Abstract
Rapid, sensitive, and selective detection of viruses is critical for applications in medical diagnostics, biosecurity, and environmental safety. In this article, we report the application of a point-defect-coupled W1 photonic crystal (PhC) waveguide biosensor to label-free optical detection of viruses. Fabricated on a silicon-on-insulator (SOI) substrate using electron-beam (e-beam) lithography and reactive-ion-etching, the PhC sensing platform allows optical detection based on resonant mode shifts in response to ambient refractive index changes produced by infiltration of target biomaterial within the holes of the PhC structure. Finite difference time domain (FDTD) calculations were performed to assist with design of the sensor, and to serve as a theoretical benchmark against which experimental results could be compared. Using Human Papillomavirus virus-like particles (VLPs) spiked in 10% fetal bovine serum as a model system, we observed a limit of detection of 1.4 nM in simple (buffer only) or complex (10% serum) sample matrices. The use of anti-VLP antibodies specific for intact VLPs with the PhC sensors provided highly selective VLP detection.
Keywords: Biosensor, Photonic Crystal, Virus
1. INTRODUCTION
Ultra-small (micro- or nanoscale) optical sensors are of considerable interest because their small active volumes potentially yield exceptionally high sensitivity, and because of their suitability for integration with microfluidics for “lab on a chip” applications. Within this context, two dimensional photonic crystal (2D PhC) optical cavities have recently emerged as attractive sensing platforms (Pal et al., 2012). 2D PhCs are structures that exhibit an engineered photonic band gap (PBG) due to the fact that the dielectric constant periodically varies spatially in two directions. Optical cavities are created by breaking the spatial translational symmetry (for instance by increasing or decreasing the radius of a central hole in the periodic lattice of air holes), and they support a single localized resonant mode or a set of closely spaced resonant modes that have frequencies within the PBG (Joannopoulos et al., 2008). A 2D PhC optical cavity allows strong localization of the electric field energy at the frequency of the resonant mode, and is characterized by a high quality factor (Q-factor) in the range of 102 to 106 (for Si) and low cavity mode volume of the order of (λ/n)3, where λ represents the optical wavelength, and n is the refractive index (RI). These characteristics make PhC optical cavities more sensitive to environmental changes than optical microcavities in other resonator structures (e.g. ring resonators, microspheres or microtoroids), which have higher Q-factors (>106) but larger mode volumes (Fan et. al, 2008). Small mode volumes such as those provided by PhC optical cavities are particularly desirable for low-copy-number biosensing, because each individual target pathogen fills a larger percent of the active sensing area, thus enhancing the device sensitivity. Thus far, preliminary efforts by our group and others have demonstrated the utility of 2D PhC optical cavities for sensing small molecules such as proteins and peptides (Lee and Fauchet, 2007; Dorfner et al., 2009; Zlatanovic et al., 2009).
We have previously reported a novel PhC design in which a 2D PhC waveguide is coupled to a 2D PhC optical cavity for sensing (Guillermain and Fauchet, 2009; Pal et al., 2010). This design uniquely allows for multiple PhC cavities to be placed in series, opening opportunities for redundant (error-corrected) sensing as well as potentially leading towards multiplexed sensors with ultra-small dimensions. We used this sensor design to demonstrate sensitive detection of protein-antibody interactions in a simple buffered solution. In this paper, we demonstrate for the first time that 2D PhC optical cavities can serve as effective nanoscale sensors for model viral pathogens, and that selective detection with low nonspecific binding is achievable in serum.
We have used human papillomavirus (HPV) as a model pathogen for our experiments. Persistent infection by HPV has been established as a cause of almost all cases of cervical cancer worldwide, with HPV16 and HPV18 accounting for the majority of the cases (Walboomers et al., 1999; Garland and Smith, 2010). Thus, detection of HPV may be a valuable tool to complement cervical cytology in regular cervical screening for the early detection of cancer (Walboomers et al., 1999). The outer viral coat of HPV consists of 72 pentamers of the structural protein L1 arranged in a skewed icosahedral lattice, which encapsulates the viral DNA along with the other structural protein L2 (Yuan et al., 1998; Chen et al., 2000). Recombinant expression of L1 protein results in the assembly of virus-like nanoparticles (VLPs), which are ~55 nm diameter structures morphologically and immunologically similar to the viral capsid (Rose et al., 1993; Yuan et al., 1998). Because they are noninfectious and therefore do not require any specialized handling, VLPs served as an authentic but harmless model system for testing our sensor’s capability of detecting virus-sized nanoparticles.
2. EXPERIMENTAL
Materials
(3-aminopropyl) dimethylethoxysilane (APDMES) and glutaraldehyde (50% aqueous solution) were purchased from Gelest, Inc. and TCI America, respectively. HPV16 virus-like nanoparticles and mouse ascites fluid containing a monoclonal antibody specific to HPV16 VLPs were generously provided by Prof. Robert C. Rose (University of Rochester). HPV16 L1 (CamVir 1) antibody was purchased from Genetex, Inc (Catalog no. GTX20069). Fetal bovine serum used in experiments was purchased from Innovative Research, Inc.
2.2 Device Fabrication
A 130 nm thick oxide hard mask layer was first thermally grown on the SOI wafers by a wet oxidation process as an intermediate layer for pattern transfer. An e-beam sensitive resist, polymethylmethacrylate (PMMA), was spin coated on the oxidized substrate and the PhC patterns were written using a JEOL JBX-9300FS e-beam system. The PhC patterns were then developed and the oxide hard mask layer was dry etched using argon assisted CHF3 gas in a reactive-ion-etcher to transfer the patterns to the hard mask. The patterns were then transferred to the device layer by etching the silicon with a gas mixture of CF4 and BCl3. The PhC device edges were either diced using a dicing saw, then polished, or cleaved to create smooth waveguide facets so that light could be coupled from an external source. Each fabricated waveguide had one to three waveguide coupled PhCs placed in series.
2.3 Optical Detection
A tunable laser source (Hewlett Packard, model 8168F, output power: −7 to 7 dBm) in the wavelength range of 1440 to 1590 nm (wavelength resolution: 0.001 nm) was used to illuminate the PhC waveguide sensors. Light from the laser source was polarized using a polarization controller and coupled into and out from the device’s waveguides through tapered, lensed fibers (Nanonics, Israel). The optical power transmitted through the device was measured either using an Indium Gallium Arsenide (InGaAs) photodiode detector (Teledyne Judson Technologies, PA, USA) or a Thorlabs DET410 InGaAs detector (Thorlabs, Inc.). A LABVIEW (National Instruments, TX, USA) script was used to acquire data at a wavelength resolution of 0.2 nm.
2.4 Sensor Surface-Functionalization
The oxidized chips were washed in piranha solution (3:1 (v/v) conc. sulfuric acid to 30% hydrogen peroxide; CAUTION: piranha solution reacts vigorously with organic materials and must be used with care) for 30 minutes, followed by thorough rinsing with ddH2O and drying under a stream of nitrogen. The chips were then incubated with a 1% (v/v) solution of (3-aminopropyl) dimethylethoxysilane (APDMES) in anhydrous toluene for 20 minutes on an orbital shaker. The chips were then repeatedly washed with anhydrous toluene, dried under a stream of nitrogen and baked at 110 °C for 30 minutes. After the chips had cooled to room temperature (approximately 5 minutes), a solution of 1.25% (v/v) glutaraldehyde in modified PBS buffer (MPBS: 10 mM NaH2PO4, 10 mM Na2HPO4, 150 mM NaCl at pH 7.2) was poured over them, and the chips were left in this solution on a shaker for 60 minutes. Afterwards, they were washed with MPBS and ddH2O and dried under a nitrogen stream. The aldehyde-functionalized chips were then subjected to antibody immobilization via amine-aldehyde coupling chemistry (Schiff base formation). The antibody of interest was spotted on the chip as a small droplet of 5 or 10 μL volume covering the photonic crystal, and allowed to immobilize at a high humidity for 60 minutes. This was done to minimize the amount of antibodies localized to planar insensitive areas which could compete with antibodies inside the photonic crystal for binding of VLPs. After antibody immobilization, the remaining aldehyde groups were blocked by incubating the chips in 0.2 mg/mL BSA solution in HBS buffer (20 mM HEPES, 150 mM NaCl, at pH 7.2) for 60 minutes. Chips used for VLP detection in buffer and in a serum background were treated identically up to the blocking step. Post-blocking, the chips used for VLP detection in buffer were washed with MPBS and incubated overnight with 5 or 10 μL of target solutions containing VLPs diluted in MPBS-0.5 buffer (MPBS-0.5: MPBS containing 0.5 M NaCl). After VLP incubation, the chips were rinsed with water and dried under nitrogen before their transmission spectra were measured. The chips used for VLP detection in serum were equilibrated with MPBS-ET-0.05 buffer (MPBS buffer, supplemented with 3 mM EDTA and 0.05% Tween-20) for 15 minutes after the blocking step, to help prevent non-specific binding of serum proteins. The chips were then incubated overnight with 5 or 10 μL of target solutions containing VLPs diluted in MPBS-0.5-ET-0.05 (MPBS containing 0.5 M NaCl, 3 mM EDTA and 0.05% Tween-20) supplemented with 10% fetal bovine serum. After VLP incubation, the chips were washed with MPBS-0.5-ET-0.05 for 15 minutes on an orbital shaker, rinsed with water, dried with nitrogen, and characterized.
3. RESULTS AND DISCUSSION
3.1 PhC Sensing Mechanism
The sensor consists of a 2D PhC silicon slab structure with a triangular lattice of air holes. A linear defect is created in the PhC by removing a single array of central air holes, resulting in a W1 waveguide that allows modes to be guided through the crystal within its photonic band gap (PBG) (Olivier, et al., 2001). Light propagation in the waveguide is confined by the PBG in the plane of periodicity and by index guiding in the direction perpendicular to the silicon slab (Liu et al., 2009). A spatial point-defect formed by modifying the radius of a single air hole next to the W1 PhC waveguide introduces a defect state in the PBG of the crystal. Hence, light is transmitted through the PhC waveguide at all wavelengths within the PBG except at the resonant wavelength of the confined cavity mode, where a dip is observed in the output transmission spectrum. By tuning the point-defect hole radius and placing the PhC structures in series, multiple transmission dips can be obtained in the output spectrum as the optical cavity mode within each PhC possesses a distinct resonant wavelength. The operating wavelength of the PhC structure is near 1.5 μm, at which silicon is transparent. Figure 1a shows an SEM image of a fabricated sensor where two such defect-coupled structures are placed adjacent to each other. When target biomaterial binds inside the optical cavity region of the PhC, the local refractive index inside the cavity changes, causing a red-shift in the resonant wavelength. The strong electric field localization in the vicinity of the spatial defect (Figure 1b; simulation parameters described below) suggests that the structure’s optical resonance is very sensitive to local refractive index changes due to analyte binding.
Figure 1.
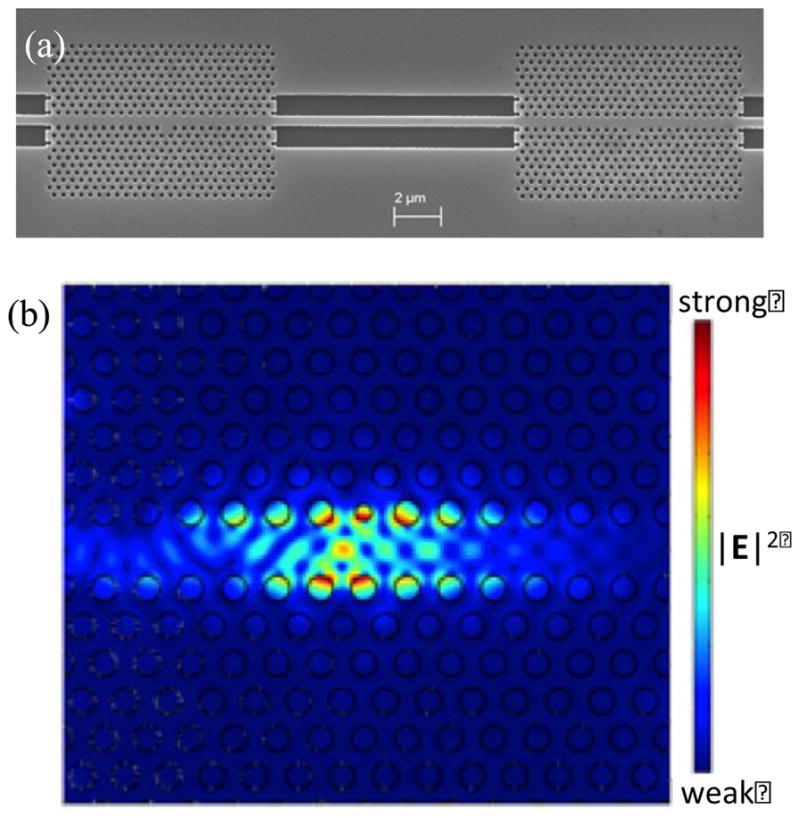
(a) SEM image of a fabricated defect-coupled PhC sensor. (b) 2D simulation results for the |E|2 field profile of the PhC structure for the lowest frequency cavity resonance.
3.2 Nanoparticle Capture Simulations
To interrogate the effect of VLP infiltration into the holes of the PhC, we simulated the transmission spectra for a PhC waveguide structure in the presence of VLP-sized nanoparticles. The simulations were performed for a 2D geometry using a freely available finite-difference-time-domain (FDTD) software package (MEEP, for spectrum computation [Prather et al., 2009; Ardavan et al., 2010]) and a commercially available finite-element method software package (COMSOL, to simulate steady-state electric field profiles). In the simulations, the defect hole had a radius of 0.2a (a = lattice constant), and the surrounding air hole radii were fixed at 0.3a. An effective index method has been shown to yield reasonably accurate results when using 2D calculations to simulate the behaviors of a slab photonic crystal structure (Painter et al., 1999). Therefore, an effective RI of 3.18 was used for silicon, while the expected RI of 1.0 was used for air. At all interfaces between silicon and air, a thin layer (corresponding to 14 nm) with a RI of 1.45 was created to represent the thermally grown oxide and surface functionalization.
The VLPs used in our experiments consist of only the outer viral coat of the HPV virus, which can be approximated by a hollow sphere with an outer radius of 27.5 nm an inner radius of 17.5 nm. This model is based on the X-ray crystal structure of the L1 protein (that comprises the outer viral coat), in which the longest dimension of the protein was found to be 10 nm (Bishop et al., 2007; Mace et al., 2009). The RI of the outer viral coat was assumed to be 1.54 (similar to the RI of adsorbed protein layers), and the RI of the interior of the VLP was assumed to be 1.32 (similar to buffer) (Prime and Whitesides, 1991; Harvey et al., 1998). By taking a volume weighted sum of these refractive indices, an effective RI of 1.45 was calculated and used for the nanoparticles in the simulations. The nanoparticle diameter was assumed to be 0.145a (corresponding to a diameter of 55 nm for a=380 nm).
The simulation results presented in Figure 2a show how the presence of nanoparticles (NP) within holes of the PhC structure causes the resonant dip in the transmission spectrum to shift to a longer wavelength. The baseline scenario (zero red-shift) was the case in which no nanoparticles are present. For the other four simulated scenarios, the defect hole was infiltrated with a single nanoparticle at its center and the non-defect air holes were each infiltrated with a specified number of randomly placed nanoparticles. A red-shift of 0.26 nm in the resonant wavelength was calculated for a single nanoparticle centered in the defect hole, suggesting that this platform has the capability to detect only a single VLP if it enters the defect hole. The simulated resonance wavelength red-shift increased with an increasing number of nanoparticles present in the non-defect holes of the PhC structure. For each additional nanoparticle infiltrating every well, there was a red-shift of about 2.1 nm. This series of simulations suggested that the PhC transmission spectrum is sensitive to infiltration of nanoparticles in holes throughout the PhC. However, the largest response was produced by particle infiltration into the PhC defect hole.
Figure 2.
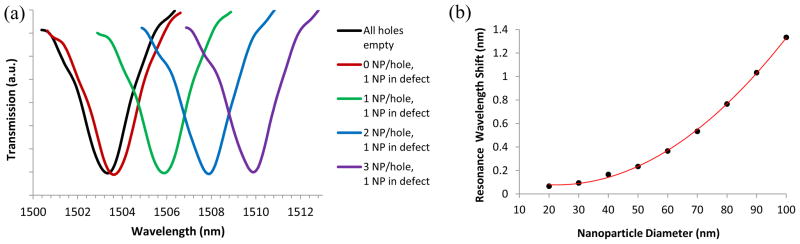
(a) Simulated resonance dips in transmission spectra in response to various nanoparticle (NP) infiltration arrangements in the defect hole and surrounding non-defect holes of the PhC waveguide structure. (b) Effect of nanoparticle diameter on the resonance wavelength shift that results from infiltration of a single nanoparticle within the defect cavity. Points denote the simulated red-shift values, whereas the continuous line denotes a second order polynomial fit (R2= 0.999) for the data.
To determine the effect of viruses of different sizes in the defect hole, we simulated the response of the PhC transmission spectrum to single nanoparticles of different diameters located at the center of the defect hole. A nanoparticle diameter range of 20–100 nm was chosen to encompass particles that are able to readily occupy the 150 nm defect hole diameter used in our current device. Figure 2b shows the effect of nanoparticle diameter on the shift in resonance wavelength. The smallest nanoparticle diameter that showed a red-shift of 0.2 nm (the maximum scan resolution used in our experiments) in the FDTD calculations was 40 nm, thus suggesting that the PhC sensing platform in our experimental set-up is capable of detecting the capture of a single virion of 40 nm size.
FDTD simulations were also performed to verify the independence of the resonance wavelengths for two PhC waveguide structures arranged in series (as in Figure 1a). The PhC geometry described above was used for both of the PhCs, with lattice constants corresponding to 380 nm and 388 nm. Four scenarios were considered: no nanoparticle infiltration, nanoparticle infiltration in the left PhC defect only, nanoparticle infiltration in the right PhC defect only, and nanoparticle infiltration in both PhC structures. Simulation results showed that the transmission dip corresponding to a particular PhC only shifted when that PhC was infiltrated, thus supporting our expectation that serially arranged PhC structures function independently.
The actual PhC structure was fabricated from p-type silicon-on-insulator (SOI) wafers (<100>) having a silicon device thickness of 450 nm and silicon dioxide buried oxide thickness of 1 μm. The structure is an array (25 × 26) of air holes having a compact dimension of 10 μm by 7 μm. The fabrication process involves PhC pattern writing using e-beam lithography and dry reactive-ion-plasma etching (described in detail in the methods section). PhC structures with three different lattice constants of 372, 380 and 388 nm were fabricated. This was accomplished by varying the air hole radii as 111, 114 and 117 nm, while the defect radii were varied as 73, 75 and 77 nm. The spectral properties of the PhC structures are measured by exciting transverse-electric-like modes, as there is no bandgap for transverse-magnetic-like modes for these structures beneath the light cone (Prather et al., 2009). Separate resonant dips were observed in the optical transmission spectrum, corresponding to the unique resonant wavelengths of the fabricated PhCs (see Figure S1a in supplemental information).
3.3 VLP Sensing
For the VLP experiments, sensors having one to three waveguide-coupled PhC structures in series were functionalized with the requisite anti-VLP antibody as described above, then incubated in a solution containing HPV VLPs in a humidity chamber at room temperature for 16 hours. The sensors were then washed with distilled deionized water to remove the unbound targets, and dried in a stream of nitrogen gas prior to performing optical measurements. The optical transmission spectra for a series of three waveguide-coupled PhCs before and after surface functionalization and treatment with a 5.8 nM HPV VLP solution are shown in supplemental information Figure S1a. An SEM image of a PhC sensor after VLP treatment is also shown in supplemental information (Figure S1b).
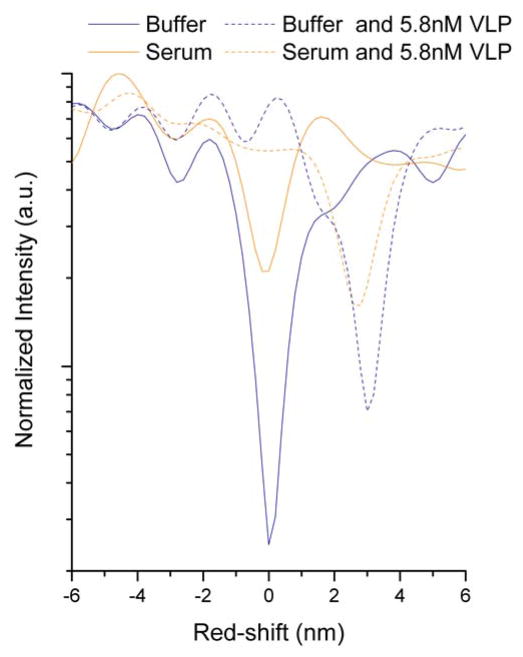
Sensor response in serum indicated that a suitably functionalized 2D PhC is highly capable of rejecting nonspecific binding by components of a complex sample matrix (Figure 3). The total red-shift observed for a sensor exposed to 10% fetal bovine serum showed a similar shift to that of a sensor exposed to buffer. Incubating sensors with 5.8 nM VLP in either buffer or 10% fetal bovine serum resulted in red-shifts that were significantly larger than their respective controls, and of similar magnitude regardless of sample matrix.
Figure 3.
Spectra obtained for individual 2D PhC sensor chips incubated with serum (orange) or buffer (blue) either with (dashed line) or without VLPs (solid line). The red-shifts for all spectra are normalized to the buffer control (shown at 0 red-shift).
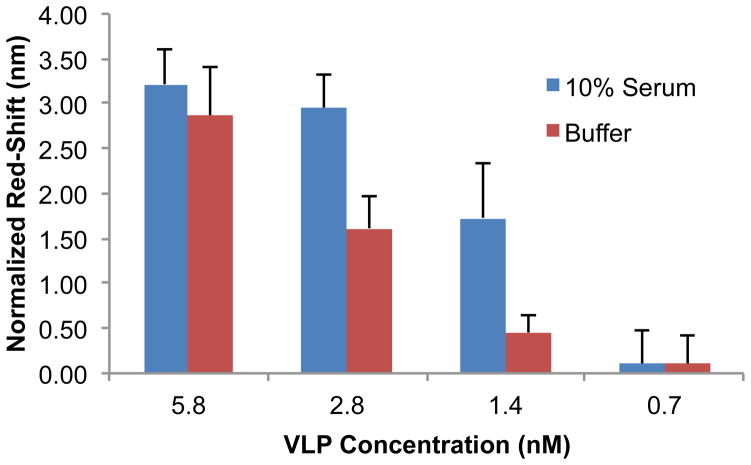
The dose-dependent response of the PhC sensors was evaluated independently in buffer and 10% fetal bovine serum for VLP concentrations ranging from 0.7 to 5.8 nM (Figure 4). The normalized red-shift values were calculated by subtracting the sensor response for the control (antibody functionalized PhC sensors treated with buffer or 10% serum) from the sensor response at different VLP concentrations. The reported error bars are the square root of the sum of squares of the control sensors’ standard deviation and the experimental sensors’ standard deviation. The sensor response in both buffer and serum was found to rise with increasing VLP concentration. The normalized red-shifts were 2.9 ± 0.5 nm and 0.1 ± 0.3 nm for VLP concentrations of 5.8 nM and 0.7 nM in buffer, respectively. The normalized sensor response in 10% fetal bovine serum was (3.2 ± 0.4) nm and (0.1 ± 0.4) nm for VLP concentrations of 5.8 nM and 0.7 nM, respectively. The reported data represent experimental results repeated on separate days, with the total number of sensors tested for each concentration in the range of 3 to 9. Observed variation in the sensor response is likely the result of fabrication artifacts leading to variations in Q factors of the PhCs, as well as variations in VLP capture at the sensitive defect and less–sensitive non-defect wells. The sensitivity of the PhC sensor for HPV-VLP sensing was calculated from the expression S = Δλ/Cmax (where Cmax is the maximum tested VLP concentration), and was found to be 0.1 nm/nM in both buffer and serum. The lowest VLP concentration detectable on our PhC device was determined to be 1.4 nM for both buffer and 10% serum backgrounds by statistically analyzing the red-shift value with respect to the control sensor (two-tailed unequal variance t-test yielded a P-value ≤ 0.05, suggesting that the red-shift for this concentration was statistically significant with respect to the control).
Figure 4.
Experimental normalized red-shifts with the PhC sensors in buffer and 10% fetal bovine serum for VLP concentrations ranging from 5.8 to 0.7 nM. The normalized red-shifts were calculated by subtracting a control shift (no target) from each experimental concentration. The error bars represent the root-sum-of-squares of the standard deviations of the red-shifts for the target and control chips.
3.4 Selectivity Results
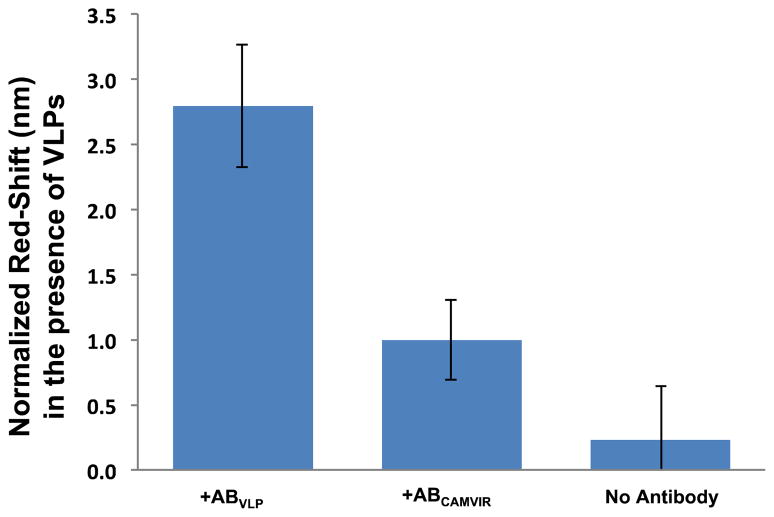
To examine selectivity, we compared the VLP-dependent response of the PhC sensor for devices functionalized with VLP-specific antibodies, with antibodies capable of binding HPV16 L1 protein but not intact VLPs (CamVir 1), and with no antibody (bovine serum albumin blocked). A VLP concentration of 5.8 nM was employed in these experiments to ensure stringent control experimental conditions. As shown in Figure 5, the response of the sensor lacking antibodies was negligible with respect to the control and smaller than the red-shift observed with the VLP-specific antibody functionalized devices (+ABVLP), thus indicating that the antibody-VLP recognition is required for detection. Sensors functionalized with HPV16 L1 CamVir 1 antibodies (+ABCamVir) showed a small normalized resonance shift, suggesting either that a small amount of unassembled L1 protein was present in the VLP solution, or that there is some cross-reactivity between CamVir 1 antibodies and VLPs. Importantly, the optical response with the CamVir 1 antibody is statistically significantly different (two-tailed unequal variance t-test yielded a P-value ≤ 0.05) from the response with the VLP-specific antibody for the same VLP concentration. This result suggests that the PhC sensors are capable of selective detection of intact HPV-VLPs, provided the receptor antibodies have the desired specificity.
Figure 5.
Comparison of the experimental control-subtracted red-shifts observed for sensors functionalized with VLP-specific antibodies, with antibodies specific for HPV16 L1 protein but not intact VLPs (Cam Vir1), and without antibodies (BSA blocked only), following exposure to VLPs. The error bars represent the standard deviation for each condition.
4 CONCLUSION
We have developed a point-defect coupled 2D PhC waveguide sensor capable of label-free recognition-mediated detection, and have demonstrated the use of the device for sensing a model viral pathogen (HPV virus-like nanoparticles) for the first time. The PhC sensors have an experimental sensitivity of 0.1 nm/nM, and the limit of detection was found to be 1.4 nM in both buffered and serum backgrounds. Specificity tests indicated that the selectivity of the PhC sensor for HPV-VLPs is determined by the selectivity of the capture antibody with which the device is functionalized. Thus, these results are strongly encouraging with regard to further development of the 2D PhC sensor. There are at least two significant challenges remaining, however. First, our current method for functionalization of the device results in attachment of antibodies to the entire chip surface, rather than just the active area of the device. Selective functionalization within the defect area will be essential in order to prevent nonproductive capture of targets on the surface. Second, as has been noted in the context of other nanoscale optical sensors, the diffusion limit of rare targets constitutes a significant barrier to overcome in order to obtain true ultrasensitivity (Sheehan and Whitman, 2005). One potentially attractive strategy for achieving this is to guide the VLPs into the optical cavity region through microfluidic channels using optical forces. Efforts along these lines are ongoing in our laboratories (Baker et al., 2011; Heiniger et al.,2011).
A significant advantage of this PhC design is its multiplexing capability. In the current paper, we have focused on testing the PhC sensors in detecting a single type of virus. As each PhC waveguide sensor can potentially be functionalized with a different receptor molecule, it is possible to detect multiple pathogenic viruses, or, alternatively, different strains of the same virus, on the same chip. Additionally, the large free spectral range (FSR) of the point-defect coupled waveguide design (as compared to other resonator structures) makes it possible to extend the above design for high throughput applications since arrays of PhC waveguides can be conveniently fabricated on a single chip (Mandal et al.,2009).
Supplementary Material
2-D Photonic Crystals are optical sensors with an exceptionally small sensing volume
We conducted FDTD calculations that suggest single virus detection is possible
Detection of human papillomavirus virus-like particles (VLPs) was demonstrated
High selectivity was obtained in buffer and in serum
Acknowledgments
This work was supported by the National Institute of Health (1 R01A108077-01). Device fabrication was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (Grant ECS 03-35765). We thank Dr. Elisa Guillermain for acquiring the SEM image shown in Figure 1, and Professor Robert Rose for generous donation of materials. We also thank the Center for Integrated Research Computing at the University of Rochester for providing the computing resources used in this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Oskooi Ardavan F, Roundy David, Ibanescu Mihai, Bermel Peter, Joannopoulos JD, Johnson Steven G. Comput Phys Commun. 2010;181:687–702. [Google Scholar]
- Baker JE, Sriram R, Fauchet PM, Miller BL. Proc SPIE. 2011;7888:78880M–78880M-6. [Google Scholar]
- Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. J Biol Chem. 2007;282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Mol Cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- Dorfner D, Zabel T, Hürlimann T, Hauke N, Frandsen L, Rant U, Abstreiter G, Finley J. Biosens Bioelectron. 2009;24:3688–3692. doi: 10.1016/j.bios.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. Anal Chim Acta. 2008;620:8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland SM, Smith JS. Drugs. 2010;70:1079–1098. doi: 10.2165/10898580-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Guillermain E, Fauchet PM. CLEO/QELS. 2009;2009:1–2. [Google Scholar]
- Harvey AH, Gallagher JS, Sengers JMHL. J Phys Chem Ref Data. 1998;27:761–774. [Google Scholar]
- Heiniger AT, Miller BL, Fauchet PM. 2011;7888:78880L–78880L-8. [Google Scholar]
- Joannopoulos JD, Johnson SG, Winn JN, Meade RD. Photonic Crystals: Molding the Flow of Light. 2. Princeton University Press; 2008. [Google Scholar]
- Liu K, Yuan XD, Ye WM, Zeng C. Opt Commun. 2009;282:4445–4448. [Google Scholar]
- Lee MR, Fauchet PM. Opt Express. 2007;15:4530–4535. doi: 10.1364/oe.15.004530. [DOI] [PubMed] [Google Scholar]
- Mace CR, Rose RC, Miller BL. Proc SPIE. 2009;7553:71670H–71670H-8. [Google Scholar]
- Mandal S, Goddard JM, Erickson D. Lab Chip. 2009;9:2924. doi: 10.1039/b907826f. [DOI] [PubMed] [Google Scholar]
- Olivier S, Rattier M, Benisty H, Weisbuch C, Smith C, De La Rue R, Krauss T, Oesterle U, Houdré R. Phys Rev B. 2001;63:1–4. doi: 10.1364/ol.26.001019. [DOI] [PubMed] [Google Scholar]
- Painter O, Vučkovič J, Scherer A. J Opt Soc Am B: Opt Phys. 1999;16:275–285. [Google Scholar]
- Pal S, Guillermain E, Sriram R, Miller B, Fauchet PM. Proc SPIE. 2010;7553:755304-1–755304-10. [Google Scholar]
- Pal S, Guillermain E, Sriram R, Miller BL, Fauchet PM. Biosens Bioelectron. 2011;26:4024–4031. doi: 10.1016/j.bios.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Fauchet PM, Miller BL. Anal Chem. 2012;84:8900–8908. doi: 10.1021/ac3012945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather DW, Sharkawy A, Shi S, Murakowski J, Schneider G. Photonic Crystals, Theory, Applications and Fabrication. 1. Wiley; 2009. [Google Scholar]
- Prime K, Whitesides G. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- Rose RC, Bonnez W, Reichman RC, Garcea RL. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan PE, Whitman LJ. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- Walboomers JMM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJF, Peto J, Meijer CJLM, Muñoz N. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Shane E, Oliver CN. J Chromatogr, A. 1998;816:21–28. doi: 10.1016/s0021-9673(98)00065-x. [DOI] [PubMed] [Google Scholar]
- Zlatanovic S, Mirkarimi LW, Sigalas MM, Bynum MA, Chow E, Robotti KM, Burr GW, Esener S, Grot A. Sens Actuators B. 2009;141:13–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.