Abstract
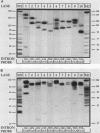
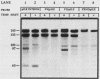
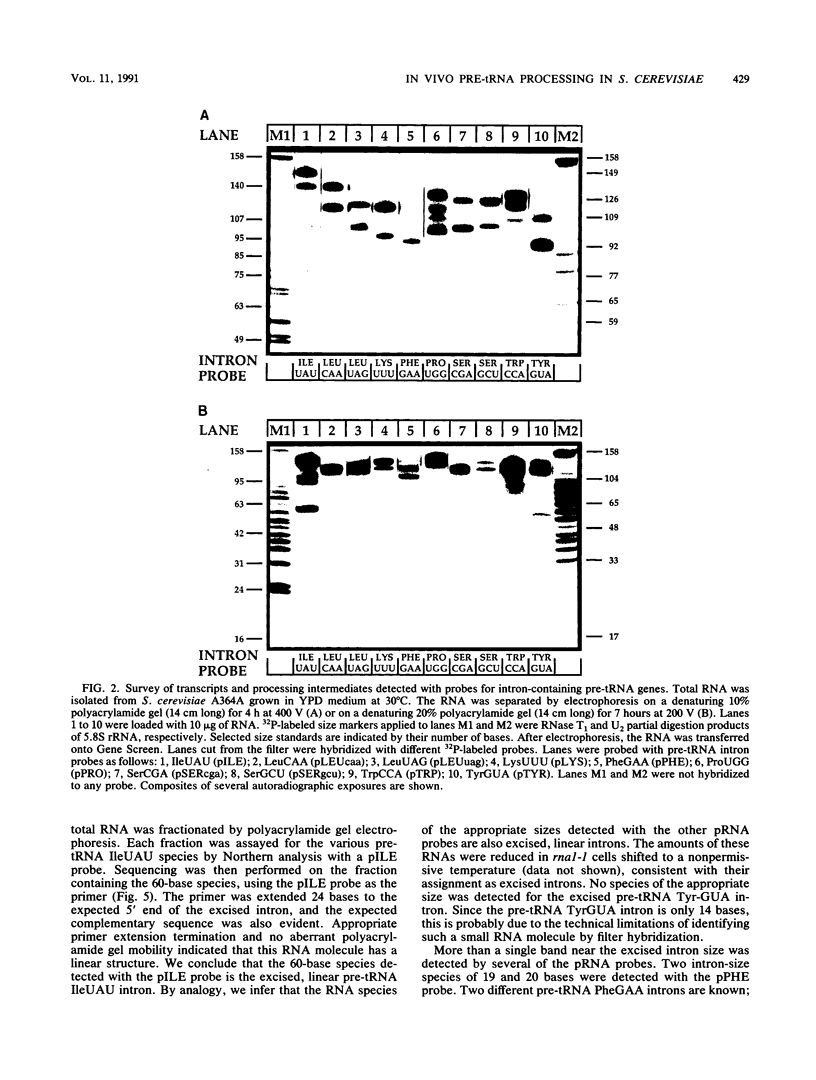
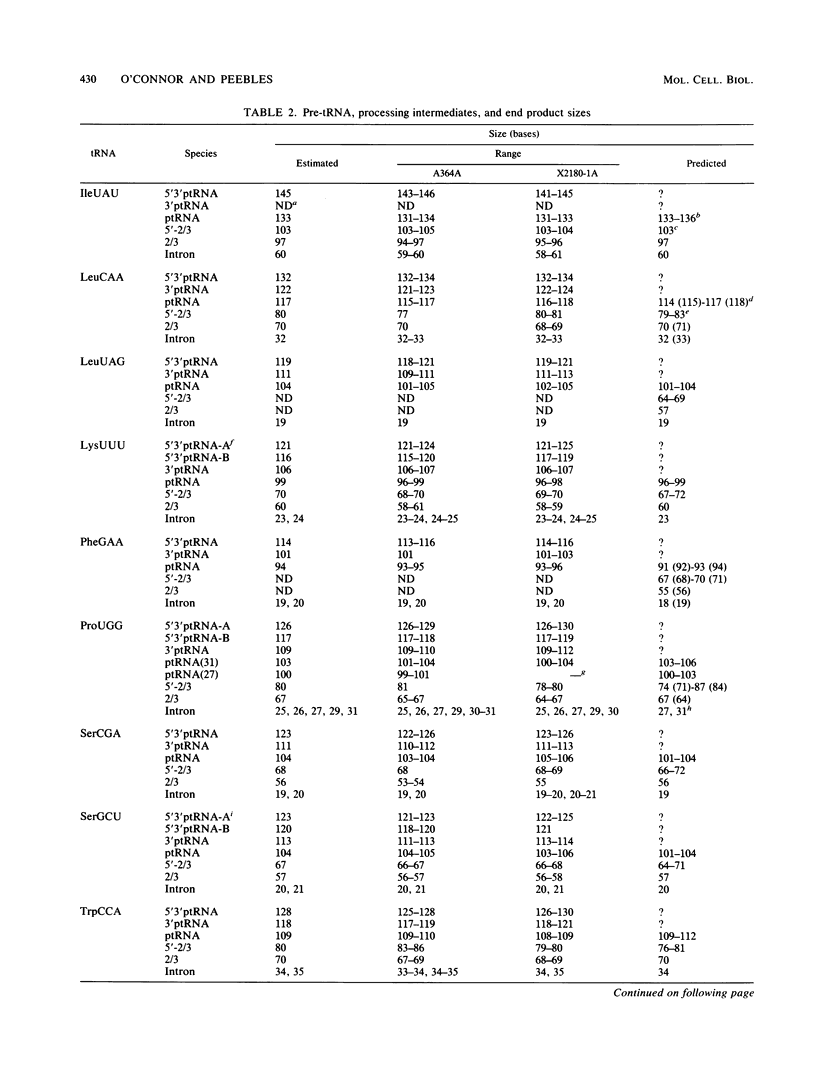
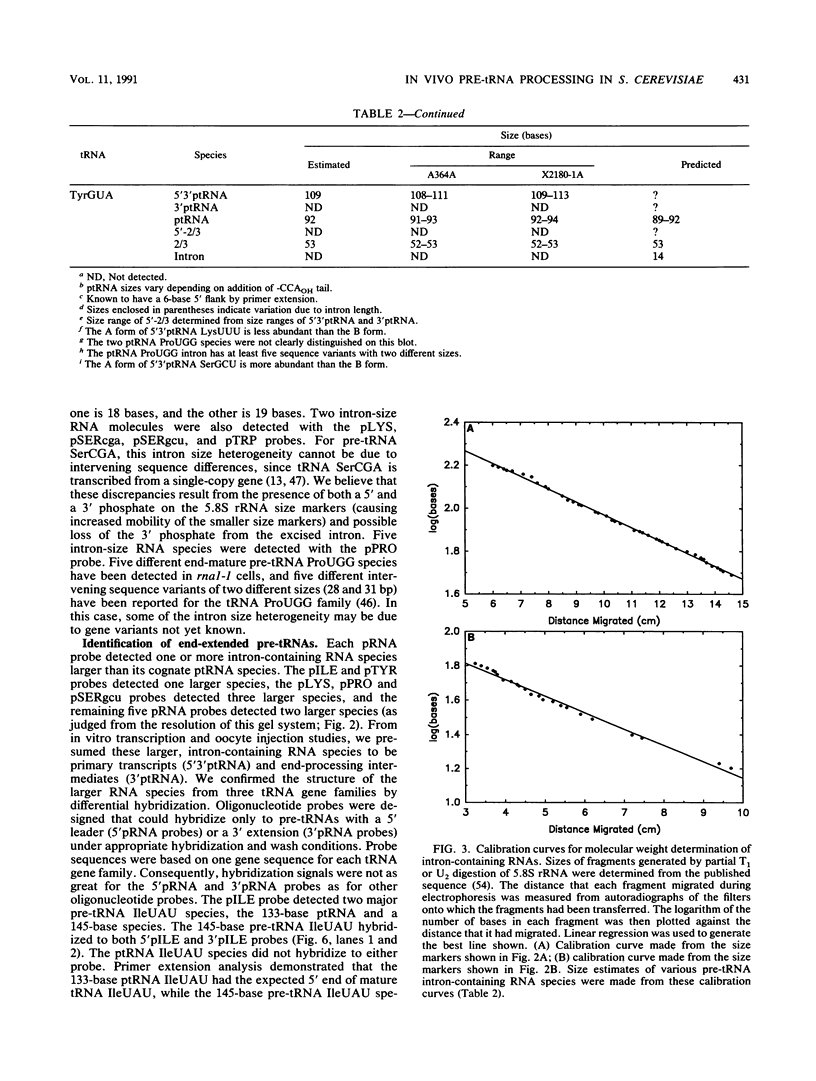
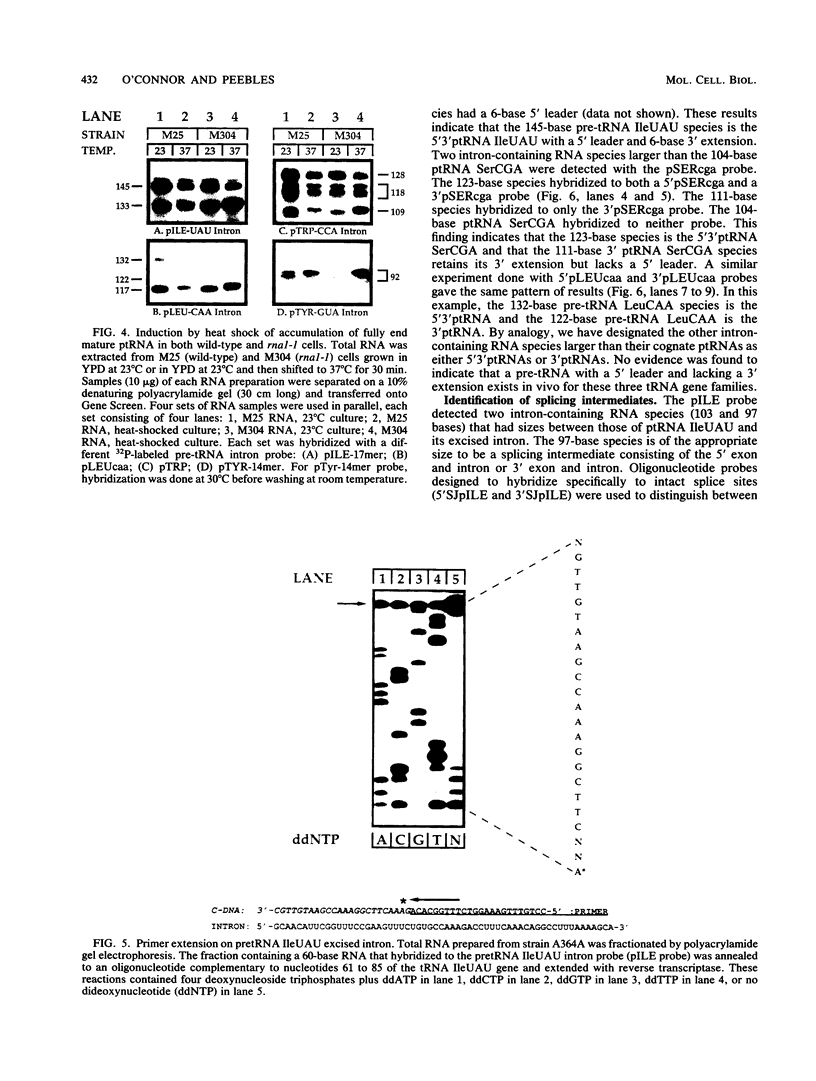
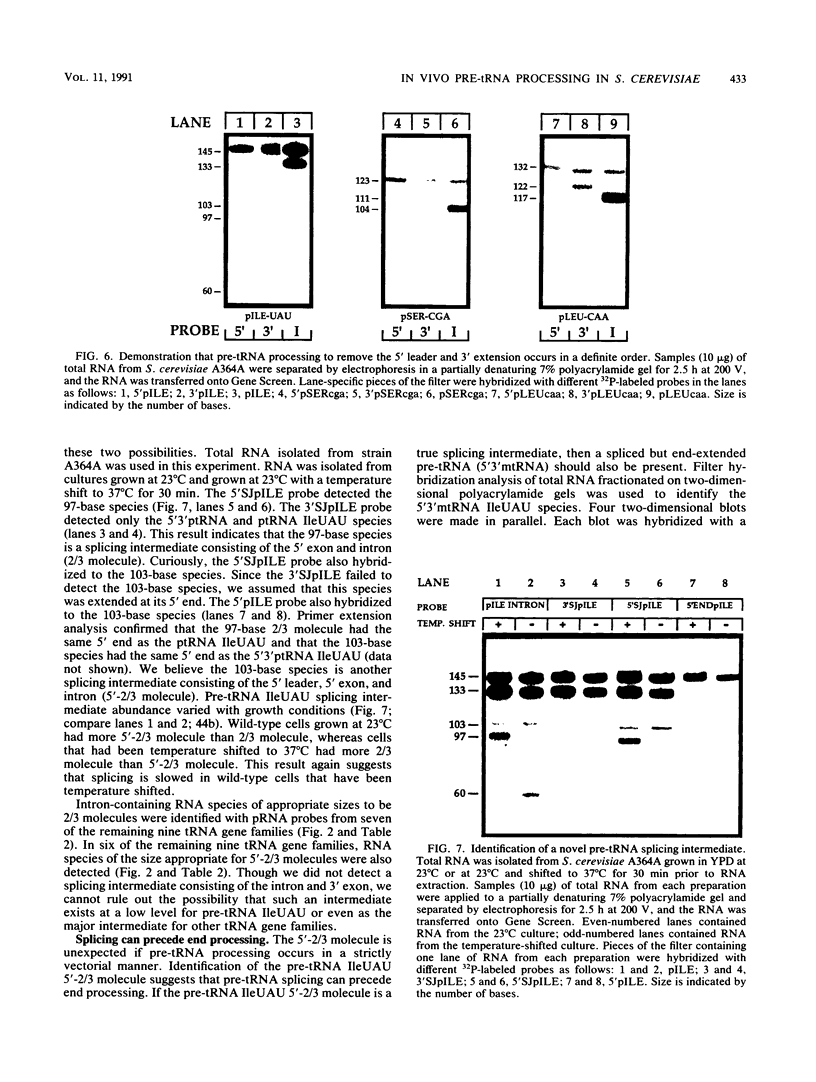
We have surveyed intron-containing RNAs of the yeast Saccharomyces cerevisiae by filter hybridization with pre-tRNA intron-specific oligonucleotide probes. We have classified various RNAs as pre-tRNAs, splicing intermediates, or excised intron products according to apparent size and structure. Linear, excised intron products were detected, and one example was isolated and sequenced directly. Additional probes designed to detect other precursor sequences were used to verify the identification of several intermediates. Pre-tRNA species with both 5' leader and 3' extension, with 3' extension only, and with mature ends were distinguished. From these results, we conclude that the processing reactions used to remove the 5' leader and 3' extension from the transcript are ordered 5' end trimming before 3' end trimming. Splicing intermediates containing the 5' exon plus the intron were detected. The splice site cleavage reactions are probably ordered 3' splice site cleavage before 5' splice site cleavage. Surprisingly, we also detected a splicing intermediate with the 5' leader and a spliced product with both 5' leader and 3' extension. Evidently, splicing and end trimming are not ordered relative to each other, splicing occurring either before or after end trimming.
Full text
PDF



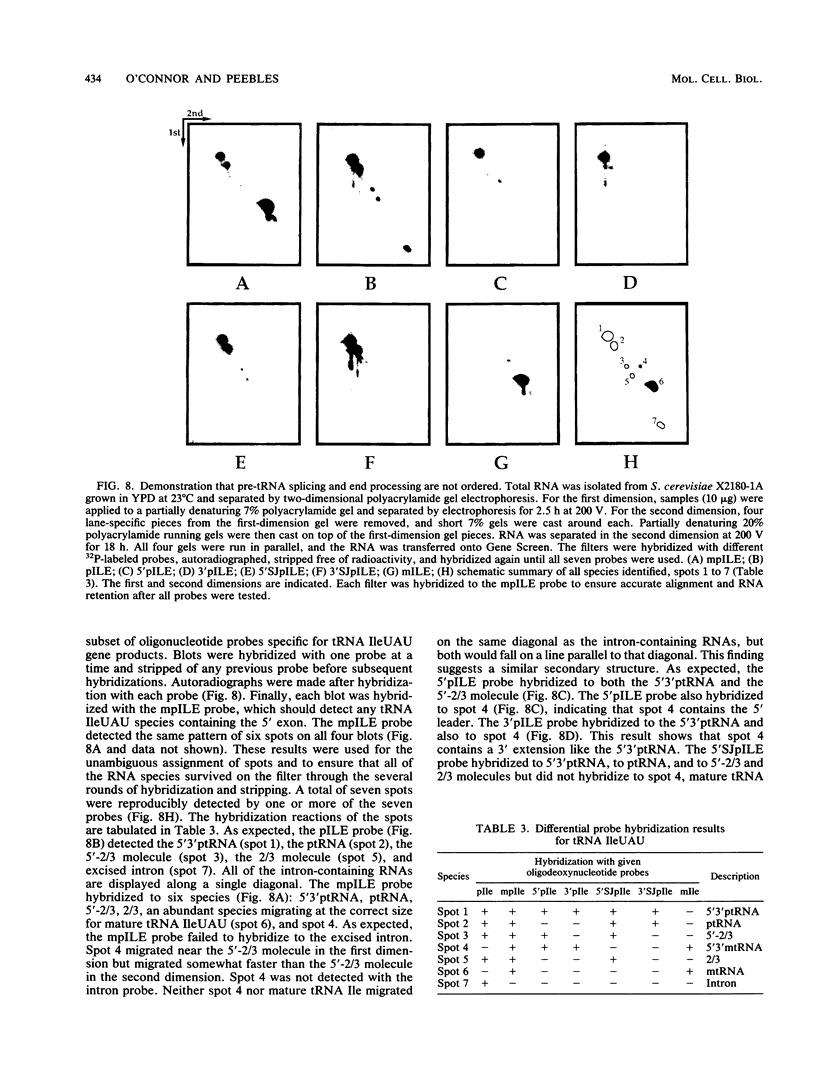
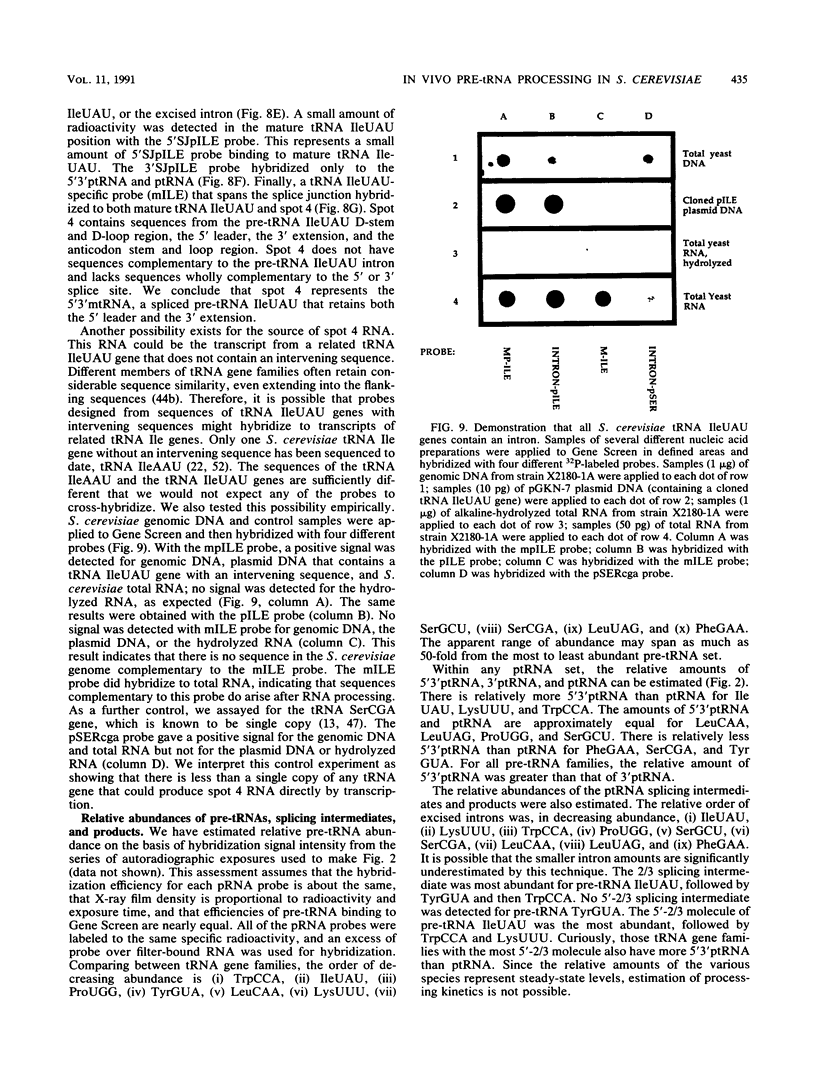
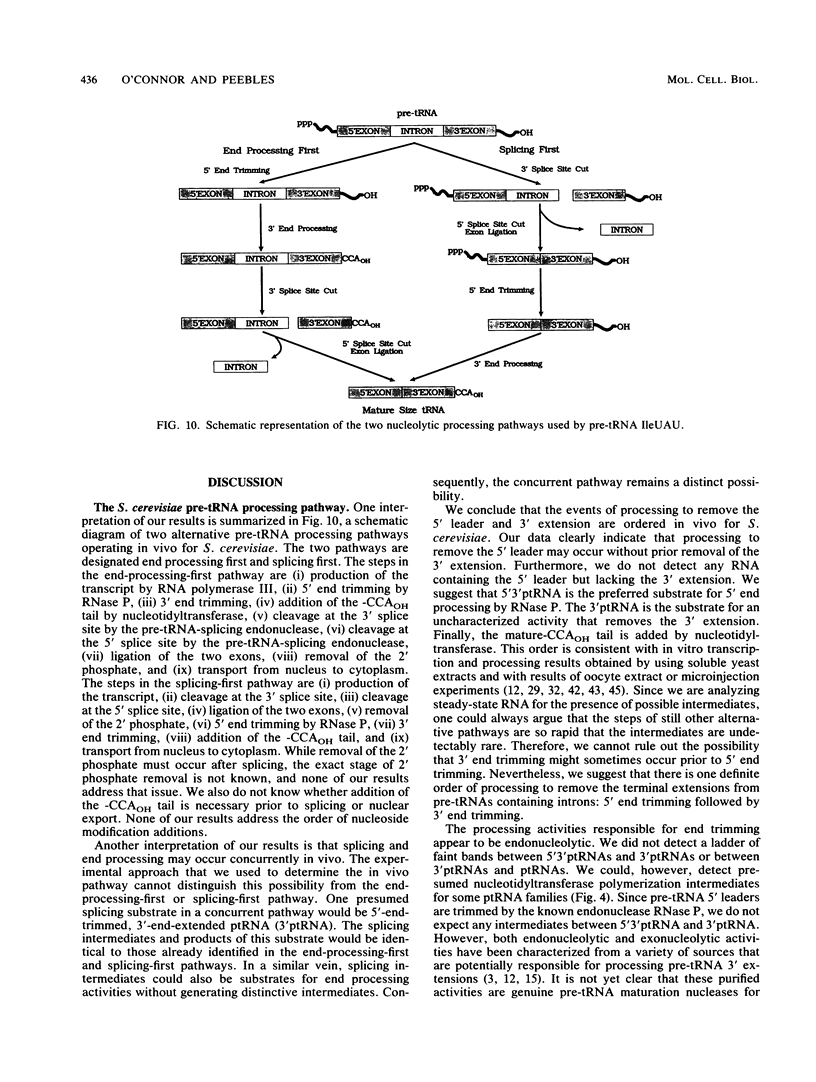



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blatt B., Feldmann H. Characterization of precursors to tRNA in yeast. FEBS Lett. 1973 Dec 1;37(2):129–133. doi: 10.1016/0014-5793(73)80441-7. [DOI] [PubMed] [Google Scholar]
- Castaño J. G., Tobian J. A., Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3' terminus of human pre-tRNA-Met(i) (3' pre-tRNase). J Biol Chem. 1985 Jul 25;260(15):9002–9008. [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987 Oct;105(4):1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Culbertson M. R., Winey M. Split tRNA genes and their products: a paradigm for the study of cell function and evolution. Yeast. 1989 Nov-Dec;5(6):405–427. doi: 10.1002/yea.320050602. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Black P., Nishikura K. Intranuclear location of the tRNA splicing enzymes. Cell. 1981 Jan;23(1):89–93. doi: 10.1016/0092-8674(81)90273-7. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Olson M. V. Transcription and processing of cloned yeast tyrosine tRNA genes microinjected into frog oocytes. Nature. 1979 Mar 8;278(5700):137–143. doi: 10.1038/278137a0. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. Reactions at the 3' terminus of transfer ribonucleic acid. A single enzyme catalyzes the incorporation of adenosine monophosphate and cytidine monophosphate into transfer ribonucleic acid. J Biol Chem. 1970 Aug 25;245(16):4225–4227. [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Etcheverry T., Salvato M., Guthrie C. Recessive lethality of yeast strains carrying the SUP61 suppressor results from loss of a transfer RNA with a unique decoding function. J Mol Biol. 1982 Jul 15;158(4):599–618. doi: 10.1016/0022-2836(82)90251-0. [DOI] [PubMed] [Google Scholar]
- Fradin A., Gruhl H., Feldmann H. Mapping of yeast tRNAs by two-dimensional electrophoresis on polyacrylamide gels. FEBS Lett. 1975 Feb 1;50(2):185–189. doi: 10.1016/0014-5793(75)80485-6. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Dingermann T., Cooley L., Söll D. Processing of precursor tRNAs in Drosophila. Processing of the 3' end involves an endonucleolytic cleavage and occurs after 5' end maturation. J Biol Chem. 1985 Jan 10;260(1):449–454. [PubMed] [Google Scholar]
- Ganguly S., Sharp P. A., RajBhandary U. L. Saccharomyces cerevisiae SUP53 tRNA gene transcripts are processed by mammalian cell extracts in vitro but are not processed in vivo. Mol Cell Biol. 1988 Jan;8(1):361–370. doi: 10.1128/mcb.8.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Greer C. L. Assembly of a tRNA splicing complex: evidence for concerted excision and joining steps in splicing in vitro. Mol Cell Biol. 1986 Feb;6(2):635–644. doi: 10.1128/mcb.6.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C. L., Peebles C. L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983 Feb;32(2):537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Greer C. L., Söll D., Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):76–84. doi: 10.1128/mcb.7.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. J., Chalker D. L., Sandmeyer S. B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol. 1988 Dec;8(12):5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Rauhut R., Vijayraghavan U., Abelson J. Accumulation of pre-tRNA splicing '2/3' intermediates in a Saccharomyces cerevisiae mutant. EMBO J. 1990 Apr;9(4):1245–1252. doi: 10.1002/j.1460-2075.1990.tb08232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Banks F. A yeast mutant which accumulates precursor tRNAs. Cell. 1978 Jun;14(2):211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Furukawa A. H., Pham H. D., Martin N. C. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell. 1982 Mar;28(3):543–550. doi: 10.1016/0092-8674(82)90209-4. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Kurjan J. tRNA synthesis: identification of in vivo precursor tRNAs from parental and mutant yeast strains. Nucleic Acids Res. 1981 Feb 25;9(4):1019–1029. doi: 10.1093/nar/9.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Schultz L. D., Shapiro R. A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980 Mar;19(3):741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Johnson J. D., Ogden R., Johnson P., Abelson J., Dembeck P., Itakura K. Transcription and processing of a yeast tRNA gene containing a modified intervening sequence. Proc Natl Acad Sci U S A. 1980 May;77(5):2564–2568. doi: 10.1073/pnas.77.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Knapp G., Beckmann J. S., Johnson P. F., Fuhrman S. A., Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978 Jun;14(2):221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Knapp G., Ogden R. C., Peebles C. L., Abelson J. Splicing of yeast tRNA precursors: structure of the reaction intermediates. Cell. 1979 Sep;18(1):37–45. doi: 10.1016/0092-8674(79)90351-9. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Laten H., Gorman J., Bock R. M. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1978 Nov;5(11):4329–4342. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Engelke D. R. Partial characterization of an RNA component that copurifies with Saccharomyces cerevisiae RNase P. Mol Cell Biol. 1989 Jun;9(6):2536–2543. doi: 10.1128/mcb.9.6.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N., DaLio A., Strobel M., Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Mar 25;16(6):2537–2552. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R. Y., Bell J. B., Roy K. L. Dihydrouridine-deficient tRNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Feb 11;10(3):889–902. doi: 10.1093/nar/10.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S. M., Phizicky E. M. A highly specific phosphatase from Saccharomyces cerevisiae implicated in tRNA splicing. Mol Cell Biol. 1990 Mar;10(3):1049–1055. doi: 10.1128/mcb.10.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Cortese R. Transcription of cloned tRNA genes and the nuclear partitioning of a tRNA precursor. Cell. 1979 Dec;18(4):1165–1172. doi: 10.1016/0092-8674(79)90229-0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., De Robertis E. M., Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980 Mar 13;284(5752):143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Nishikura K., De Robertis E. M. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol. 1981 Jan 15;145(2):405–420. doi: 10.1016/0022-2836(81)90212-6. [DOI] [PubMed] [Google Scholar]
- Ogden R. C., Beckman J. S., Abelson J., Kang H. S., Söll D., Schmidt O. In vitro transcription and processing of a yeast tRNA gene containing an intervening sequence. Cell. 1979 Jun;17(2):399–406. doi: 10.1016/0092-8674(79)90166-1. [DOI] [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. V., Page G. S., Sentenac A., Piper P. W., Worthington M., Weiss R. B., Hall B. D. Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature. 1981 Jun 11;291(5815):464–469. doi: 10.1038/291464a0. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983 Feb;32(2):525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Ogden R. C., Knapp G., Abelson J. Splicing of yeast tRNA precursors: a two-stage reaction. Cell. 1979 Sep;18(1):27–35. doi: 10.1016/0092-8674(79)90350-7. [DOI] [PubMed] [Google Scholar]
- Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. A self-splicing RNA excises an intron lariat. Cell. 1986 Jan 31;44(2):213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- Phizicky E. M., Schwartz R. C., Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986 Feb 25;261(6):2978–2986. [PubMed] [Google Scholar]
- Pixa G., Dirheimer G., Keith G. Sequence of tRNA Ile IAU from brewer's yeast. Biochem Biophys Res Commun. 1984 Mar 30;119(3):905–912. doi: 10.1016/0006-291x(84)90859-3. [DOI] [PubMed] [Google Scholar]
- Rauhut R., Green P. R., Abelson J. Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J Biol Chem. 1990 Oct 25;265(30):18180–18184. [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988 Nov 18;55(4):719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Rubin G. M. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974 Jan 3;41(1):197–202. doi: 10.1111/j.1432-1033.1974.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Standring D. N., Venegas A., Rutter W. J. Yeast tRNA3Leu gene transcribed and spliced in a HeLa cell extract. Proc Natl Acad Sci U S A. 1981 Oct;78(10):5963–5967. doi: 10.1073/pnas.78.10.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986 Jul;6(7):2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucka R., Feldmann H. Structure of a Saccharomyces cerevisiae gene encoding minor (AGY)tRNA(Ser). Nucleic Acids Res. 1988 Apr 25;16(8):3583–3583. doi: 10.1093/nar/16.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely E., Belford H. G., Greer C. L. Intron sequence and structure requirements for tRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 1988 Sep 25;263(27):13839–13847. [PubMed] [Google Scholar]
- Tanner N. K., Hanna M. M., Abelson J. Binding interactions between yeast tRNA ligase and a precursor transfer ribonucleic acid containing two photoreactive uridine analogues. Biochemistry. 1988 Nov 29;27(24):8852–8861. doi: 10.1021/bi00424a025. [DOI] [PubMed] [Google Scholar]
- Tobian J. A., Drinkard L., Zasloff M. tRNA nuclear transport: defining the critical regions of human tRNAimet by point mutagenesis. Cell. 1985 Dec;43(2 Pt 1):415–422. doi: 10.1016/0092-8674(85)90171-0. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Hopper A. K. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol Cell Biol. 1988 Dec;8(12):5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Culbertson M. R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988 Apr;118(4):609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Edelman I., Culbertson M. R. A synthetic intron in a naturally intronless yeast pre-tRNA is spliced efficiently in vivo. Mol Cell Biol. 1989 Jan;9(1):329–331. doi: 10.1128/mcb.9.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mendenhall M. D., Cummins C. M., Culbertson M. R., Knapp G. Splicing of a yeast proline tRNA containing a novel suppressor mutation in the anticodon stem. J Mol Biol. 1986 Nov 5;192(1):49–63. doi: 10.1016/0022-2836(86)90463-8. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Rosenberg M., Santos T. Impaired nuclear transport of a human variant tRNAiMet. Nature. 1982 Nov 4;300(5887):81–84. doi: 10.1038/300081a0. [DOI] [PubMed] [Google Scholar]
- van Tol H., Stange N., Gross H. J., Beier H. A human and a plant intron-containing tRNATyr gene are both transcribed in a HeLa cell extract but spliced along different pathways. EMBO J. 1987 Jan;6(1):35–41. doi: 10.1002/j.1460-2075.1987.tb04715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl W. H., Wills N., Broach J. R. A general screen for mutant of Saccharomyces cerevisiae deficient in tRNA biosynthesis. Genetics. 1989 Sep;123(1):55–68. doi: 10.1093/genetics/123.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]