Abstract
Background
Global DNA hypomethylation is an early molecular event in carcinogenesis. Whether methylation measured in peripheral blood mononuclear cells(PBMCs) DNA is a clinically reliable biomarker for early detection or cancer risk assessment is to be established.
Methods
From an original sample-set of 753 male and female adults(aged 64.8±7.3years),PBMCs DNA methylation was measured in 68 subjects with history of cancer at time of enrollment and 62 who developed cancer during follow-up. Age-and sex-matched controls for prevalent and incident cancer cases(n=68 and n=58,respectively)were also selected. Global DNA methylation was assessed by LC/MS. Methylenetetrahydrofolate reductase (MTHFR) 677C>T genotype and plasma folate concentrations were also determined for the known gene-nutrient interaction affecting DNA methylation.
Results
Cancer subjects had significantly lower PBMCs-DNA methylation than controls [4.39(95%CIs 4.25–4.53) vs. 5.13(95%CIs 5.03–5.21)%mCyt/(mCyt+Cyt), P<0.0001]. A DNA methylation threshold of 4.74% clearly categorized cancer patients from controls so that those with DNA methylation <4.74% showed an increased prevalence of cancer than those with higher levels (91.5% vs. 19%;P <0.001). Subjects with cancer at follow-up had, already at enrollment, reduced DNA methylation compared to controls [4.34(95%CIs 4.24–4.51) vs. 5.08(95%CIs 5.05–5.22)%mCyt/(mCyt+Cyt),P<0.0001].Moreover, MTHFR677C>T genotype and folate interact for determining DNA methylation, so that MTHFR677TT carriers with low folate had the lowest DNA methylation and concordantly showed a higher prevalence of cancer history (OR=7.04,95%CIs 1.52–32.63, P=0.013).
Conclusions
Genomic PBMCs-DNA methylation may be a useful epigenetic biomarker for early detection and cancer risk estimation.
Impact
This study identifies a threshold for PBMCs-DNA methylation to detect cancer-affected from cancer–free subjects and an at-risk condition for cancer based on genomic DNA methylation and MTHFR677C>T-folate status.
Keywords: DNA methylation, epigenetics, cancer, biomarkers, peripheral blood mononuclear cells, MTHFR, gene-nutrient interactions, folate
Introduction
DNA methylation is an epigenetic phenomenon that affects the regulation of gene expression and genome integrity (1). Aberrant DNA methylation is a critical mechanism for carcinogenesis (2, 3). Global DNA hypomethylation is an almost universal finding in many human cancer tissues (4) and cancer precursor cells (5), and many studies in rodent models demonstrated that global DNA hypomethylation can, by itself, induce cancer (6). However, it is yet unclear whether this common epigenetic feature can be a clinically functional biomarker or even a screening tool to identify patients affected by cancer disease. A stimulating scientific debate is indeed ongoing to clarify the usefulness of genomic methylation status in DNA obtained from an easily accessible tissue in humans such as peripheral blood mononuclear cells (PBMCs) as a suitable biomarker even for cancer tissue of different origin (7).
Differently from mutations and loss of heterozygosity, DNA methylation is a reversible phenomenon (8, 9) and it can be modified by nutrients such as folate (10) as well as influenced by the common 677C>T polymorphism in methylenetetrahydrofolate reductase (MTHFR) gene (11, 12). An interaction between folate status and the MTHFR variant can affect global DNA methylation (11) and thereby potentially modify the risk of cancer. The polymorphic MTHFR677C>T favors the reduction of the 5-methyltetrahydrofolate (5-methylTHF) proportion in tissue folate pool (11) and, at the low folate status, individuals carrying the MTHFR677C>T homozygous variant genotype (677TT) have a decreased global DNA methylation (11, 12) which may induce molecular modifications in the cell eventually leading towards the development of cancer (13). Most recently, DNA methylation has been tested in blood as a circulating tumor cell DNA marker (14) and a number of studies evaluated the possible role of circulating white blood cells DNA methylation in different types of cancer as a potential marker to define the risk for malignancies of different tissue origin (7, 15–18).
We, therefore, set out to determine whether global hypomethylation in PBMCs DNA can indicate the presence of cancer or impending cancer development.
Materials and Methods
Study subjects
From an original sample-set of 753 subjects of both sexes and mean age of 64.8±7.3 years, who were recruited from a single geographical area in Italy with a similar socio-economic background in which they were previously engaged from 1996 to 2004 for a cardiovascular risk factors study (19, 20), we retrospectively evaluated the clinical history for cancer diagnosis and selected 68 subjects who were diagnosed of cancer at the time of enrollment to the study and 62 subjects who developed cancer during an eight-year follow-up period, along with 68 age- and sex-matched controls at enrollment and 58 age- and sex-matched controls, at follow-up. Clinical history of the subjects referring to cancer disease was obtained by search within the national population register, and by an ambulatory or a telephone-administered survey performed by a physician. The causes of death including cancer were obtained from death certificates kept at the Italian Institute of Statistics (ISTAT). This study was approved by the University Hospital of Verona Ethical Review Boards and informed consent was obtained from all subjects after full explanation of the study.
Specimen characteristics
Samples of peripheral venous blood were drawn in EDTA-containing BD Vacutainer® tubes (Franklin Lakes, NJ USA) from each subject after an overnight fast, before proceeding for plasma as well as DNA extraction. Plasma was accurately stored at −80°C before analyses.
Assay methods
DNA was extracted from PBMCs by Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) and global DNA methylation, expressed as percent (%) 5-methylcytosine (mCyt)/(mCyt + Cyt), was determined using a liquid chromatography/mass spectrometry (LC/MS) method, as we previously described (11, 21). Percent relative standard deviations for method precision of 1.9 (within-day) and 1.7 (day-to-day) (21). Plasma folate concentrations were measured by an automated chemiluminescence method (Chiron Diagnostics, East Walpole, MA, USA) having an intra-assay and inter-assay coefficients of variation of 5% and 9%, respectively. Total plasma homocysteine (tHcy) was measured by a high-performance liquid chromatography (HPLC) method with fluorescent detection, as previously described (11). High-sensitivity C-reactive protein (hs-CRP) was measured by a particle-enhanced nephelometric immunoassay with commercially available methods in a BNII Behring Nephelometer Analyzer (Dade Behring Inc, Newark, DE), as previously described (22). The analysis of the MTHFR677C>T polymorphism was performed by PCR followed by HinfI digestion (19, 23).
Statistical analyses
The distribution of continuous variables in groups are expressed as mean ± SD. Statistical analysis was performed with log-transformed data for all skewed variables and geometric means (antilogarithms of the transformed means) are presented with 95% confidence intervals (CIs). Statistical significance for differences in continuous variables was tested by Student's unpaired t-test or ANOVA variant analysis, with polynomial contrasts for linear trend when appropriate. Categorical variables were analyzed using a χ2 test, with χ2 for linear trend when appropriate. Adjustment for confounding variables was performed by general linear model or logistic regression analysis. Interaction terms were estimated by means of general linear models. Receiver Operating Characteristic (ROC) curve analysis was performed to identify the most efficient cutoff value of DNA methylation for cancer diagnosis as well as incident cancer occurrence during follow-up. The plasma folate concentrations corresponding to the 50° percentile in controls (12 nmol/L) were used as the threshold for either low (<12 nmol/L) or high folate (≥12 nmol/L). Statistical significance refers to a two-tailed analysis where P<0.05.
Results
Analysis of prevalent/historic cancer cases at baseline
The prevalence of cancer in this cohort was 9.2% which is similar to that reported for the whole population in the same Italian geographical area. Cancer types were hematological malignancies (20%), bladder cancer (19%), gastrointestinal cancers (13%), prostate (12%), breast (9%), kidney (7%), lung (6%), and larynx cancers (4%).
Cancer cases and controls as compared to other subjects within the study cohort showed a higher prevalence of coronary artery disease affected subjects (88.4% vs. 69.8%, P<0.001), slightly higher age (64.52±10.89 vs. 59.74±7.27, P<0.001) and prevalence of smokers (60% vs. 73%, P=0.01) and subjects of male sex (83.7% vs. 75%, P=0.039). No differences were instead detected in terms of plasma tHcy, folate, hs-CRP, vitamin B12 concentrations (P>0.2 for all the comparisons).
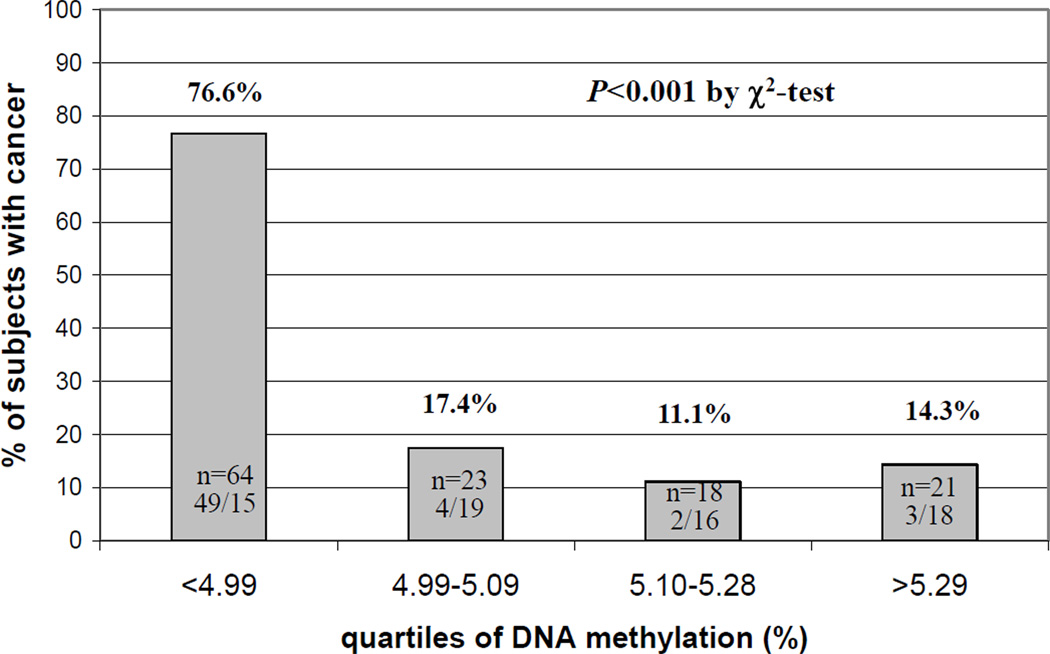
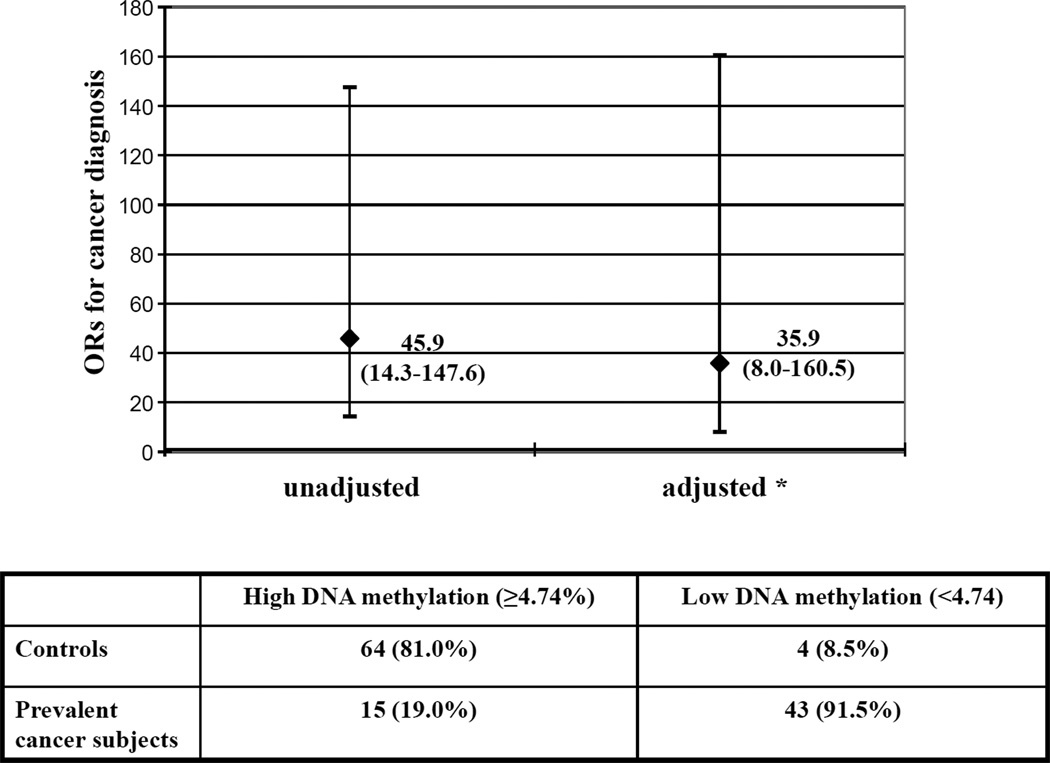
The main clinical and biochemical characteristics of patients with or without cancer history at baseline are described in Table 1. As shown, there were no significant differences between cancer cases and controls as it referred to age, sex, smoking habit, inflammatory marker hs-CRP, tHcy and vitamin B12. Plasma folate concentrations were significantly lower in cancer subjects than in controls (9.86 nmol/L, 95%CIs 8.26–11.77 vs. 13.24 nmol/L, 95%CIs 11.9–14.72, P=0.003). The frequency of homozygous mutants for the MTHFR677C>T polymorphism was significantly higher in cancer subjects compared to controls (31.03% vs. 11.70%, P=0.013). Moreover, cancer subjects who already had cancer at enrollment showed reduced global DNA methylation compared to controls who did not have cancer at enrollment (4.39%, 95%CIs 4.25–4.53 vs. 5.13 95%CIs 5.03–5.21, expressed as percent mCyt/mCyt+Cyt, P<0.0001) (Table 1). By stratifying the study population according to quartiles of DNA methylation (defined on the basis of control’s levels), subjects with cancer history were clearly more represented within the lowest quartile as compared to the others (P<0.001 by χ2-test) (Figure 1). In order to determine a precise cutoff to differentiate cases from controls, a ROC curve analysis was performed using data from the overall population, on which basis a threshold level of 4.74% for DNA methylation was chosen (AUC = 0.880 with 95%CIs 0.817–0.944) (Figure 2). Subjects with DNA methylation lower than 4.74% showed a much increased prevalence of cancer (43/47) than those with higher (≥4.74%) DNA methylation levels (91.5% vs. 19%, P<0.001 by χ2-test; OR 45.9 with 95%CI 14.3–147.6) (Figure 3). Such association persisted statistically significant even after adjustments for sex, age, smoking habit, serum concentrations of inflammatory marker hs-CRP as well as plasma folate, vitamin B12 and tHcy concentrations (OR 35.9 with 95%CIs 8.0–160.5) (Figure 3).
Table 1.
Characteristics of the subjects with history of cancer, at baseline.
| Variables | Controls (n=68) |
Cancer patients (n=68) |
P-value |
|---|---|---|---|
| age (years) | 64.10±6.69 | 65.65±7.92 | N.S. |
| sex (% males) | 83.5 | 82.7 | N.S.* |
| smoking habit (% smokers) | 71.2 | 68.7 | N.S.* |
| hs-CRP (mg/L) | 3.51 (2.75–4.26) |
3.07 (1.30–4.83) |
N.S. |
| tHcy (µmol/L) | 16.08 (14.81–17.44) |
15.46 (13.56–17.64) |
N.S. |
| vitamin B12 (pmol/L) | 375.63 (337.98–417.46) |
414.80 (361.33–476.18) |
N.S. |
| folate (nmol/L) | 13.2 (11.9–14.72) |
9.86 (8.26–11.77) |
0.003 |
| % MTHFR677TT genotype carriers |
11.70 | 31.03 | 0.013* |
| DNA Methylation (% mCyt/mCyt + Cyt) |
5.13 (5.03–5.21) |
4.39 (4.25–4.53) |
<0.0001 |
Values are expressed as means ± SD for age. Plasma folate, hs-CRP, vitamin B12, tHcy and DNA methylation status are presented as geometric means (antilogaritms of transformed means) and 95% confidence intervals are reported in parentheses with two-tailed P values. Statistical difference was evaluated by Student’s t-test except when differently indicated.
Statistical difference was evaluated by χ2 test. N.S., not statistically significant.
tHcy: total plasma homocysteine; hs-CRP: high-sensitivity C-reactive protein.
Figure 1.
Prevalence of subjects with cancer within the study population according to quartiles of global DNA methylation levels. DNA methylation refers to quantitative measurements of 5-metilcytosine (mCyt) indicated as percent (%) mCyt/(mCyt+Cyt). Number and corresponding percentage of subjects for the four groups divided into quartiles of DNA methylation are indicated within or on the top of each bar, respectively. Values of DNA methylation for each quartiles are reported at the bottom of each bar.
Figure 2.
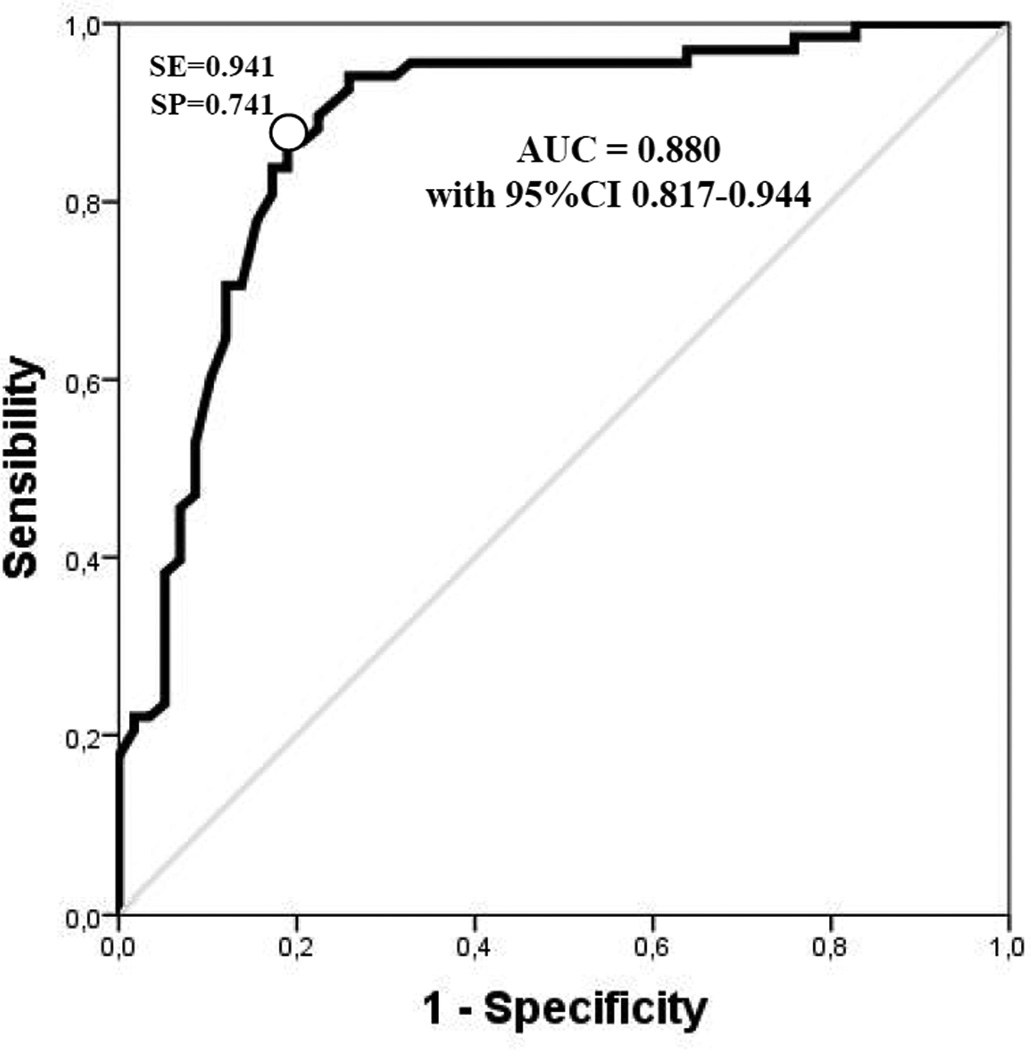
Receiver Operating Characteristic (ROC) curve for DNA methylation and cancer diagnosis. AUC, Area Under Curve = 0.880 with 95%CIs 0.817–0.944. The white point on the ROC curve corresponds to the 4.74% methylation and % sensibility (SE) and % specificity (SP) are indicated at that point (SE=0.941; SP=0.741).
Figure 3.
Association between low levels of DNA methylation [below the cutoff of 4.74% mCyt/(mCyt+Cyt)] and cancer history estimated by means of unadjusted and adjusted Odds Ratio (ORs). The table under the Figure indicates the number of subjects with history of cancer (prevalent cancer subjects) and of controls with corresponding percentages indicated in parentheses.
* the adjusted Odds Ratio analysis was performed by multivariate logistic regression model that simultaneously controlled for sex, age, smoking habit, serum concentrations of inflammatory marker high-sensitivity C-reactive protein, plasma levels of folate, vitamin B12 and plasma total homocysteine.
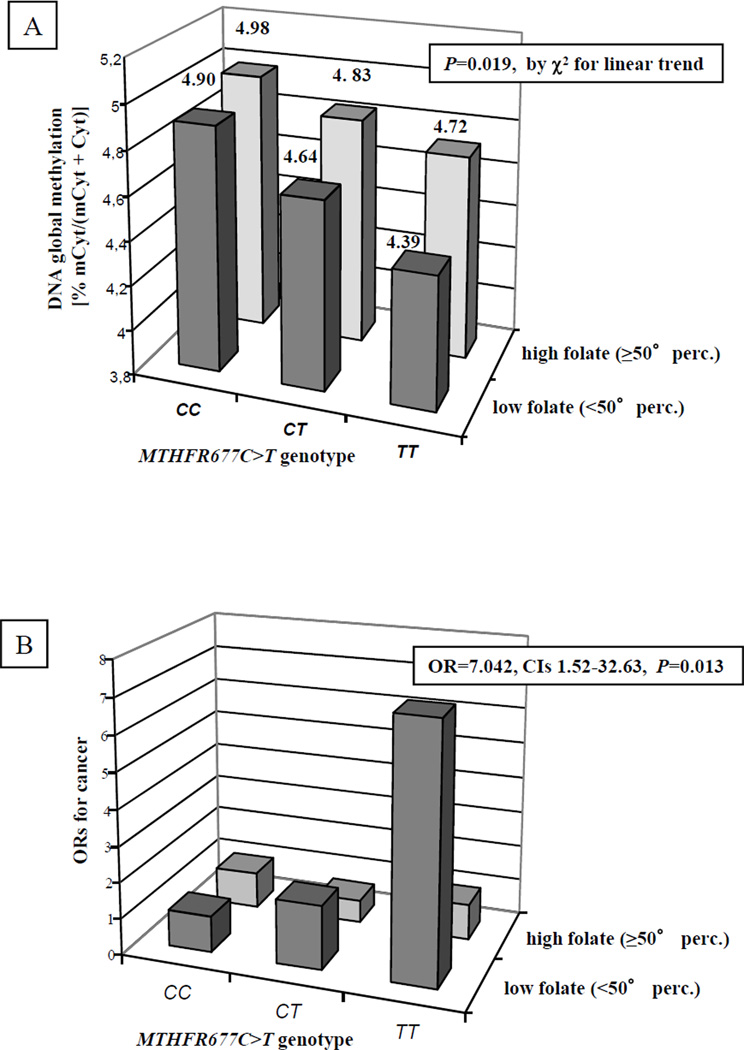
In a gene-nutrient interaction model analysis, to better define the potential significance of the DNA methylation cutoff value, the combined effect of MTHFR677C>T genotype and plasma folate concentrations in determining DNA methylation levels and in relation with cancer history was explored (Figure 4). No statistically significant interaction term was found (for DNA methylation P=0.718; for cancer history P=0.201). However, the data were indicative for an additive effect between MTHFR677C>T genotype and plasma folate concentrations in determining DNA methylation, with the subgroup of MTHFR677TT homozygous carrier associated with low plasma folate concentrations having the lowest DNA methylation levels (4.39% mCyt/mCt+Cyt for MTHFR677TT with low folate) (Figure 4, panel A). The reduction in DNA methylation levels associated with MTHFR677T allele was significant only in those subjects with low plasma folate concentrations (P=0.019 by ANOVA with polynomial contrast for linear trend), while among those with high plasma folate concentrations there was no statistically significant association (P=0.181 by ANOVA with polynomial contrast for linear trend). Consistently, the subgroup of MTHFR677TT homozygous carrier associated with low plasma folate concentrations showed also the highest prevalence of subjects with cancer history (81.2% TT vs. 52.2% CT vs. 38.1% CC, P=0.011, by χ2 for linear trend). The association between MTHFR677C>T genotype and cancer was significant only in subjects with low plasma folate concentrations (P=0.011 by χ2 for linear trend) and not among those with high plasma folate concentrations (P=0.761 by χ2 for linear trend). Considering MTHFR677CC subjects with high folate concentrations as reference group (OR=1), only MTHFR677TT subjects with low folate concentrations showed a higher risk of cancer (OR=7.04, CIs 1.52–32.63, P=0.013), while no significant association was observed for MTHFR677TT subjects with high folate (OR=1.77, CIs 0.53–5.89, P=0.35) (Figure 4, panel B).
Figure 4.
Analysis of the combined effect between plasma folate concentrations and the MTHFR677C>T genotype in determining the levels of DNA methylation [indicated as % mCyt/mCyt+Cyt)] (panel A). Plasma folate concentrations were considered low or high (when <50° or ≥50° percentile within the control group, respectively).
The reduction of DNA methylation levels associated with the MTHFR677TT genotype was significant only in subjects with low plasma folate concentrations (P=0.019 by ANOVA with polynomial contrast for linear trend), while not significant in those with high plasma folate concentrations.
Panel B shows the association between global DNA methylation according to MTHFR677C>T genotypes and plasma folate concentrations and cancer risk by means of Odds ratio (ORs). The analysis was performed by considering as the reference group MTHFR677CC either with low (<50° percentile) or high (≥50° percentile) folate concentrations with OR=1, in the group of combined MTHFR677TT and low folate concentrations the OR for cancer was 7.042, CIs 1.52–32.63, P=0.013. No significant association was observed for the MTHFR677TT and high folate group (OR=1.773, CIs 0.53–5.89, P=0.35).
Analysis of incident cancer cases during the follow-up
The main clinical and biochemical characteristics of cancer patients during follow-up as compared to controls are described in Table 2. Controls and incident cancer patients were age- and sex-matched and with similar percentage of subjects with smoking habit. As shown, DNA methylation at time of enrollment was lower in incident cancer patients as compared to controls [4.47% (95%CIs 3.92–5.12) vs. 5.07% (95%CIs 4.95–5.18), P=0.001]. Among those analyzed for DNA methylation, ten subjects died during the follow-up period: four of them died of cancer and six of other causes, mainly of cardiovascular disease. Those who died of cancer had a lower global DNA methylation compared with those who died of other causes [4.17% (95%CIs 3.48–4.96) vs. 4.89 (95%CIs 4.57–5.19), P=0.015]. The survivors had overall a significantly higher methylation compared to those who died of cancer [4.93% (95%CIs 4.79–5.01) vs. 4.17 (95%CIs 3.48–4.96), P=0.004] but not to those who died of other causes [4.93% (95%CIs 4.79–5.01) vs. 4.89% (95%CIs 4.57–5.19), P=0.79]. A further analysis was carried out, in order to determine a precise cutoff to differentiate subjects who did or did not developed cancer during follow-up, a ROC curve analysis was performed, on which basis a threshold level of 4.69% for DNA methylation was chosen (AUC = 0.834 with 95%CIs 0.714–0.954). Subjects with DNA methylation lower than 4.69% showed a much increased occurrence of incident cancer than those with higher (≥4.69%) DNA methylation levels (66.7% vs. 5.3%, P<0.001 by χ2-test; OR 36.0 with 95%CI 3.9–329.9). Such association persisted statistically significant even after adjustments for sex, age, smoking habit, serum concentrations of inflammatory marker hs-CRP as well as plasma folate, vitamin B12 and tHcy concentrations (OR 43.6 with 95%CIs 2.7–708.4).
Table 2.
Characteristics of the study subjects and control cases at follow-up.
| Variables | Controls (n=58) |
Incident cancers (n=62) |
P-value |
|---|---|---|---|
| age (years) | 60.35±10.17 | 62.52±8.98 | N.S. |
| sex (% Males) | 80.5 | 87.0 | N.S.* |
| smoking habit (% smokers) | 65.9 | 72.7 | N.S.* |
| hs-CRP (mg/L) | 3.30 (2.87–3.80) |
6.15 (2.35–16.10) |
0.047 |
| tHcy (µmol/L) | 15.71 (13.84–17.84) |
14.47 (10.65–19.63) |
N.S. |
| vitamin B12 (pmol/L) | 413.55 (346.40–493.73) |
516.56 (228.86–1165.96) |
N.S. |
| folate (nmol/L) | 5.58 (4.85–6.44) |
6.08 (2.85–17.98) |
N.S. |
| % MTHFR677TT genotype carriers |
16.0 | 15.8 | N.S.* |
| DNA Methylation (% mCyt/mCyt + Cyt) |
5.07 (4.95–5.18) |
4.47 (3.92–5.12) |
0.001 |
Values are expressed as means ± SD for age. Plasma folate, hs-CRP, vitamin B12, tHcy and DNA methylation status are presented as geometric means (antilogaritms of transformed means) and 95% confidence intervals are reported in parentheses with two-tailed P values. Statistical difference was evaluated by Student’s t-test except when differently indicated.
Statistical difference was evaluated by χ2 test. N.S., not statistically significant.
tHcy: total plasma homocysteine; hs-CRP: high-sensitivity C-reactive protein.
Discussion
DNA methylation has been extensively studied as an epigenetic mechanism for carcinogenesis as well as a potential biomarker for cancer disease. However, decades of study could not clarify the mechanistic and clinical significance of DNA methylation due to its diverse biological functions as well as its unique tissue specificity. In the present study we enlightened the value of DNA methylation as a clinical index to screen patients affected by cancer or at risk to develop cancer disease. Interestingly, global DNA methylation status of cancer subjects was invariably decreased compared with control group subjects, indicating that cancer subjects have a systemically decreased global DNA methylation. Furthermore, we also observed that global DNA methylation in PBMCs at the enrollment was lower in subject who developed cancer during the eight year follow-up period compared to those who did not develop cancer during the same period.
Our observations are consistent with previous studies which investigated the relationship between peripheral blood leukocytes DNA methylation and the risk of cancer (24). In a case control study, leukocyte DNA hypomethylation was associated with increased risk for adenoma (P for trend=0.01) and a non-significantly increased risk for cancer (P for trend=0.08). In that study, colorectal cancer subjects also had significantly lower blood folate (16). In the Colorectal Neoplasia Screening With Colonoscopy in Asymptomatic Women at Regional Navy/Army Medical Centers (CONCeRN) Study subjects, women in the second (OR, 0.72; 95%CIs 0.34–1.52) and third tertiles (OR, 0.17; 95% CIs 0.06–0.49) had lower risk of colorectal adenoma, which is a precursor of colorectal cancer, compared with women in the lowest tertile of leukocyte genomic DNA methylation (P for trend=0.002). Several other studies in which the overall white blood cell genomic DNA methylation was measured as a marker of different cancer types including bladder (25–27), stomach (17), breast (28), and head and neck cancer (18), have found an elevated risk for cancer among those in the lower quantile of genomic DNA methylation compared with those in the highest quantile, regardless of the methods to measure global DNA methylation.
Owing to the LC/MS DNA methylation assay which can precisely measure DNA methylation status, it was possible to observe cutoff values, interestingly very similar in both prevalent and incident cancer study design (4.74% and 4.69% global DNA methylation, respectively), that clearly differentiate controls from cancer subjects by DNA methylation status, indicating that this values might be considered as a clinically valuable marker for cancer screening. Our study also indicates that a possible cause of conflict results from previous studies that determined the efficacy of global DNA methylation in PBMCs as a cancer biomarker might be the methods utilized to measure DNA methylation which not always have adequate sensitivity and reproducibility.
Even though we found a significantly lowered genomic DNA methylation status in cancer patients, it is not straightforward to explain why patients who have cancer originated from different types of organs have a decreased DNA methylation status in their PBMCs, especially because epigenetic patterns are usually considered to be highly tissue specific. Furthermore, PBMCs are normal cells that are expected to maintain normal DNA methylation status compared with cancer cells which, instead, tend to have decreased global DNA methylation. We, however, may speculate about the implication of several mechanisms by which cancer subjects have decreased global DNA methylation in PBMCs; 1) cancer developed from a certain organ may alter the whole body metabolism such as energy metabolism or one-carbon metabolism that mediates methyl transfer reactions even for maintenance of a stable DNA methylation status. The global metabolic unbalance as well as that of DNA methylation reaction in cancer may subsequently induce a reduction in global DNA methylation of PBMCs, 2) circulating cancer cells or cancer products such as microRNAs or inflammatory mediators directly affect the PBMCs DNA methylation in blood, and 3) the systemic condition that accelerates cancer development concurrently reduces DNA methylation in PBMCs.
The results of this study support the hypothesis that decreased global DNA methylation status rather indicates a systemic condition prone to cancer development instead of a direct cancer effect, because: 1) in our study, global DNA methylation in PBMCs was already decreased before developing cancer and 2) findings from this study show a combined effect between MTHFR677C>T genotype and plasma folate concentrations, both of which have been regarded as cancer risk factors, for DNA methylation levels and cancer risk at the same time: if associated with low plasma folate, the MTHFR677TT homozygous mutant genotype appears to be crucial in determining the lowest DNA methylation levels and the highest risk of cancer.
Since DNA methylation is a predictor for the development of cancer, it appears that lowered DNA methylation in PBMCs imply that this epigenetic feature collectively reflects the systemic condition that can favour the development of cancer. In this regard, it would be of high interest to follow-up the patients for a longer period and to enlarge the population sample set to be able to evaluate the drop in global methylation levels according to different types of cancer as well as to timing of cancer development. Further studies are warranted in this view.
Among the most interesting findings of the present study, there was also the observation of a decreased DNA methylation status related with lower folate status and higher frequency of MTHFR677TT genotype. Furthermore, MTHFR677TT variants further increased the risk only in those subjects with low folate status (OR for cancer of 7.042, CIs 1.52–32.63, P=0.013). These observations are consistent with previous reports describing that higher cancer risk associated to the presence of MTHFR677TT genotype only for those subjects with low folate status (29) but the link with reduced methyl availability due to lower plasma folate concentrations was previously only hypothesized (29) but confirmed by the present data in which a potential mechanism for this relationship is brought into play by the correlation with lower DNA methylation status.
In conclusion, the low global DNA methylation levels in PBMCs may be a predictor of cancer. The low global DNA methylation can also predict the development of cancer in the near future. Measurements of plasma folate concentrations and genotyping for MTHFR677C>T polymorphism may give an additional impact in subjects with low DNA methylation status. In the future, larger prospective clinical studies are certainly warranted. Whether differences in DNA methylation are related to disease status or outcomes as well as the understanding of precise correlation between PBMCs and specific cancer tissue DNA methylation also need to be determined.
Acknowledgements
We are deeply thankful to Mrs. Maria Zoppi for her invaluable help for the secretariat work and Mr. Diego Minguzzi for his excellent technical assistance.
Grant support
This material is based upon work supported by the U.S. Department of Agriculture, under agreement No. 51000-074-01S. Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Dept of Agriculture. This project has been supported in part by the National Institute of Health Grants R01 AG025834 (SWC) and in part by Cariverona Foundation, Italy (SF).
Footnotes
Potential Conflicts of Interest: The authors declare no conflicts of interest related to this work.
References
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 3.Laird PW, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 4.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 5.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 6.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 7.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortez CC, Jones PA. Chromatin, cancer and drug therapies. Mutat Res. 2008;647:44–51. doi: 10.1016/j.mrfmmm.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friso S, Choi SW. Gene-nutrient interactions and DNA methylation. J Nutr. 2002;132:2382S–2387S. doi: 10.1093/jn/132.8.2382S. [DOI] [PubMed] [Google Scholar]
- 11.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- 13.Sohn KJ, Jang H, Campan M, Weisenberger DJ, Dickhout J, Wang YC, et al. The methylenetetrahydrofolate reductase C677T mutation induces cell-specific changes in genomic DNA methylation and uracil misincorporation: a possible molecular basis for the site-specific cancer risk modification. Int J Cancer. 2009;124:1999–2005. doi: 10.1002/ijc.24003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HI, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13:2378–2384. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 15.Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47–55. doi: 10.1053/j.gastro.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, et al. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–1248. doi: 10.1016/s0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 17.Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–1874. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 19.Girelli D, Friso S, Trabetti E, Olivieri O, Russo C, Pessotto R, et al. Methylenetetrahydrofolate reductase C677T mutation, plasma homocysteine, and folate in subjects from northern Italy with or without angiographically documented severe coronary atherosclerotic disease: evidence for an important genetic-environmental interaction. Blood. 1998;91:4158–4163. [PubMed] [Google Scholar]
- 20.Girelli D, Russo C, Ferraresi P, Olivieri O, Pinotti M, Friso S, et al. Polymorphisms in the factor VII gene and the risk of myocardial infarction in patients with coronary artery disease. N Engl J Med. 2000;343:774–780. doi: 10.1056/NEJM200009143431104. [DOI] [PubMed] [Google Scholar]
- 21.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74:4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 22.Friso S, Girelli D, Martinelli N, Olivieri O, Lotto V, Bozzini C, et al. Low plasma vitamin B-6 concentrations and modulation of coronary artery disease risk. Am J Clin Nutr. 2004;79:992–998. doi: 10.1093/ajcn/79.6.992. [DOI] [PubMed] [Google Scholar]
- 23.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nature Genetics. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 24.Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One. 2012;7:e34615. doi: 10.1371/journal.pone.0034615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cash HL, Tao L, Yuan JM, Marsit CJ, Houseman EA, Xiang YB, et al. LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int J Cancer. 2012;130:1151–1159. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16:1682–1689. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]