity Resin (Thermo Scientific, Rockford, IL, USA). The column was then washed with 20 bed volumes solubilization buffer. Recombinant protein was eluted in 1 mL fractions of solubilization buffer containing 300 mM imidazole.
E. coli bearing pTOB14 or pTOB15 plasmids were grown overnight at 37°C and used to inoculate 1L LB containing ampicillin the following morning. When the culture density reached 0.6 OD600, protein expression was induced with 0.4 mM IPTG for 24 hrs. Cells were harvested and then sonicated in STE buffer [20 mM Tris-Cl (pH 8.0), 500 mM NaCl, and 0.1 mM EDTA]. The lysate was ultracentrifuged as above and the resulting supernatant applied to a column containing 2.5 mL bed volumes of chitin beads (New England Biolabs, Ipswich, MA, USA). The column was then washed with 20 mL STE buffer. To elute, 5 mL cleavage buffer [20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 0.1 mM EDTA, and 50 mM dithiothreitol] were loaded onto the column and allowed to incubate overnight at 4°C. Recombinant proteins were then eluted. All proteins were quantified using UV/VIS spectrophotometry from the absorption peak of a protein scan using predicted extinction coefficients. Proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Bio-Rad 4–20% Tris-HCl ready-made gels (Bio-Rad, Hercules, CA, USA).
Continuous Fluorometric Assay
MazF activity was assayed as previously described (Wang & Hergenrother, 2007) with modifications, using the following labeled oligonucleotude substrate obtained from IDT (Coraville, IA, USA): 6-carboxyfluorescein (6-FAM)-5’-rArArGrArGrUrUrGrCrArCrArG-3’-Black Hole Quencher 1 (BHQ1). Reaction mixtures consisted of 5 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 250 nM MazF, and varying amounts of substrate, MrpC, and MrpC2 where appropriate. Rates were monitored as a function of fluorescence emitted upon cleavage of substrate. Experiments were conducted at 32°C with shaking, using a Synergy 2 HTI plate reader (BioTek, Winooski, VT, USA) equipped with 485 ± 15 nm excitation filter and 530 ± 15 nm emission filter. For kinetics, data were fitted to eq 1:
where Vmax is the apparent maximum rate and Km is the apparent Michaelis-Menton constant.
Quantification of Transcript Level Changes During Development
To determine mRNA transcript levels of target genes, RNA was extracted at time points during development from DK1622, LS3118,or LS3160. Samples collected at different time points after induction of development were subjected to RNA extraction using the RNASNAP method followed by standard ethanol precipitation (Stead et al., 2012). Quantification of transcript levels was done using the iScript One-Step RT-PCR Kit With SYBR Green (Bio-Rad, Hercules, CA, USA) and carried out with the iCycler iQ Realtime PCR Detection System (Bio-Rad, Hercules, CA, USA) with the following reaction cycle: 50°C for 10 minutes, 95°C for 5 minutes, then 45 cycles of 95°C for 10 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. Meltcurve analysis was carried out at 55–95°C for 10 seconds, increasing the temperature 0.5°C each cycle for 80 cycles. Relative fold-changes in transcript levels were calculated using the ΔΔCT method and expressed as log2 inductions (Livak & Schmittgen, 2001), normalized against the housekeeping gene, rpoC (MXAN_3078). Primers sequences are listed in table S2.
Supplementary Material
Acknowledgements
The authors would like to thank Dan Wall for providing strain DK8611.
Research reported in this publication was supported by the National Institute of General Medical Science of the National Institutes of Health under award number R01GM095826 and by the Natural Science Foundation under award number MCB0742976.
Footnotes
The authors declare no conflict of interest.
References
- Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal "addiction module" regulated by guanosine [corrected] 3',5'-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol. 2009;191:6029–6039. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton TO, Daugherty LE, Dailey TA, Dailey HA. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry. 2009;48:6705–6711. doi: 10.1021/bi900850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J Bacteriol. 2003;185:6083–6094. doi: 10.1128/JB.185.20.6083-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos JM, Geisselsoder J, Zusman DR. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Chen H, Keseler IM, Shimkets LJ. Genome size of Myxococcus xanthus determined by pulsed-field gel electrophoresis. J Bacteriol. 1990;172:4206–4213. doi: 10.1128/jb.172.8.4206-4213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- Durfee T, Nelson R, Baldwin S, Plunkett G, 3rd, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csorgo B, Posfai G, Weinstock GM, Blattner FR. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Inclan YF, Vlamakis HC, Zusman DR. FrzZ, a dual CheY-like response regulator, functions as an output for the Frz chemosensory pathway of Myxococcus xanthus. Mol Microbiol. 2007;65:90–102. doi: 10.1111/j.1365-2958.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- Julien B, Kaiser AD, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci U S A. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Wegener-Feldbrugge S, Sogaard-Andersen L. Two intercellular signals required for fruiting body formation in Myxococcus xanthus act sequentially but non-hierarchically. Mol Microbiol. 2012;86:65–81. doi: 10.1111/j.1365-2958.2012.08173.x. [DOI] [PubMed] [Google Scholar]
- Lee B, Holkenbrink C, Treuner-Lange A, Higgs PI. Myxococcus xanthus developmental cell fate production: heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J Bacteriol. 2012;194:3058–3068. doi: 10.1128/JB.06756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, Shi W. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol Microbiol. 2005;55:206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- Mrazek J, Xie S. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics. 2006;22:3099–3100. doi: 10.1093/bioinformatics/btl551. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye S. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol Microbiol. 2005;58:367–379. doi: 10.1111/j.1365-2958.2005.04826.x. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye S. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol. 2006;60:1205–1217. doi: 10.1111/j.1365-2958.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- Park JH, Yamaguchi Y, Inouye M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011;585:2526–2532. doi: 10.1016/j.febslet.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- Perry JA, Cvitkovitch DG, Levesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. 2009;299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenbacher FP, Suzuki M, Hurley JM, Montville TJ, Kirn TJ, Ouyang M, Woychik NA. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J Bacteriol. 2012;194:3464–3474. doi: 10.1128/JB.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- Stead MB, Agrawal A, Bowden KE, Nasir R, Mohanty BK, Meagher RB, Kushner SR. RNAsnap: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 2012;40:e156. doi: 10.1093/nar/gks680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Shi W. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. Journal of bacteriology. 2001;183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Inouye S. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc Natl Acad Sci U S A. 2003;100:8782–8787. doi: 10.1073/pnas.1533026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Inouye S, Inouye M. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene. 1996;183:153–157. doi: 10.1016/s0378-1119(96)00546-x. [DOI] [PubMed] [Google Scholar]
- Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NR, Hergenrother PJ. A continuous fluorometric assay for the assessment of MazF ribonuclease activity. Anal Biochem. 2007;371:173–183. doi: 10.1016/j.ab.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichart D, Querfurth N, Dreger M, Hengge-Aronis R. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J Bacteriol. 2003;185:115–125. doi: 10.1128/JB.185.1.115-125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman JW, Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1977;129:798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog Mol Biol Transl Sci. 2009;85:467–500. doi: 10.1016/S0079-6603(08)00812-X. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zambrano MM, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Zhu L, Inoue K, Yoshizumi S, Kobayashi H, Zhang Y, Ouyang M, Kato F, Sugai M, Inouye M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J Bacteriol. 2009;191:3248–3255. doi: 10.1128/JB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol. 2008;69:559–569. doi: 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.