Abstract
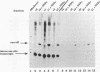
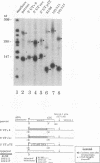
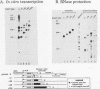
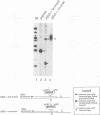
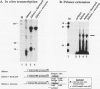
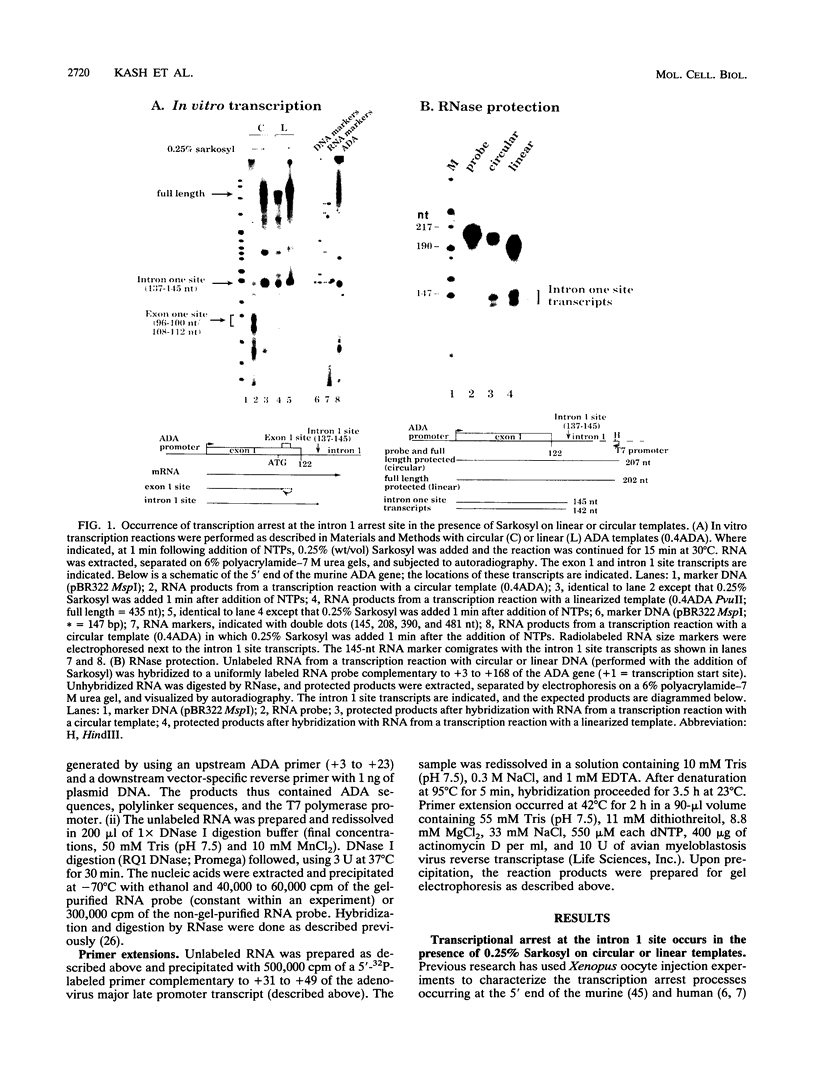
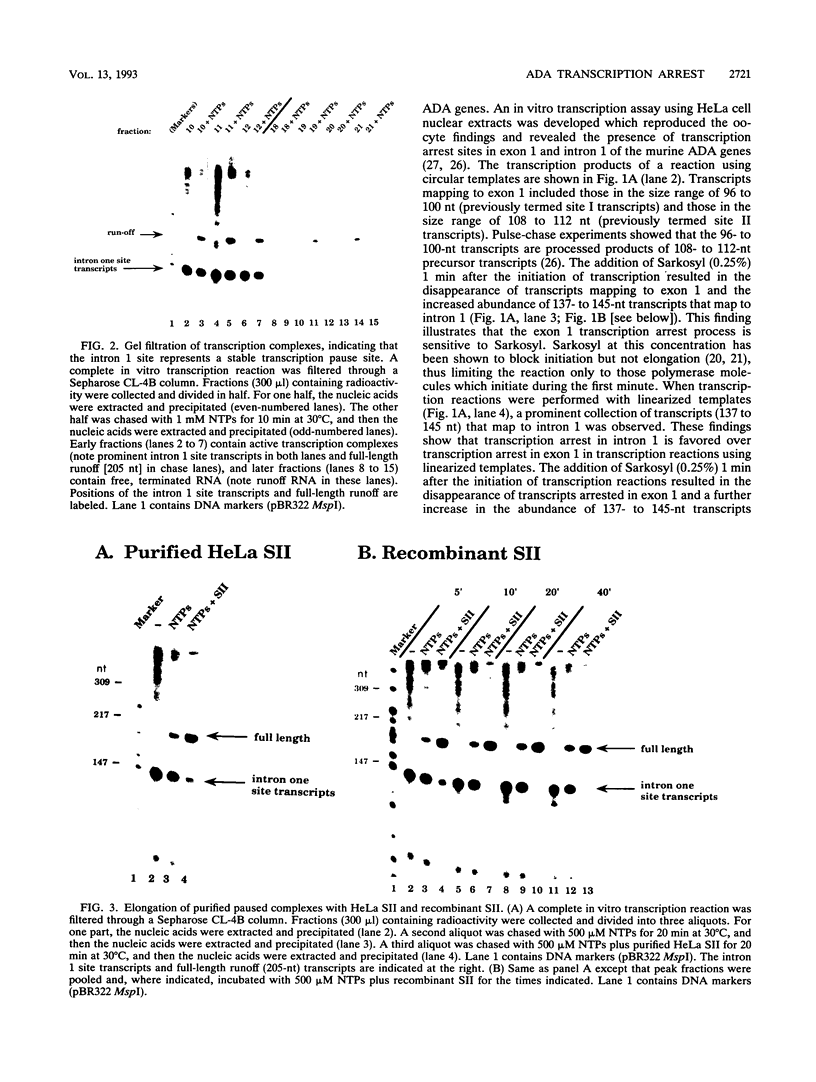
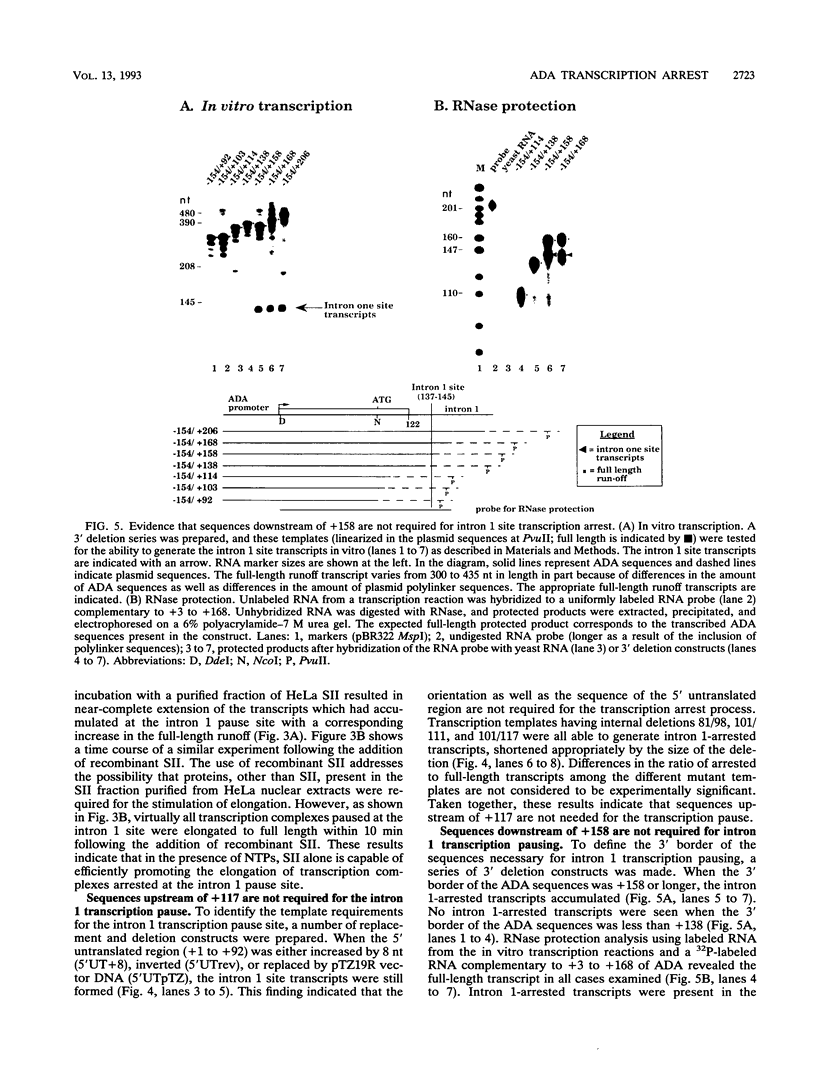
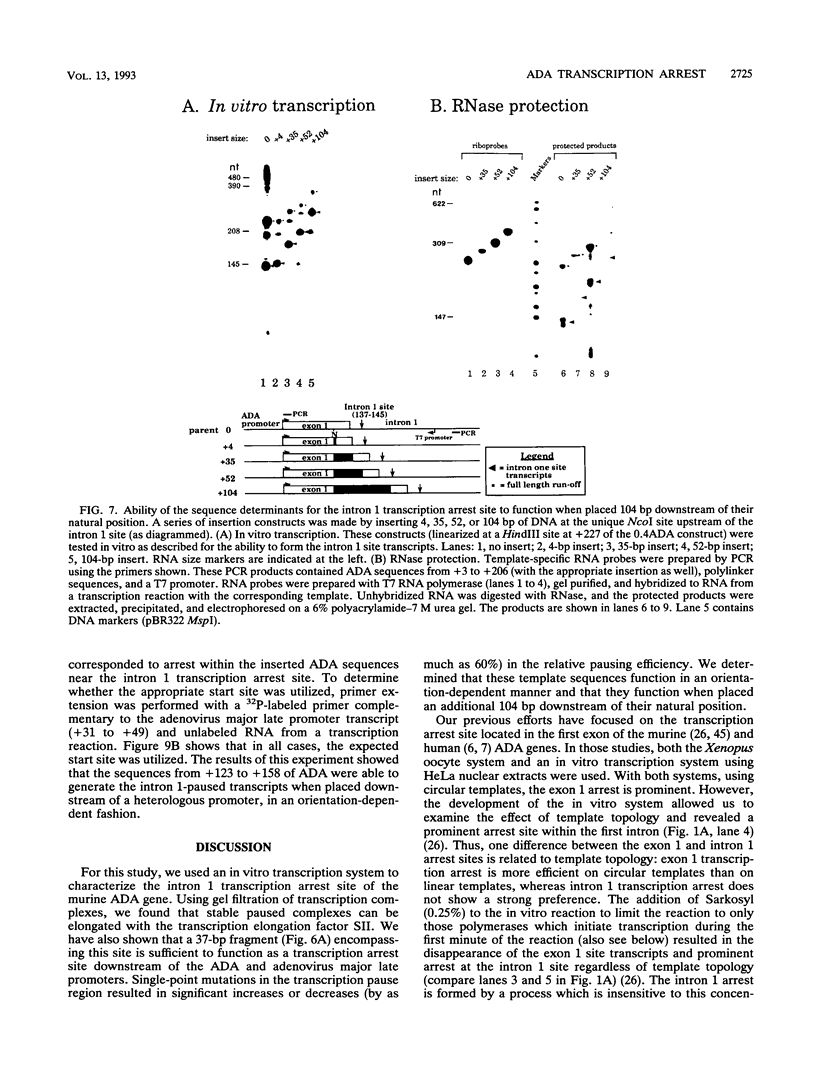
Transcription arrest plays a role in regulating the expression of a number of genes, including the murine adenosine deaminase (ADA) gene. We have previously identified two prominent arrest sites at the 5' end of the ADA gene: one in the first exon and one in the first intron (J. W. Innis and R. E. Kellems, Mol. Cell. Biol. 11:5398-5409, 1991). Here we report the functional characterization of the intron 1 arrest site, located 137 to 145 nucleotides downstream of the cap site. We have determined, using gel filtration, that the intron 1 arrest site is a stable RNA polymerase II pause site and that the transcription elongation factor SII promotes read-through at this site. Additionally, the sequence determinants for the pause are located within a 37-bp fragment encompassing this site (+123 to +158) and can direct transcription arrest in an orientation-dependent manner in the context of the ADA and adenovirus major late promoters. Specific point mutations in this region increase or decrease the relative pausing efficiency. We also show that the sequence determinants for transcription arrest can function when placed an additional 104 bp downstream of their natural position.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Asher E., Aloni Y. Transcription of minute virus of mice, an autonomous parvovirus, may be regulated by attenuation. J Virol. 1984 Oct;52(1):266–276. doi: 10.1128/jvi.52.1.266-276.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bengal E., Flores O., Krauskopf A., Reinberg D., Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991 Mar;11(3):1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Chen Z., Harless M. L., Wright D. A., Kellems R. E. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990 Sep;10(9):4555–4564. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Innis J. W., Sun M. H., Wright D. A., Kellems R. E. Sequence requirements for transcriptional arrest in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1991 Dec;11(12):6248–6256. doi: 10.1128/mcb.11.12.6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsky J. M., Maa M. C., Ramamurthy V., Kellems R. E. Adenosine deaminase gene expression. Tissue-dependent regulation of transcriptional elongation. J Biol Chem. 1989 Aug 25;264(24):14561–14565. [PubMed] [Google Scholar]
- Chomyn A., Martinuzzi A., Yoneda M., Daga A., Hurko O., Johns D., Lai S. T., Nonaka I., Angelini C., Attardi G. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol Cell Biol. 1989 Nov;9(11):5254–5259. doi: 10.1128/mcb.9.11.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O., Ha I., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990 Apr 5;265(10):5629–5634. [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring N. B., Kessler M., Aloni Y. Parameters affecting the elongation block by RNA polymerase II at the SV40 attenuator 1 in vitro. Biochemistry. 1992 Sep 8;31(35):8369–8376. doi: 10.1021/bi00150a034. [DOI] [PubMed] [Google Scholar]
- Grayhack E. J., Yang X. J., Lau L. F., Roberts J. W. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985 Aug;42(1):259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- Haley J. D., Waterfield M. D. Contributory effects of de novo transcription and premature transcript termination in the regulation of human epidermal growth factor receptor proto-oncogene RNA synthesis. J Biol Chem. 1991 Jan 25;266(3):1746–1753. [PubMed] [Google Scholar]
- Hart C. M., Roberts J. W. Rho-dependent transcription termination. Characterization of the requirement for cytidine in the nascent transcript. J Biol Chem. 1991 Dec 15;266(35):24140–24148. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985 Jul 5;260(13):8163–8172. [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation in SV40 as a mechanism of transcription-termination by RNA polymerase B. Nucleic Acids Res. 1984 Feb 10;12(3):1401–1414. doi: 10.1093/nar/12.3.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Parisi M. A., Bennett J. L., Clayton D. A. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991 May 16;351(6323):236–239. doi: 10.1038/351236a0. [DOI] [PubMed] [Google Scholar]
- Innis J. W., Kellems R. E. A heat-labile factor promotes premature 3' end formation in exon 1 of the murine adenosine deaminase gene in a cell-free transcription system. Mol Cell Biol. 1991 Nov;11(11):5398–5409. doi: 10.1128/mcb.11.11.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis J. W., Moore D. J., Kash S. F., Ramamurthy V., Sawadogo M., Kellems R. E. The murine adenosine deaminase promoter requires an atypical TATA box which binds transcription factor IID and transcriptional activity is stimulated by multiple upstream Sp1 binding sites. J Biol Chem. 1991 Nov 15;266(32):21765–21772. [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kephart D. D., Marshall N. F., Price D. H. Stability of Drosophila RNA polymerase II elongation complexes in vitro. Mol Cell Biol. 1992 May;12(5):2067–2077. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf A., Bengal E., Aloni Y. The block to transcription elongation at the minute virus of mice attenuator is regulated by cellular elongation factors. Mol Cell Biol. 1991 Jul;11(7):3515–3521. doi: 10.1128/mcb.11.7.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A., Meulia T., Brunvand M., Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992 Nov;6(11):2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Krystal G., Birrer M., Way J., Nau M., Sausville E., Thompson C., Minna J., Battey J. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb N. J., Fernandez A., Tourkine N., Jeanteur P., Blanchard J. M. Demonstration in living cells of an intragenic negative regulatory element within the rodent c-fos gene. Cell. 1990 May 4;61(3):485–496. doi: 10.1016/0092-8674(90)90530-r. [DOI] [PubMed] [Google Scholar]
- Lattier D. L., States J. C., Hutton J. J., Wiginton D. A. Cell type-specific transcriptional regulation of the human adenosine deaminase gene. Nucleic Acids Res. 1989 Feb 11;17(3):1061–1076. doi: 10.1093/nar/17.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Kraus K. W., Wolfner M. F., Lis J. T. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992 Feb;6(2):284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- London L., Keene R. G., Landick R. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol Cell Biol. 1991 Sep;11(9):4599–4615. doi: 10.1128/mcb.11.9.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maa M. C., Chinsky J. M., Ramamurthy V., Martin B. D., Kellems R. E. Identification of transcription stop sites at the 5' and 3' ends of the murine adenosine deaminase gene. J Biol Chem. 1990 Jul 25;265(21):12513–12519. [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N. F., Price D. H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992 May;12(5):2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Jeanteur P., Lebleu B. Sequence requirements for premature transcription arrest within the first intron of the mouse c-fos gene. Mol Cell Biol. 1991 May;11(5):2832–2841. doi: 10.1128/mcb.11.5.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok M., Maderious A., Chen-Kiang S. Premature termination by human RNA polymerase II occurs temporally in the adenovirus major late transcriptional unit. Mol Cell Biol. 1984 Oct;4(10):2031–2040. doi: 10.1128/mcb.4.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V., Maa M. C., Harless M. L., Wright D. A., Kellems R. E. Sequence requirements for transcriptional arrest in exon 1 of the murine adenosine deaminase gene. Mol Cell Biol. 1990 Apr;10(4):1484–1491. doi: 10.1128/mcb.10.4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport J., Reinberg D., Zandomeni R., Weinmann R. Purification and functional characterization of transcription factor SII from calf thymus. Role in RNA polymerase II elongation. J Biol Chem. 1987 Apr 15;262(11):5227–5232. [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., Roeder R. G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987 Mar 5;262(7):3322–3330. [PubMed] [Google Scholar]
- Reinberg D., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J Biol Chem. 1987 Mar 5;262(7):3331–3337. [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992 Feb 25;267(6):3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Reines D., Wells D., Chamberlin M. J., Kane C. M. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987 Jul 20;196(2):299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- Resnekov O., Aloni Y. RNA polymerase II is capable of pausing and prematurely terminating transcription at a precise location in vivo and in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(1):12–16. doi: 10.1073/pnas.86.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S., Bentley D. L. Distinct modes of transcription read through or terminate at the c-myc attenuator. EMBO J. 1992 Mar;11(3):1085–1093. doi: 10.1002/j.1460-2075.1992.tb05147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990 Nov;10(11):6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Sancar A., Little J. W., Rupp W. D. The uvrB gene of Escherichia coli has both lexA-repressed and lexA-independent promoters. Cell. 1982 Mar;28(3):523–530. doi: 10.1016/0092-8674(82)90207-0. [DOI] [PubMed] [Google Scholar]
- Sekimizu K., Kobayashi N., Mizuno D., Natori S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry. 1976 Nov 16;15(23):5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- Sellitti M. A., Pavco P. A., Steege D. A. lac repressor blocks in vivo transcription of lac control region DNA. Proc Natl Acad Sci U S A. 1987 May;84(10):3199–3203. doi: 10.1073/pnas.84.10.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Kuroda M. I., Yanofsky C., Henner D. J. Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon. J Bacteriol. 1986 May;166(2):461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SivaRaman L., Reines D., Kane C. M. Purified elongation factor SII is sufficient to promote read-through by purified RNA polymerase II at specific termination sites in the human histone H3.3 gene. J Biol Chem. 1990 Aug 25;265(24):14554–14560. [PubMed] [Google Scholar]
- Skarnes W. C., Tessier D. C., Acheson N. H. RNA polymerases stall and/or prematurely terminate nearby both early and late promoters on polyomavirus DNA. J Mol Biol. 1988 Sep 5;203(1):153–171. doi: 10.1016/0022-2836(88)90099-x. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Toohey M. G., Jones K. A. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes Dev. 1989 Mar;3(3):265–282. doi: 10.1101/gad.3.3.265. [DOI] [PubMed] [Google Scholar]
- Watson R. J. Expression of the c-myb and c-myc genes is regulated independently in differentiating mouse erythroleukemia cells by common processes of premature transcription arrest and increased mRNA turnover. Mol Cell Biol. 1988 Sep;8(9):3938–3942. doi: 10.1128/mcb.8.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest D. K., Hawley D. K. In vitro analysis of a transcription termination site for RNA polymerase II. Mol Cell Biol. 1990 Nov;10(11):5782–5795. doi: 10.1128/mcb.10.11.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Transcription attenuation. J Biol Chem. 1988 Jan 15;263(2):609–612. [PubMed] [Google Scholar]