Abstract
Diabetes damages retinal mitochondrial DNA (mtDNA), and compromises the mtDNA transcription. In the transcription and replication of mtDNA, nuclear-encoded transcription factor A (TFAM) is considered as a key activator, and we have shown that in diabetes while retinal TFAM gene expression is increased, its mitochondrial levels are decreased. This study investigates the role of mitochondrial outer and inner membrane transport systems in the transfer of TFAM into the mitochondria in diabetes, and how reversal of hyperglycemia affects the ability of TFAM to reach to the mitochondria. Components of membrane transport system, Tom70, Tom40, Tim23 and Tim44, were analyzed in the retina from streptozotocin-induced diabetic rats maintained in poor control (PC) or in good control (GC) for 8 months, or in PC for 4 months followed by in GC for 4 months (Rev). The binding of TFAM with Tom70 and Tim44 was determined by co-immunoprecipitation, and that with mtDNA by ChIP. Retinal expressions of Tom70, Tom40 and Tim44 were significantly decreased in diabetes, and the binding of TFAM with Tom70, Tim44 and mtDNA were impaired. Reversal of hyperglycemia had no beneficial effect on decreased binding of TFAM and Tom proteins and mtDNA. Thus, subnormal membrane transport system in diabetes impairs the transfer of TFAM into the mitochondria, and decreased TFAM-mtDNA binding results in subnormal mitochondria transcription. These processes continue to be dysfunctional even after the hyperglycemic insult is terminated. Strategies targeting mitochondrial membrane transport proteins could have potential in improving mitochondrial biogenesis and slowing/halting the progression of diabetic retinopathy.
Keywords: Diabetic retinopathy, Metabolic memory, Mitochondria, mtDNA biogenesis
Introduction
Diabetic retinopathy, a microvascular complication of diabetes, is the leading cause of acquired blindness in young adults [1, 2]. Increased oxidative stress is considered to play an important role in the impairment of retinal metabolism, and this precedes the apoptosis of capillaries cells, resulting in the histopathology characteristic of diabetic retinopathy [3-5]. However the exact pathway responsible for the development of diabetic retinopathy is not entirely identified. In diabetes, retinal mitochondria are dysfunctional, their DNA (mtDNA) is damaged, the transcription of proteins encoded by mtDNA is subnormal, and these abnormalities lead to a vicious cycle of superoxide generation [6-8]. MtDNA transcription and replication requires a number of nuclear encoded proteins, and mitochondrial transcription factor A (TFAM) is considered as a key activator of mitochondrial transcription and replication [9, 10]. We have shown that although the gene expression of retinal TFAM is increased in diabetes, its protein expression in the mitochondria, the site of its action is decreased [8, 11].
The majority of mitochondrial proteins are encoded in the nucleus, synthesized in the cytosol, and transported to the mitochondria by a complex mechanism that involves membrane transport complexes [12, 13]. Translocase outer membrane complex (Tom) is the main gate in the outer mitochondrial membrane and it contains receptors that recognize mitochondrial proteins. Tom70 serves as a specific dock for cytosolic chaperone and carries key mitochondrial proteins, while Tom40, a channel-forming protein, permits the transport of pre-protein to the inter membrane space [13, 14]. Subsequently, proteins cross the inner mitochondria membrane by translocases of inner mitochondrial membrane complex (Tim). Tim complex contains a channel forming protein, Tim23, which transports positive charged proteins to the matrix [13, 15]. The proteins are pulled into the matrix by heat shock proteins 70 (Hsp70) that docks on Tim44 [15, 16], and once in the matrix, proteins are folded by Hsp60 [17, 18]. We have shown that in diabetic retinopathy several components of mitochondrial transport system are decreased [19]. However, their role in the biogenesis of retinal mtDNA is not clear.
Prior exposure to hyperglycemia has been shown to have long-lasting consequences that contribute to the progression of diabetic retinopathy even after cessation of hyperglycemic insult; both clinical and experimental studies have suggested a ‘metabolic memory’ phenomenon [20-23]. Our previous studies have demonstrated that reversal of hyperglycemia does not benefit diabetes-induced impairments in the mitochondria biogenesis and the membrane transport machinery continues to be dysfunctional [11, 19]. The effect of reversal of hyperglycemia on the transport of TFAM into the mitochondria remains unexplored.
The aim of this study is to investigate the role of mitochondrial membrane transport system in the transfer of TFAM into the mitochondria in the development of diabetic retinopathy, and also in the resistance of retinopathy to arrest after termination of hyperglycemia. The role of mitochondrial transporter system in mitochondrial biogenesis is also investigated in isolated retinal endothelial cells, the site of histopathology associated with diabetic retinopathy.
Methods
Rats
Wistar rats (male, body weight 200g) were randomly assigned into normal or diabetic groups (streptozotocin-induced). Diabetic rats were either allowed to remain in poor metabolic control for ~8 months (PC), or in PC for ~4 months followed by good metabolic control for ~4 additional months (Rev) or were kept in good metabolic control (GC) for ~8 months. Each group had 10 or more rats. The rats in poor metabolic control received 1-2 IU insulin 4-5 times a week to prevent ketosis and weight loss and the animals in GC received insulin twice daily (6-7 IU total) to maintain its blood glucose and a steady gain in body weight (Table I). At the end of the desired experimental duration, the animals were euthanized by CO2 inhalation, and retina were immediately isolated. These procedures are routinely used in our laboratory [7, 11, 19, 22-24]. Treatment of animals conformed to the Association for Research in Vision and Ophthalmology's Resolution on Treatment of Animals in Research and the institutional guidelines.
Table I.
Severity of hyperglycemia in rats maintained in various glycemic control
| Body weight (g) | Glycated hemoglobin (%) | |
|---|---|---|
| Normal | 480±56 | 6.4±1.4 |
| PC (8 month of PC) | 387±40* | 11.3±1.3* |
| Rev (4 mos PC→4 mos GC) | 367±50*→496±66# | 10.8±1.6*→6.7±1.0# |
| GC (8 months of GC) | 460±61# | 6.7±0.9# |
The values are presented as mean ±SD of seven or more rats in each group
P < 0.05 compared to normal
P <0.05 compared to PC.
Retinal endothelial cells
Retinal endothelial cells, isolated from bovine eyes (BRECs), were cultured on polystyrene culture plates coated with 0.1% gelatin in a humidified incubator at 37°C, in an atmosphere of 5% CO2 and 95% air [6, 11]. The cells from the 4th-6th passage were incubated in Dulbecco's modified Eagle medium (DMEM) containing 2% heat-inactivated fetal bovine serum, 10% Nu serum, 50μg/ml heparin, 1μg/ml endothelial growth factor supplemented with 5mM or 20mM glucose for 4 days. Osmotic control included cells incubated in 20mM mannitol instead of 20mM glucose.
In order to confirm that human retinal endothelial cells express similar patterns of mitochondrial membrane transport system, some of the key experiments were repeated in the human retinal endothelial cells (HRECs) obtained from human retina obtained from Cell System, Kirkland, WA. The cells were cultured in DMEM containing fetal bovine serum (10%), bovine pituitary endothelial growth factor (15μg/ml) and ITS (insulin/transferrin/selenium), Gluta Max and antibiotic/antimycotic (1% each), as previously reported by us [25]. Cells from passages 4th-6th were incubated in the incubation medium containing 1% fetal bovine serum, 9% Nu-serum, 0.5μg/ml endothelial growth factor, 0.5% ITS, and 1% each Gluta Max and antibiotic/antimycotic, with 5mM glucose or 20mM glucose for 4 days. At the end of the incubation (4 days), RNA was extracted and analyzed for gene expressions.
Isolation of mitochondria
Mitochondria were isolated using mitochondria isolation kit from Invitrogen (Carlsbad, CA, USA) [7, 11, 26]. Mitochondria pellet was washed with PBS and resuspended in mitochondrial isolation buffer (250mM sucrose, 2mM EDTA, 25mM Tris-HCl pH 7.4). Protein was determined by the bicinchoninic acid protein assay (Sigma-Aldrich, St. Louis, MO). Mitochondria prepared by this method were largely free of nuclear contaminations [7, 26].
Gene expression
Total RNA was extracted from the retina or isolated cells with Trizol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Gene expressions of Tom70, Tom40, Tim23, Hsp60 and Tim44 were quantified by real-time RT-PCR using the SYBR green assay reagent (qPCR) and gene-specific primers (Table II). Rat Tim44 and human Tim44 and TFAM gene expressions were estimated using TaqMan primers (primer NM-017267.1, AF026030.1, NM-003201.1, respectively). Relative amplification was quantified by normalizing the gene-specific amplification to that of β-actin in each sample for SYBR green assays, and to that of 18s rRNA for TaqMan assay. Changes in mRNA abundance were calculated using the ΔΔCt method [8, 27].
Table II.
Primers for the target genes
| Target | Sequence | |
|---|---|---|
| Rat | ||
| D-loop | forward | 5′-CCTCCGTGAAATCAACAACC-3′ |
| reverse | 5′-TAAGGGGAACGTATGGACGA-3′ | |
| Cox II | forward | 5′-TGAGCCATCCCTTCACTAGG-3′ |
| reverse | 5′-TGAGCCGCAAATTTCAGAG-3′ | |
| Tom40 | forward | 5′-TGGGTCAAACGCTGGTCGCG-3′ |
| reverse | 5′-CCCATCGTTCGAGGACGCCG-3′ | |
| Tom70 | forward | 5′- TGTCGTCCGCTCCTGCTGCT-3′ |
| reverse | 5′-GCGTGGAGAGGAAGCCGGGA-3′ | |
| Tim23 | forward | 5′-CGCGGGTCTTCCCGCTGTTG-3′ |
| reverse | 5′-GGCTCCGAAAAAGCCGGCCA-3′ | |
| Hsp60 | forward | 5′-TGGAGTAGCCGTGTTGAAGGTTGG-3′ |
| reverse | 5′-CCGAAGTAGAGCACAGCCCCCT-3′ | |
| β-actin | forward | 5′-AGCGAGCCGGAGCCAATCAG-3′ |
| reverse | 5′ -TGCGCCGCCGGGTTTTATAGG-3′ | |
| BRECs | ||
| Tom70 | forward | 5′ -CGACCAGCAAGGAGGCGGTG-3′ |
| reverse | 5′-ATCCCACTCCCGGAACCGGG-3′ | |
| Tim44 | forward | 5′-GGGAACTGGCGTCCACCTGC- 3′ |
| reverse | 5′-CCGATGCAGCCCCCTACCCT-3′ | |
| β-actin | forward | 5′-TGTTCCCTTCCACAGGGTGT-3′ |
| reverse | 5′-TCCCAGTTGGTAACAATGCCA-3′ | |
| HRECs | ||
| Tom70 | forward | 5′-GAAGTGATTTCCGCCCCTCC-3′ |
| reverse | 5′-CGCCACAATCACCAAACAGC-3′ | |
| β-actin | forward | 5′-AGCCTCGCCTTTGCCGATCCG-3′ |
| reverse | 5′ -TCTCTTGCTCTGGGCCTCGTC-3′ |
Protein expression
Protein (25-80μg) was separated on a 4-16% SDS-PAGE, transferred to a nitrocellulose membrane, and blocked with 5% non-fat milk for 1 hour. The membranes were incubated with antibodies against the protein of interest (Tom40, Tim23, Hsp60 from Santa Cruz Biotechnology, Santa Cruz, CA or Tom70, Tim44 from Abcam, Cambridge, MA) overnight at 4°C. β-actin (Sigma-Aldrich) was used as the loading control for homogenate and cytosol fractions, and cytochrome c oxidase subunit IV (Cox IV from Molecular Probe, Eugene, OR) for mitochondria fraction.
Interaction of TFAM with the membrane transport proteins
Interaction of TFAM with Tom70 or Tim44 was determined by co-immunoprecipitation technique. Protein (150μg) was incubated overnight at 4°C with 1μg of TFAM antibody (Santa Cruz Biotechnology) followed by 1 hour with 20μl of Protein A/G plus agarose immunoprecipitation beads reagent (prewashed and suspended in lysis buffer). The beads were washed four times, and the proteins were separated on a SDS-PAGE. The membranes were immunoblotted with anti-Tim44 or Tom70 antibodies, and then the membranes were stripped to detect the expression of TFAM, the loading control [8, 11, 28].
Total Tom complex
Retina was homogenated in MOPS buffer (44mM Sucrose, 20mM MOPS, 1mM EDTA and protease inhibitors), centrifuged at 500g for 3 minute, and the resultant supernatant was centrifuged at 12,000 g for 20 minute at 4°C. Pellet rich in mitochondria was re-suspended in 30μl ACBT- buffer (75mM bis-Tris, 1.5M aminocapriatic acid, pH7.5) and solubilized using 5μl lauryl-β-D-maltoside (10% solution) for 10 minutes at 4°C. Samples were centrifuged at 20,000g for 20 minutes, the pellet was discarded and the supernatant was used for analysis. Samples were stained with blue native sample buffer (750mM aminocapriatic acid, 0.5mM EDTA, 50mM bis-Tris pH7.0 with Coomassie brilliant blue G250) and analyzed on a native gel using the procedure described by others [29]. Proteins were transferred using modified transfer buffer (25mM Tris-HCl pH 9.4, 40mM aminocapriatic acid and 20% methanol), blocked overnight with 5% milk in PBS, and immunoblotted with anti-Tom40 antibody (Santa Cruz Biotechnology).
Binding of TFAM with mtDNA
The binding of TFAM with mtDNA was assessed by chromatin immunoprecipitation (ChIP) using ChIP Assay Kit (Millipore, Temecula, CA), as previously reported by us [25, 30]. Rat retina was crosslinked with 1% paraformaldehyde for 15 minutes, and the fixed sample was resuspended in lysis buffer containing 1% SDS, 10mM EDTA, 50mM Tris-HCl, pH 8.1 (ChIP Assay Kit, Millipore) and protease inhibitors. The samples were sonicated on ice four to six times for 10 seconds each, the supernatant was diluted in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2mM EDTA, 16.7mM Tris-HCl, and 167mM NaCl, pH 8.1), and pre-cleared with protein A agarose/salmon sperm DNA for 30 minutes. A small aliquot of protein-DNA complex (15-20μg) was analyzed for starting chromatin input, and the remaining complex was immunoprecipitated with anti-rabbit TFAM antibody or rabbit normal IgG. The immunoprecipitate collected with protein A agarose/salmon sperm DNA was washed with low-salt buffer, high-salt buffer, and LiCl buffer. This was followed by washing the immunoprecipitate twice with Tris-EDTA buffer and eluted twice with 1% SDS containing 0.1 mM NaHCO3. Elutes and input were heated at 65°C for 6 hours to reverse the formaldehyde crosslinking, and digested with protease K at 45°C for 1 hour. DNA fragments were recovered by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation, and resuspended in 20μl free nuclease water. Expressions of the mitochondrial DNA's D-loop region and cytochrome oxidase subunit II (Cox II, mtDNA-encoded) were quantified by qPCR using specific primers (Table II). Normal rabbit IgG was used as the negative antibody control and DNA from the input as the internal control.
Co-localization of TFAM with Tom70 and with Tim44
The cells grown on 12-mm coverslips, coated with 0.1% gelatin, were incubated in 5mM or 20mM glucose for 4 days, fixed with cold methanol at -20°C for 15 minutes, and permeabilized in 0.25% Triton X-100 for 10 minutes. The cells were blocked in 5% BSA for 1 hour and incubated with either mouse anti-TFAM and rabbit anti-Tom70 antibodies or with mouse anti-TFAM and rabbit anti-Tim44 antibodies for 2 hours. The slides were washed with PBS, incubated with FITC conjugated anti-mouse or Texas red conjugated anti-rabbit secondary antibodies for 1 hour in the dark. After washing the cells with PBS, the coverslips were mounted with Vecta Shield (Vector Laboratories) [5, 28].
Statistical analysis
Data are expressed as mean±SD. Statistical analysis was calculated using Sigma Stat software. The Shapiro-Wilk test was used to test for normal distribution of the data. For variables with normal distribution Student T-test was used for comparing two groups and ANOVA followed by Bonferroni was applied for multiples groups. For data that did not present normal distribution Mann-Whitney U were used to compare two groups and Kruskal-Wallis followed by Dunn's for multiples groups was used. P value of <0.05 was considered statistically significant.
Results
Effect of diabetes on the transport of retinal TFAM into the mitochondria
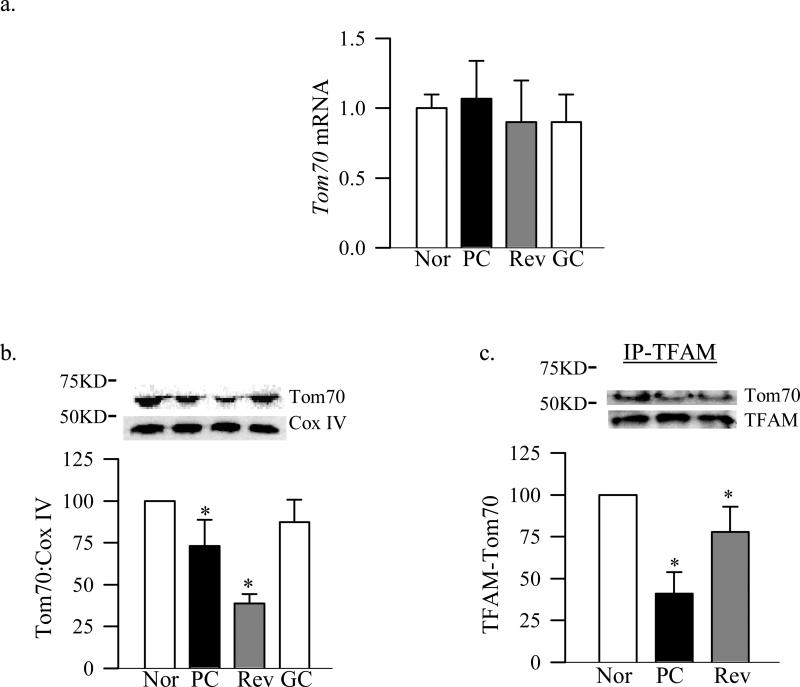
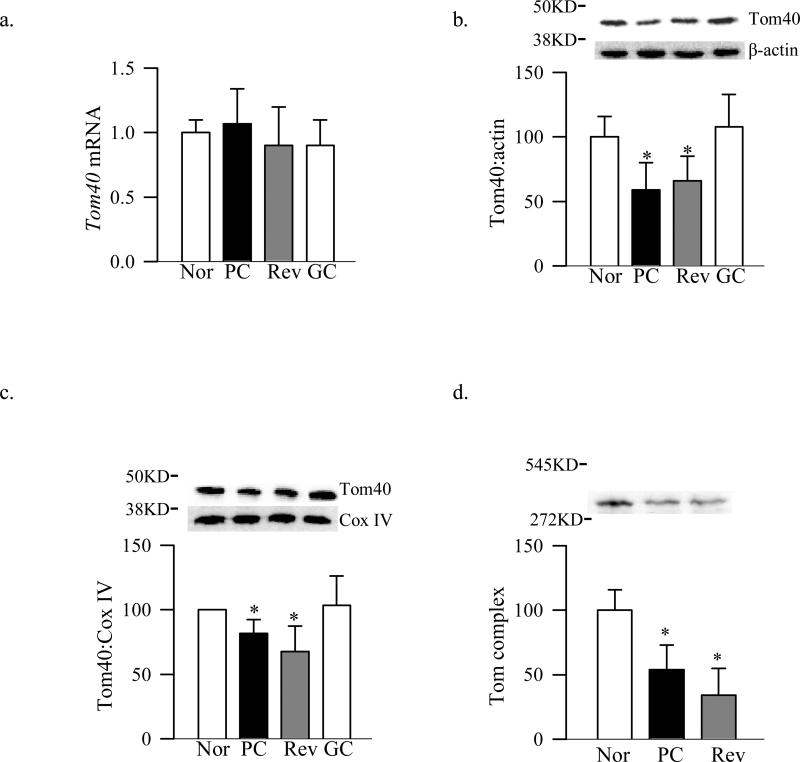
Tom70 is the main receptor of the Tom complex for the proteins which are chaperoned by Hsp70 [14], and Tom40 is the main channel of Tom complex [12]. Figure 1 shows that diabetes had no effect on retinal Tom70 gene expression, but its accumulation in the mitochondria was decreased by 25%. In addition Tom70 binding with TFAM was decreased by 50% compared to the age-matched normal rats. Diabetes had no effect on the gene expression of Tom40, but its total protein expression and accumulation in the mitochondria were significantly decreased (Figure 2a-c). To investigate the effect of diabetes on the total Tom complex, we used Blue Native Page method, and figure 2d shows that the expression of Tom complex (molecular weight of 440KD) was decreased by 60% in the retina from diabetic rats compared to that from age-matched non-diabetic rats.
Figure 1.
Effect of diabetes and cessation of hyperglycemia on Tom70 and its binding with TFAM. Tom70 (a) gene expression was quantified by qPCR using specific rat primers and β-actin as an internal control, and (b) protein expression in the mitochondria was assessed by western blot analysis using Cox IV as a loading control and Precision Plus standard protein markers (10-250KD, BioRad, Hercules, CA) . (c) Binding of TFAM with Tom70 was performed by immunoprecipitating TFAM, followed by western blot analysis for Tom70. TFAM was used as the loading control. The values are represented as mean ±SD from five to seven animals in each group: Nor- normal, PC- poor glycemic control for ~8 months, Rev- poor glycemic control for ~ 4 months followed by ~4 months good control and GC- good glycemic control for ~8 months. *P<0.05 compared with normal.
Figure 2.
Retinal Tom40 in diabetes. Tom 40 (a) gene expression was quantified by qPCR, and its protein expressions (b) in the retinal homogenate (c) and in the mitochondria were analyzed by Western blotting technique using β-actin and Cox IV as internal controls for the homogenate and mitochondria respectively. (d) Expression of the Tom complex, a 440KD complex, was quantified by Blue Native Page technique using molecular weights 14KD-545KD, non-denaturing, from Sigma-Aldrich. The values are represented as mean ±SD from five to eight animals in each group.
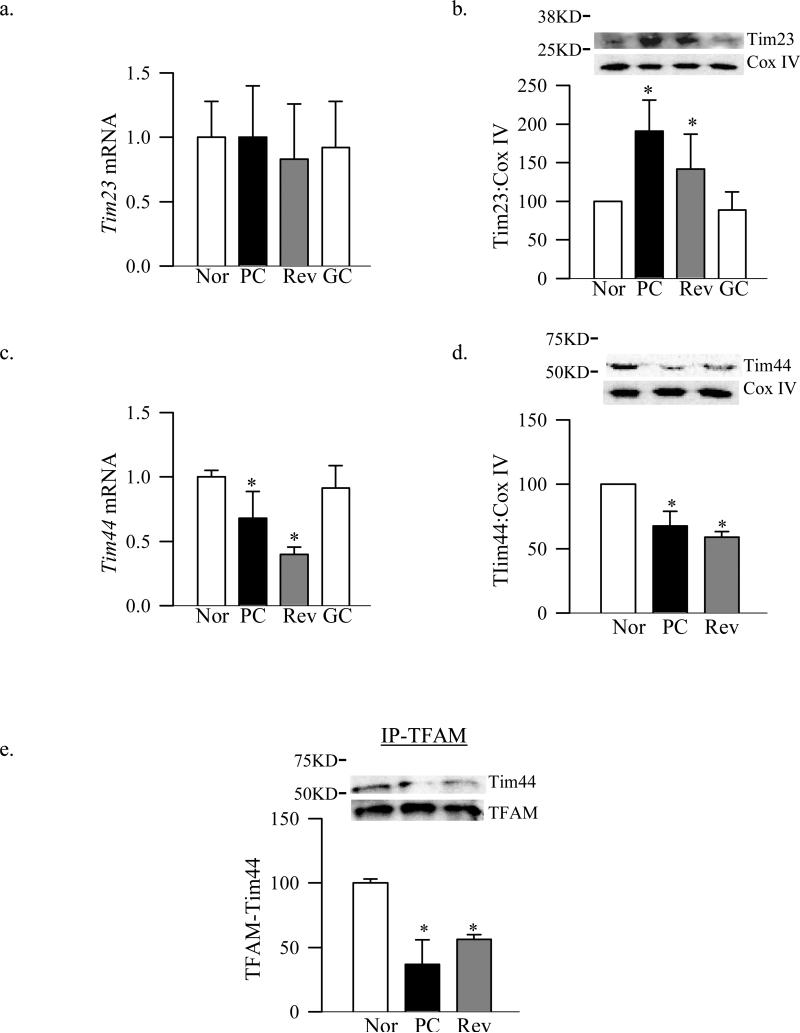
Since TFAM has to be transported from the outer membrane into the inner membrane, the expression of the channel forming protein Tim23 and the mitochondrial matrix docking protein Tim44 were analyzed. Despite no change in the gene expression of Tim23, its expression in mitochondria was increased by 80% in diabetic rats (Figure 3a&b). But, in contrast, Tim44 gene expression, its accumulation in the mitochondria and binding with TFAM were decreased by 40-60% compared to the values from normal rats (Figure 3c-e)
Figure 3.
Effect of diabetes on the expression of retinal Tim complex and the binding of TFAM with Tim44. Tim 23 (a) gene and (b) protein expressions were quantified by qPCR and by western blot techniques using Tim44 antibody from Abcam (~51KD band). The same sample was utilized to quantify Tim44 (c) gene and (d) mitochondrial expressions. (e) Binding of TFAM with Tim44 was performed by immunoprecipitating TFAM, followed by western blot analysis for Tim44 using TFAM as a loading control within samples. The values are represented as mean ±SD from four to eight animals in each group. Nor- normal, PC- poor glycemic control for ~8 months, Rev- poor glycemic control for ~ 4 months followed by ~4 months good control and GC- good glycemic control for ~8 months. *P<0.05 compared with normal.
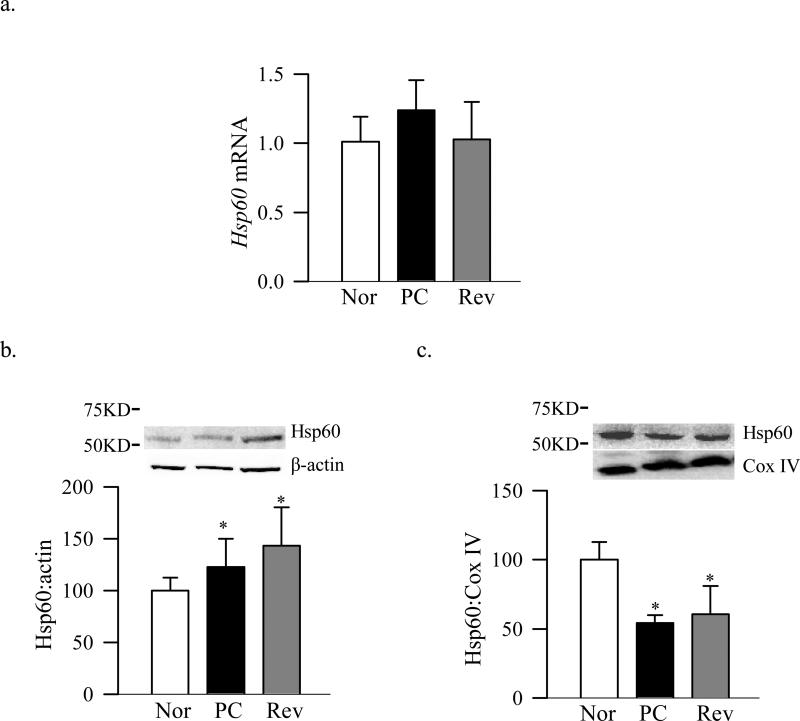
Figure 4 shows that the mRNA levels of the chaperone which is responsible for folding of proteins in the mitochondrial matrix, Hsp60, was not changed in diabetes (Figure 4a), however, the distribution of this chaperone in the mitochondria was altered. While retinal protein expression of Hsp60 increased in the cytosol (Figure 4b), its mitochondrial accumulation significantly decreased in diabetes (Figure 4c).
Figure 4.
Effect of diabetes and the termination of hyperglycemia on the retinal Hsp60. (a) Hsp60 gene expression was quantified by qPCR using β-actin as internal control. Protein expressions of Hsp60 in the (b) cytosol and (c) mitochondria were analyzed by Western blot technique using β-actin and Cox IV as a loading control respectively. The values are represented as mean ±SD from four to six animals in each group. *P<0.05 compared with normal.
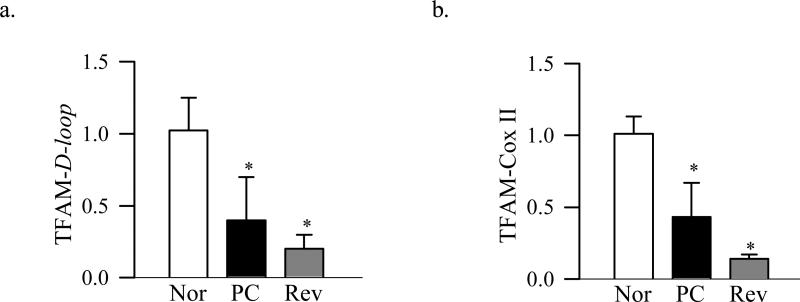
TFAM binds to the D-loop region of the mtDNA to regulate transcription of the mitochondrial genome, and binds to other regions in a non-sequence specific manner to stabilize mtDNA [32]. Results presented in figure 5 show that diabetes decreases the binding of TFAM with mtDNA at the D-loop region and also at the Cox II region by over 40% suggesting that diabetes compromises both, the transcription and the stability of mtDNA.
Figure 5.
TFAM binding with the retinal mtDNA. Binding of TFAM with (a) the D-loop and (b) Cox II regions of the mtDNA was assessed by ChIP assay using TFAM antibody, followed by quantification of the D-loop and Cox II regions by qPCR. Ct values were normalized to input DNA and rabbit IgG was used as a negative antibody control. The values are represented as mean ±SD from five to nine animals in each group. Nor- normal, PC- poor glycemic control for ~8 months, Rev- poor glycemic control for ~ 4 months followed by ~4 months good control and GC- good glycemic control for ~8 months. *P<0.05 compared with normal.
Reinstitution of good control and the transport of retinal TFAM into the mitochondria
The expression of Tom70 gene remained normal, but its abundance in the mitochondria and binding with TFAM continued to be subnormal compared to the values obtained from normal rats (Figure 1a-c). Protein expression of Tom40, its accumulation in the mitochondria and the overall expression of the Tom complex also continued to be subnormal even after 4 months of good control that had followed by 4 months of poor control (Figure 2). In addition, Tim23 mitochondrial expression remained elevated, while gene and mitochondrial accumulation continued to be subnormal (Figure 3b-d). This was accompanied by continued abnormalities in the gene and protein expressions of Hsp60 (Figure 4a-c), and subnormal binding of TFAM with the D-loop and at the Cox II regions (Figure 5a&b); the values obtained from the rats in Rev group were not statistically different from those obtained from rats in PC group.
However, when the rats were maintained in good glycemic control for the entire 8 months (GC group), gene and protein expressions of Tom70, Tom40, Tim23 and Tim44 were similar to those obtained from the rats in the normal group, but were significantly different from those obtained from rats in PC or Rev groups (Figures 1-3).
Effect of high glucose on the transport of TFAM into the mitochondria in retinal endothelial cells
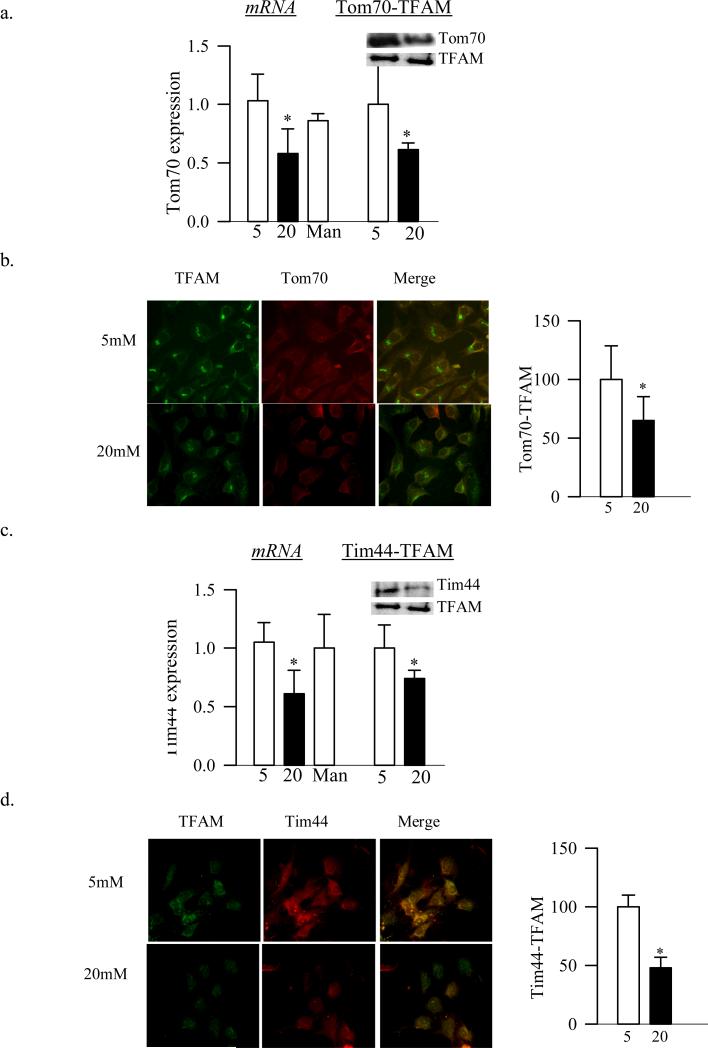
Four days of high glucose exposure of retinal endothelial cells decreased the gene expression of Tom70 by 40%, and its binding to TFAM, as quantified by both immunopreciptation and by fluorescence microscopy, by ~40% compared to the cells exposed to normal glucose (Figure 6a&b). As in the rat retina, gene expression of Tim44 and also the binding of TFAM with Tim44 were attenuated by 30-50% by high glucose compared to the cells in normal glucose (Figures 6c&d).
Figure 6.
Effects of high glucose on Tom70 and Tim44, and their binding to TFAM in bovine retinal endothelial cells. (a) Tom70 gene expression and its binding to TFAM were quantified by qPCR and by immunopreciptation techniques respectively. (b) BRECs grown in cover-slips, and incubated in 5mM or 20mM glucose for 4 days were fixed and incubated with mouse anti-TFAM, and rabbit anti-Tom70. The cells were then incubated with FITC-conjugated anti-mouse and Texas Red-conjugated anti-rabbit antibodies. The cells were examined under a Zeiss ApoTome at 40X objective. The co-localization was calculated using Axio vision software. (c) Gene expression and its binding of Tim44 with TFAM were quantified by qPCR and by immunopreciptation techniques. (d) Binding of Tim44 with TFAM was quantified by microscopy using FITC conjugated TFAM antibody, and Texas Red-conjugated Tim44. The values are presented as means ± SD. Each measurement was made in duplicate in three-four different cells preparations. 5=5mM glucose, 20=20mM glucose and Man=20mM mannitol. *p<0.05 compared to 5mM glucose.
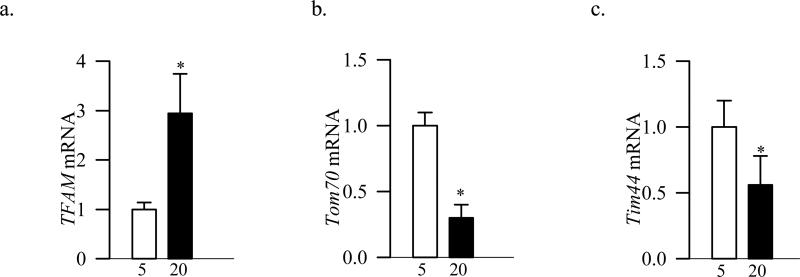
Consistent with the results obtained from BRECs, the mRNA levels of TFAM was increased by almost three folds in HRECs incubated in high glucose (Figure 7a), but those of Tom70 and Tim44 were significantly decreased compared to HRECs incubated in normal glucose medium (Figure 7b&c).
Figure 7.
Effects of high glucose on TFAM, Tom70 and Tim44 in HRECs. Gene expressions of (a) TFAM , (b) Tom70 and (c) Tim44 were quantified by qPCR techniques using human specific primers. Internal standards included β-actin for Tom70 and 18s rRNA for TFAM and Tim44. Each measurement was made in duplicate in two different cells preparations. 5=5mM glucose and 20=20mM glucose. *p<0.05 compared to 5mM glucose.
Discussion
Mitochondrial superoxide radicals are considered to serve as a common link between biochemical mechanisms that underlay the diabetic complications [32]. Our previous work has shown that in the pathogenesis of diabetic retinopathy, mitochondrial number is decreased, mtDNA is damaged and its transcription and replication machinery is impaired [7, 8, 11, 26]. Moreover, the mitochondrial accumulation of TFAM, one of the key mitochondrial transcriptional factors, is subnormal, resulting in decreased transcription of the key proteins required in the electron transport chain function, and this possibly initiates a vicious cycle of superoxide accumulation [7, 8, 11]. Here, we have investigated the mechanism responsible for decreased TFAM accumulation in the mitochondria; the results demonstrate that the mitochondrial membrane transport system is dysfunctional and the binding of TFAM with the membrane transport proteins is impaired. Furthermore, the binding of TFAM at the promoter region of the mtDNA, and at other non-specific region is also decreased resulting in subnormal transcription and compromised stability. This makes mtDNA more vulnerable to the damage, and the process continues even after termination of hyperglycemic insult.
Mitochondria contain about 1500 different proteins, but only 13 of them are synthesized by mtDNA, and the rest are encoded by nuclear DNA, synthesized in the cytosol and transported to the mitochondria [9, 12, 33]. The newly synthesized mitochondrial proteins require a complex mechanism to reach to their site of action. The cytosolic Hsp70 serves as a key chaperone helping with the transport of proteins to the mitochondria membrane, and this chaperone also ensures the correct folding of proteins [13]. Hsp70 docks on the mitochondria surface by its interaction with the Tom70 receptor and releases the pre-protein by an active process [14]. We have recently shown that in diabetes the binding of Hsp70 to TFAM is decreased [8, 11]. Furthermore, pre-proteins carried by Hsp70 are transferred from the mitochondrial surface receptor to a common Tom40 translocation pore to be transported to the inner membrane space [13]. Now we show that, despite no change in the gene expression of retinal Tom70 and Tom40, their mitochondrial levels are significantly decreased in diabetes, and the overall Tom complex becomes subnormal. These changes in the Tom complex are accompanied by subnormal binding of TFAM with Tom70 suggesting that impaired Tom complex in diabetes could be responsible for subnormal levels of TFAM in the mitochondria, resulting in impaired mtDNA biogenesis. In support, abnormalities in Tom40 complex have been implicated in mitochondrial dysfunction associated with neurodegenerative diseases [34, 35].
Proteins cross the inner membrane using Tim complex through Tim23 channel, and are pulled into the matrix by a mitochondrial Hsp70, which docks on Tim44 protein [15, 16]. Our current data show that, despite increase in Tim23, Tim44 remains subnormal in diabetic retinopathy. These results are consistent with our previous study, and those of other showing downregulation of mitochondrial transport proteins in diabetic complications [19, 27, 36, 37]. Thus, decreased Tim44 in the mitochondria further decreases the availability of Tim44 to transport TFAM to the mitochondria. However, the effect of diabetes on the post-translational modifications of TFAM [38, 39], which may contribute to its decreased transport into the mitochondria, cannot be ruled out. However, we need to acknowledge that though Tim23 and Tim44 are the members of the Tim complex, the results also indicate that diabetes affects them in a variant manner; while Tim23 is increased in retinal mitochondria, Tim44 expression is decreased. The reason for such variant is unclear, but could include their localization in the mitochondria, as Tim44 is localized in the matrix and Tim23 is organized in the inner membrane [40], or the differences in the vulnerability of these two proteins to post-translational modifications which are favored in diabetic environment [41].
Once inside the mitochondria matrix, Hsp60 helps in proper folding of the proteins [17, 18]. In stress conditions Hsp60 is released from the mitochondria to the cytosol, but the function of Hsp60 in the cytosol is not clear [42]. In the retina of diabetic rodents, the gene expression of Hsp60 is increased but its protein is decreased in mitochondria [43], and the binding of Hsp60 with TFAM is impaired [11]. Here we show that, while diabetes increases the levels of Hsp60 in the retinal cytosol, mitochondrial levels are decreased, suggesting that the folding of TFAM in the mitochondria matrix could also be compromised, further compromising TFAM-mediated mtDNA biogenesis and stability.
TFAM, after reaching into the mitochondria matrix, induces the replication of mtDNA and facilitates the formation of nucleoid structures to protect the mtDNA from oxidative damage [31, 44]. We have shown that in the pathogenesis of diabetic retinopathy TFAM protein accumulation in the mitochondria is decreased despite increase in its gene expression [8, 11], and here our results demonstrate that TFAM becomes less active, as evidenced by its decreased binding to the D-loop and Cox II regions. This decreased binding also makes mtDNA more vulnerable to the damage. In support, our previous reports have shown increased retinal mtDNA damage in diabetes [7, 8, 28], and others have shown relationship between changes in binding of TFAM to the mtDNA and neurodegenerative disease [45].
Termination of hyperglycemia does not halt the progression of diabetic retinopathy [20-23], and the alteration in the mitochondria protein transport machinery and in mtDNA biogenesis, induced by the hyperglycemic insult, persists for a period of good control that has followed hyperglycemic insult [11, 19]. Our data presented here clearly show that 4 months of good glycemic control in rats, which has followed 4 months of poor control, does not provide any benefit to the impaired mitochondrial protein transporters, and the binding of TFAM with mtDNA remains compromised. These data suggest that mitochondrial membrane proteins have important role in the continued dysfunctional mitochondria biogenesis, and in the progression of diabetic retinopathy. We need to acknowledge that the levels of Tom40 and the binding of TFAM with mtDNA are slightly lower in the Rev group compared to the PC group. The exact mechanism responsible for such discrepancy is not clear, but could include sustained attack of increased ROS, which the retina continues to experience even after termination of the hyperglycemic insult [11, 19, 22-24, 30, 41]. On the other hand, if good glycemic control is initiated soon after the induction of diabetes, alterations in the transporter proteins and transport of TFAM are not observed, and the mitochondria biogenesis remains normal, supporting the importance of early and continued good glycemic control for diabetic patients.
The microvasculature of the retina is the site of pathology associated with diabetic retinopathy, and retinal capillary cells exposed to high glucose can mimic the dysfunction observed in the retina [46-48]. Here we show that Tom70 gene expression is decreased significantly in retinal endothelial cells incubated with high glucose, and its binding with TFAM becomes subnormal. Consistent with the results from diabetic animals, Tim44 is down-regulated in high glucose and its binding to TFAM is also affected. Similar glucose-induced alterations in TFAM and mitochondria membrane transport proteins Tom70 and Tim44 in retinal endothelial cells from humans further confirms their role in diabetic retinopathy. These results suggest that the impairment in mitochondria biogenesis observed in endothelial cells in high glucose conditions could be attributed to the decrease of mitochondrial transporters proteins.
In conclusion, our results demonstrate that the downregulation of mitochondrial transport system impairs mitochondrial biogenesis by impeding the transport of TFAM to the mitochondria. This contributes to the decrease in transcription of mtDNA-encoded proteins, and electron transport chain becomes dysfunctional resulting in mitochondria damage. The system continues to be damaged even after reversal of the hyperglycemic insult, suggesting its role in the metabolic memory associated with continued progression of diabetic retinopathy. Future molecular strategies targeting mitochondria transporter proteins may be promising for improving mitochondrial biogenesis that could halt the development of diabetic retinopathy.
Acknowledgements
We thank Doug Putt for technical assistance. This study was supported in parts by grants from the National Institutes of Health, Juvenile Diabetes Research Foundation, the Thomas Foundation, Midwest Eye Banks, and Research to Prevent Blindness.
References
- 1.Chew EY. Epidemiology of diabetic retinopathy. Hosp Med. 2003;64:396–399. doi: 10.12968/hosp.2003.64.7.2275. [DOI] [PubMed] [Google Scholar]
- 2.Frank RN. Diabetic Retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 3.Kern TS, Tang J, Mizutani M, Kowluru R, Nagraj R, Lorenzi M. Response of capillary cell death to aminoguanidine predicts the development of retinopathy: Comparison of diabetes and galactosemia. Invest Ophthalmol Vis Sci. 2000;41:3972–3978. [PubMed] [Google Scholar]
- 4.Kowluru RA, Kern TS, Engerman RL. Abnormalities of retinal metabolism in diabetes or experimental galactosemia. IV. Antioxidant defense system. Free Rad Biol & Med. 1997;22:587–592. doi: 10.1016/s0891-5849(96)00347-4. [DOI] [PubMed] [Google Scholar]
- 5.Kowluru RA, Tang J, Kern TS. Abnormalities of retinal metabolism in diabetes and experimental galactosemia. VII. Effect of long-term administration of antioxidants on the development of retinopathy. Diabetes. 2001;50:1938–1942. doi: 10.2337/diabetes.50.8.1938. [DOI] [PubMed] [Google Scholar]
- 6.Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Inves Ophthalmol Vis Sci. 2003;44:5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- 7.Madsen-Bouters SA, Mohammad G, Kanwar M, Kowluru RA. Role of mitochondrial DNA damage in the development of diabetic retinopathy, and the metabolic memory phenomenon associated with its progression. Antioxid Redox Signal. 2010;13:797–805. doi: 10.1089/ars.2009.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos JM, Tewari S, Goldberg AFX, Kowluru RA. Mitochondria biogenesis and the development of diabetic retinopathy. Free Rad Biol Med. 2011;51:1849–1860. doi: 10.1016/j.freeradbiomed.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 11.Santos JM, Kowluru RA. Role of mitochondria biogenesis in the metabolic memory associated with the continued progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2011;52:8791–8798. doi: 10.1167/iovs.11-8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endo T, Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta. 2010;1803:706–714. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 14.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 15.Cui W, Josyula R, Li J, Fu Z, Sha B. Membrane binding mechanism of yeast mitochondrial peripheral membrane protein TIM44. Protein Pept Lett. 2011;18:718–725. doi: 10.2174/092986611795445996. [DOI] [PubMed] [Google Scholar]
- 16.Koehler CM. Protein translocation pathways of the mitochondrion. FEBS Lett. 2000;476:27–31. doi: 10.1016/s0014-5793(00)01664-1. [DOI] [PubMed] [Google Scholar]
- 17.Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Knowlton AA. HSP60, Bax, apoptosis and the heart. J Cell Mol Med. 2005;9:51–58. doi: 10.1111/j.1582-4934.2005.tb00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong Q, Kowluru RA. Diabetic retinopathy and damage to mitochondrial structure and transport machinery. Inves Ophthalmol Vis Sci. 2011;52:8739–8746. doi: 10.1167/iovs.11-8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diabetes Control and Complications Trial Research Group The effect of intensive treatment of diabetes on the development of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 21.Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. N Engl J Med. 2000;342:381–389. doi: 10.1056/NEJM200002103420603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowluru RA. Effect of re-institution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes. 2003;52:818–823. doi: 10.2337/diabetes.52.3.818. [DOI] [PubMed] [Google Scholar]
- 23.Kowluru RA, Kanwar M, Kennedy A. Metabolic memory phenomenon and accumulation of peroxynitrite in retinal capillaries. Exp Diabetes Res. 2007;2007:2196. doi: 10.1155/2007/21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowluru RA, Kanwar M, Kennedy A. Metabolic memory phenomenon and accumulation of peroxynitrite in retinal capillaries. Exp Diabetes Res. 2007;2007:2196. doi: 10.1155/2007/21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowluru RA, Kanwar M. Oxidative stress and development of diabetic retinopathy: Contributary role of matrix metalloproteinase-2. Free Rad Biol Med. 2009;46:1677–1683. doi: 10.1016/j.freeradbiomed.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tewari S, Santos JM, Kowluru RA. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxid Redox Signal. 2012;17:492–504. doi: 10.1089/ars.2011.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110:1306–1313. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowluru RA, Mohammad G, Dos Santos JM, Zhong Q. Abrogation of MMP9 gene protects against the development of retinopathy in diabetic mice by preventing mitochondrial damage. Diabetes. 2011;60:3023–3033. doi: 10.2337/db11-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvaruso MA, Smeitink J, Nijtmans L. Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods. 2008;46:281–287. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011;60:1304–1313. doi: 10.2337/db10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malarkey CS, Bestwick M, Kuhlwilm JE, Shadel GS, Churchill ME. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 33.Endo T, Yamamoto H, Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci. 2003;116:3259–3267. doi: 10.1242/jcs.00667. [DOI] [PubMed] [Google Scholar]
- 34.Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura A, Sakurai T, Yamada M, Koumura A, Hayashi Y, Tanaka Y, et al. Antibodies Against the Tom40 Subunit of the Translocase of the Outer Mitochondrial Membrane Complex and Cognitive Impairment in Alzheimer's Disease. J Alzheimers Dis. 2012;29:373–377. doi: 10.3233/JAD-2011-111343. [DOI] [PubMed] [Google Scholar]
- 36.Wada J, Makino H, Kanwar YS. Gene expression and identification of gene therapy targets in diabetic nephropathy. Kidney Int. 2002;61:S73–S78. doi: 10.1046/j.1523-1755.2002.0610s1073.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wada J, Hashimoto I, Eguchi J, Yasuhara A, Kanwar YS, et al. Therapeutic approach for diabetic nephropathy using gene delivery of translocase of inner mitochondrial membrane 44 by reducing mitochondrial superoxide production. J Am Soc Nephrol. 2006;17:1090–1101. doi: 10.1681/ASN.2005111148. [DOI] [PubMed] [Google Scholar]
- 38.Suarez J, Hu Y, Makino A, Fricovsky E, Wang H, Dillmann WH. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am J Physiol Cell Physiol. 2008;296:C1561–C1568. doi: 10.1152/ajpcell.00076.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antelman J, Manandhar G, Yi YJ, Li R, Whitworth KM, Sutovsky M, et al. Expression of mitochondrial transcription factor A (TFAM) during porcine gametogenesis and preimplantation embryo development. J Cell Physiol. 2008;217:529–543. doi: 10.1002/jcp.21528. [DOI] [PubMed] [Google Scholar]
- 40.Bauer MF, Gempel K, Reichert AS, Rappold GA, Lichtner P, Gerbitz KD, Neupert W, Brunner M, Hofmann S. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J Mol Biol. 1999;289:69–82. doi: 10.1006/jmbi.1999.2751. [DOI] [PubMed] [Google Scholar]
- 41.Madsen-Bouterse SA, Mohammad G, Kowluru RA. Glyceraldehyde 3 phosphate dehydrogenase in retinal microvasculature: Implications for the development and progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2010;51:1765–1772. doi: 10.1167/iovs.09-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voos W, Röttgers K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta. 2002;159:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- 43.Mohammad G, Kowluru RA. Novel role of mitochondrial matrix metalloproteinase-2 in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2011;52:3832–3841. doi: 10.1167/iovs.10-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohgaki K, Kanki T, Fukuoh A, Kurisaki H, Aoki Y, Ikeuchi M, et al. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- 45.Rothfuss O, Gasser T, Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2010;38:e24. doi: 10.1093/nar/gkp1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy S, Sala R, Cagliero E, Lorenzi M. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci USA. 1990;87:404–408. doi: 10.1073/pnas.87.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madsen-Bouters SA, Zhong Q, Mohammad G, Ho YS, Kowluru RA. Oxidative damage of mitochondrial DNA in diabetes, and its protection by manganese superoxide dismutase. Free Rad Resaerch. 2010;44:313–321. doi: 10.3109/10715760903494168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowluru RA, Chan PS. Metabolic memory in diabetes - from in vitro oddity to in vivo problem: Role of Apoptosis. Brain Res Bull. 2010;87:297–302. doi: 10.1016/j.brainresbull.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]