Abstract
Analysis of the reproductive physiology of anautogenous mosquitoes at the molecular level is complicated by the simultaneity of ovarian maturation and the digestion of a blood meal. In contrast to anautogenous mosquitoes, autogenous female mosquitoes can acquire greater nutrient stores as larvae and exhibit higher ovarian production of ecdysteroids at adult eclosion. These features essentially replace the role of a blood meal in provisioning the first batch of eggs and initiating egg development. To gain insight into the process of ovary maturation we first performed a transcript analysis of the obligatory autogenous mosquito Georgecraigius atropalpus (formerly Ochlerotatus atropalpus). We identified ESTs using suppressive subtractive hybridization (SSH) of transcripts from ovaries at critical times during oogenesis in the absence of blood digestion. Preliminary expression studies of genes such as apolipophorin III (APO) and oxysterol binding protein (OSBP) suggested these genes might be cued to female nutritional status. We then applied our findings to the medically important anautogenous mosquito Aedes aegypti. RNAi-based analyses of these genes in Ae. aegypti revealed a reduction in APO transcripts leads to reduced lipid levels in carcass and ovaries and that OSBP may play a role in overall lipid and sterol homeostasis. In addition to expanding our understanding of mosquito ovarian development, the continued use of a comparative approach between autogenous and anautogenous species may provide novel intervention points for the regulation of mosquito egg production.
Keywords: oogenesis, Ochlerotatus atropalpus, autogeny, anautogeny, SSH, oxysterol binding protein, gonadotropin inducible transcription factor, apolipophorin III
1. Introduction
Most mosquito species are obligatory blood feeders because the nutrients in blood are required for egg production. This “anautogenous” form of reproduction is observed in a number of the major vectors of blood-borne pathogens that cause human diseases such as the yellow fever mosquito Aedes aegypti and the African malaria mosquito Anopheles gambiae. Other mosquito species utilize an “autogenous” form of reproduction in which females are capable of producing their first batch of eggs without ingesting a protein meal as an adult (Clements, 1992). Autogeny would seem a preferred form of reproduction when animal hosts for adult blood feeding are limited. A number of behavioral and developmental traits are often associated with autogenous mosquitoes (O’Meara, 1979, 1985). Larvae tend to take longer to complete development, adults are less likely to disperse, and mating does not require the extensive flight activity needed for mating swarms.
The nutritional condition of anautogenous females is complex because it is determined by a combination of larval-derived reserves and the nutrients acquired from sugar and blood feeding by adults. The nutritional condition of autogenous females, however, is determined almost exclusively by larval-derived teneral reserves. In a study of the obligatory autogenous rock pool mosquito Georgecraigius atropalpus (Coquillett) (formerly Ochlerotatus atropalpus), Telang and Wells (2004) found that body size and egg load of newly emerged females were positively correlated with the quality of larval diet.
Reproduction by female mosquitoes is dependent on nutrient resources but modulated by hormones. This process is likewise more complex in anautogenous females than autogenous females because in the former a blood meal serves as both raw material for yolk and as an activator of hormone secretion by diverse reproductive tissues. Earlier research has shown that larval nutrition significantly affects the endocrinology of egg development in both anautogenous Ae. aegypti and autogenous Gc. atropalpus (Telang et al., 2006). Newly emerged Ae. aegypti, which have lower larval-derived protein reserves, produce higher amounts of juvenile hormone (JH) than similarly staged Gc. atropalpus. While JH primes fat body machinery for vitellogenin synthesis, increased synthesis of this yolk protein does not begin in anautogenous females until a blood meal triggers the release of ecdysteroids from ovaries leading to a corresponding rise in hemolymph ecdysteroid levels. In contrast, newly emerged Gc. atropalpus females, which have higher protein reserves and develop their first batch of eggs without a blood meal, initiate ovarian ecdysteroid production upon emergence (reviewed in Telang et al., 2006).
The study of Gc. atropalpus thus far suggests that it offers key advantages for understanding the molecular biology of mosquito egg development. First, regardless of the quality of larval nourishment, our strain of Gc. atropalpus is invariably autogenous for the first ovarian cycle and, upon completion of this first cycle, is anautogenous for all subsequent cycles (Telang and Wells, 2004). Second, by altering the quality of the larval diet, females can be obtained that are large with sizeable nutrient reserves and high fecundity or small with meager nutrient reserves and low fecundity. These attributes have allowed us to measure both significant differences in ecdysteroid production and titer as a function of nutritional state and many parameters of egg development without the added complexity introduced by blood meal ingestion. This understanding of Gc. atropalpus development has made it possible to undertake focused transcriptome studies of key tissues at varying time points during oogenesis following eclosion in autogenous females. In contrast to studies of oogenesis in anautogenous species, the gene transcript pools observed in newly emerged Gc. atropalpus should be less complex and directly related to egg production because no blood digestion or management of resultant nutrients or toxins is occurring.
In this study, we first characterized sets of genes identified specifically in ovaries of autogenous Gc. atropalpus that are likely involved in nutrient mobilization, ovarian ecdysteroid production and the regulation of these processes during oocyte maturation. We then mined genomic databases to identify homologs of these genes belonging to the medically important and anautogenous vector Aedes aegypti. We conducted gene expression studies in Ae. aegypti as well as RNAi experiments to elucidate roles of specific genes in nutrient mobilization and egg development in this mosquito. We also used an in vitro enzyme immunoassay for ecdysteroid to test the hypothesis that ovarian production of this hormone can be decreased using an RNAi approach. Lastly, we hypothesized that specific genes may play a role in lipid mobilization during oogenesis. We used biochemical assays to determine whole body lipid mobilization from reserves to ovaries in RNAi-treated females.
2. Materials and methods
2.1. Insects
Georgecraigius atropalpus (formerly Ochlerotatus atropalpus) was reared using established procedures (Telang and Wells, 2004). Eggs were hatched overnight in partially deoxygenated tap water. Larvae were maintained in uncrowded conditions and fed a 10% solution of bovine liver powder (VWR). All stages were maintained at 27 ±1°C with a daily photoperiod of 16 h light and 8 h dark. By manipulating the amount of food available throughout larval development, we are able to rear large females with large nutrient reserves (rich or A-diet females) or small females with small nutrient reserves (poor or C-diet females). Regardless of the quality of larval nourishment, our strain of Gc. atropalpus is invariably autogenous during the first ovarian cycle and, upon completion of this first cycle, is anautogenous for all subsequent cycles (Telang and Wells, 2004). Newly emerged females were maintained on water and 10% sucrose solution. Only females of known ages post eclosion (PE) were used for experiments. After completion of their first oogenic cycle, females were maintained on sucrose solution without being provided access to a blood meal. Eggs from non-blood fed females were used to propagate the autogenous colony.
A colony of Aedes aegypti (L.) (Rockefeller strain) was maintained under similar insectary conditions. Larval Ae. aegypti was fed a diet of ground tropical fish flakes (TetraMin/TetraFin Fish Flakes), Bovine Liver Powder (VWR), and Rodent diet (Teklad Global, Harlan Laboratories) in a 10:1:10 mixture. Adult females were fed ad libitum on 10% sucrose for 48 h prior to a blood meal. Females 4 days PE were fed on porcine blood with 3.8% sodium citrate added as preservative and supplemented with ATP to a final concentration of 1mM immediately before use. Blood feeding was performed using Parafilm-lined glass feeders attached to a circulating water bath maintained at 37°C.
2.2. cDNA preparation and subtractive hybridization
We produced two cDNA populations for this study. The first population was prepared using transcripts isolated from the ovaries of Gc. atropalpus females 18h post emergence. These animals are in the autogenous phase of reproduction with ovaries at the late second or early third Christophers stage (Christophers, 1911; Clements, 1992). Ovaries from sixty autogenous females were dissected and placed directly into RNAlater™ (Ambion). Total RNA was extracted and treated with DNase I using a RNeasy mini kit (Qiagen). Messenger RNA (~ 2 μg) was then isolated with a GenElute mRNA miniprep kit (Sigma) and quantified using Ribogreen reagent (Molecular Probes) on a Fluostar Galaxy platereader (BMG). The second population of transcripts was prepared in the same manner starting with sixty ovaries from females that had recently completed their autogenous phase of reproduction but not taken a blood meal to begin their first round of anautogenous egg production. The ovaries of these females are in arrest at Christophers stage I. The resulting mRNA from both populations was converted to cDNA using MMLV reverse transcriptase (Clontech). Differentially expressed genes in the ovaries of the autogenous females were then obtained by subtracting the cDNAs prepared from this population against those obtained from the anautogenous mosquitoes using a PCR-Select cDNA subtraction kit according to manufacturer’s instructions (Clontech). This kit utilizes the technique of suppressive subtractive hybridization (SSH) whereby novel and rarely expressed cDNAs in the target or “tester” population are selectively amplified while abundant cDNAs and those held in common with a reference or “driver” population are suppressed (Diatchenko et al., 1996).
Subtracted products were amplified on a Perkin-Elmer GeneAmp PCR System 2400 thermocycler using Advantage2 Polymerase reagents (BD Bioscience).
2.3. Cloning of cDNAs and EST sequencing
Following the SSH procedure, cDNAs were ligated into the pCR4-TOPO vector (Invitrogen) and transformed into TOP10 competent cells (Invitrogen). Transformants were plated on LB agar plates under ampicillin selection and grown overnight at 37°C. Selected colonies were grown in 2 mL LB-ampicillin media overnight at 37°C and their plasmid DNA harvested using a QIAprep Spin Miniprep kit (Qiagen). Sequencing reactions were prepared using ABI Prism BigDye Terminator Cycle Sequencing reagents then sent to the University of Wisconsin Biotechnology Center (Madison, WI) for electrophoresis and sequence data acquisition. Electropherograms were analyzed using a sequence analysis software program (Lasergene, DNASTAR). Potential protein sequence homologs were identified using the translated BLAST (tblastx) algorithm restricted to arthropod databases at the National Center for Biotechnology Information (NCBI). A cutoff for assigning significant amino acid sequence homology was set at an E value of 10-4.
2.4. Reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR was used to determine tissue distribution of certain Gc. atropalpus transcripts identified by the SSH procedure. Females derived from larvae fed a rich diet (A-diet) were collected when their ovaries had reached the late second or early third Christophers stage (Clements, 1992). Head, thorax, abdominal wall, ovaries, and gut were dissected in saline. Ovaries from females derived from larvae fed a poor diet (C-diet) were also collected at the same Christophers stage to investigate gene expression in relation to nutritional status. Another group of females were allowed to deposit their first (autogenous) batch of eggs. Ovaries from these newly anautogenous females were also dissected to examine gene expression as a function of the change in reproductive status. Dissected tissues were placed directly in RNAlater™ (Ambion), incubated at 4°C overnight and then stored at -80°C until further processing. Total RNA was extracted from tissues using an RNeasy kit (Qiagen). RNA integrity was checked on a 1% agarose gel then quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). Total RNA (1 μg) was used to synthesize cDNA using an iScript cDNA kit (Bio-Rad). Primers corresponding to genes of interest were designed using PrimerSelect (Lasergene, DNASTAR). PCR was conducted with Titanium Taq DNA polymerase (Clontech) using the gene-specific primers in the following program: 94°C (2 mins) followed by 30 cycles of 94°C (30 s), 50-65°C (30 s), 72°C (30 s). Amplification of Gc. atropalpus actin was used as an endogenous control for cDNA quantity as well as to confirm integrity of tissue and cDNA preparations. All reaction products were electrophoresed on a 1% (w/v) agarose/TAE (40 mM Tris/acetate, 2 mM EDTA, pH 8.0) gel to confirm product size. PCR products for candidate genes (including the reference gene actin) were gel purified, cloned, sequenced and identified using a tblastx search (NCBI). Tissues from three separate cohorts were analyzed using RT-PCR.
2.5. Quantitative real-time PCR analysis of gene transcripts (qPCR) in Gc. atropalpus
A subset of genes analyzed by RT-PCR was also analyzed in qPCR experiments. To determine transcript abundance in relation to female nutritional status and reproductive phase, we prepared cDNA templates using mRNA derived from ovaries of A-diet and C-diet animals, as well as ovaries from anautogenous females. In addition, we examined the expression of particular transcripts from the ovaries of autogenous females at different times during a gonotrophic cycle. For this experiment, ovaries were dissected from autogenous females at specific times after emergence (0-6, 24 and 48 h). At these particular time points, follicles were staged as Christopher’s stage 1, 2/3, and 4 respectively (Clements, 1992). These time points encompass the interval over which ovarian ecdysteroid production is known to rise and fall (Telang et al., 2006). Total RNA isolation and cDNA synthesis proceeded as described above. For qPCR, template cDNA-specific master mixes were prepared with IQ™ SYBR® Green Supermix (Biorad), nuclease-free water, the appropriate primers (250 nM final concentration of each primer per 10 μl reaction volume) and template cDNA. Primers were designed using PrimerSelect (Lasergene, DNASTAR). Three reactions per cDNA template and a non-template control (NTC) were performed in parallel to control for pipetting error. The triplicate tissue cDNA reactions were averaged for inclusion in our statistical model. To estimate the number of transcripts in the various samples, a standard curve was derived using purified PCR products for the gene of interest as template. qPCR master mixes were prepared as above with serially diluted PCR product (1 ng, 100 pg, 10 pg, 1 pg, 100 fg or 10 fg/PCR) to which primers for the gene of interest were added. The standard line was then used to calculate the transcript copy number in each of the qPCR reactions as described by Paton et al. (2000). This method has been successfully used to analyze genes expressed in ovaries of Ae. aegypti in which transcript quantities rise and fall during a gonotrophic cycle (Sieglaff et al., 2005). Real-time PCR was conducted on a Chromo4™ Real-Time Detector (Biorad) using the program: 95°C for 3 min, 95°C for 20s, 55-65°C for 20s, 72°C for 20s for 45 cycles, followed by melting curve analysis. Reaction products were electrophoresed on a 1% agarose/TAE gel to confirm product size. Results from the Chromo4™ detector were processed and quantified using Opticon Monitor 3 software (Biorad). Real-time PCR results from multiple cohorts were analyzed; data shown represent results for tissues from groups of females in which data for at least two out of three separate cohorts were in agreement.
2.6. RNAi-mediated knockdown experiments in Aedes aegypti females
Select genes isolated from the SSH procedure were used in RNAi-mediated knockdown experiments in Ae. aegypti females. The EST sequences for Georgecraigius atropalpus oxysterol binding protein (OatrOSBP) and apolipoprotein 3 (OatrApo) were used to identify protein sequence homologs in Aedes aeqypti(AaegOSBP and AaegApo) using the tblastx algorithm restricted to arthropod databases at the NCBI. Primers were designed to GenBank sequences for putative AaegOSBP (Accession # XM_001651481) and AaegApo (Accession # XM_001659474). AaegOSBP and AaegApo cDNA were amplified with forward and reverse primers containing the T7 sequence at their 5’ ends. The primer sequences are as follows: OSBP forward primer (5’-TCTGCCAAGGGAGTGAAGTG -3’), OSBP reverse primer (5’- GTATGGCGGGTAGGGACGAC -3’), Apo forward primer (5’- TTTGGCGCGTGTAACTCG -3’), Apo reverse primer (5’-TCTTCTGCAACGTGTCAACC -3’). PCR products were purified (QIAquick, Qiagen) and served as templates for dsRNA synthesis using the HiScribe kit (NEB). Briefly, 1 μg of template cDNA was transcribed in vitro by using T7 RNA polymerase. The dsRNA was purified using an RNeasy kit (Qiagen) and eluted in RNase-free water. As a control, dsRNA was generated for the enhanced green-fluorescent protein, dsEGFP (pEGFP-1 vector, Clontech). All dsRNAs were concentrated to 4 μg/μl using a lyophilizer. Female Ae. aegypti mosquitoes were given access to sugar and males post-eclosion and were injected 48 h PE with 2 μg of dsRNA (0.5 μl of a 4 μg/μl solution) using a positive-displacement microinjector (Tritech). For injections, groups of females were cold anaesthetized and kept on ice until ready for injection. Mosquitoes were transferred to a cold block set to 2°C and mounted on a dissecting microscope. Female mosquitoes were injected with the OSBP or Apo dsRNA, dsEGFP or water. Injected females were allowed to recover for 48 h in a humid environment and were then provided a blood meal. RNAi knockdown experiments were conducted on two independent cohorts of injected females.
2.7. Effect of RNAi on ovarian ecdysteroid production in Aedes aegypti females
At 24 h PBM, ovaries from dsRNA-treated females were dissected for our in vitro assay of ovarian ecdysteroid production. Briefly, four ovary pairs from treated females were dissected in saline solution then transferred to and incubated in 60 μl of buffered medium in a polypropylene tube cap for 6 h at 27°C as previously described (Telang et al., 2006). After incubation, 50 μl of medium were collected, stored at -80°C, and later analyzed for ecdysteroid content using an enzyme immunoassay (EIA) (Kingan, 1989; Margam et al., 2006). For each experiment, triplicates of four ovary pairs were analyzed for each dsRNA treatment in the same EIA. The EIA is based on ecdysteroid (in either standard or sample) competing with a known amount of peroxidase-labeled conjugated ecdysone for the ecdysteroid antiserum. Our EIA uses an ecdysteroid antiserum with high affinity toward both ecdysone and 20HE (T. Kingan, pers comm.). EIA plates (96-well, flat-bottom, Costar) were incubated overnight at 4°C with 0.5 μg/well of an Fc-specific goat anti-rabbit IgG antibody (Jackson Immunoresearch). A concentrated stock of Fc-specific antisera was diluted in PBS (10 mM sodium phosphate, 150 mM NaCl, pH 7.5) to obtain a final coating volume of 90 μl/well. The next day the IgG coating solution was discarded followed by addition of EIA buffer (20 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA, and 0.1% (w/v) bovine serum albumin at pH 7.4; 320 μl/well) used as blocking solution. Plates were incubated for 2 h at room temperature with constant mixing on an orbital shaker. EIA blocking solution was discarded and plates were washed twice with 320 μl of PBS/0.05% Tween 20 (PBST) at 5 min per wash on an orbital shaker. All samples were tested at a final dilution of 1:25 using EIA buffer. Fifty μl of diluted sample or 20-hydroxyecdysone standards (Sigma; 0.5, 1, 2.5, 5, 10, 20, and 40 pg) were added to wells in duplicate, while 100 μl of EIA/BSA buffer was used as a blank to account for non-specific binding. Anti- 20HE rabbit polyclonal antisera was diluted 1:100,000 in EIA/BSA buffer and added to wells (50 μl /well) containing samples and standards followed by addition of 50 μl 20HE- horseradish peroxidase (20HE-HRP) conjugate (1:15,000 in EIA/BSA buffer). Plates were incubated for 5 min on an orbital shaker to mix contents and then incubated overnight at 4 °C to allow the competitive reaction to equilibrate. Next day, the contents were discarded and plate was washed three times with PBST, with 5 min incubation on a shaker for each wash. After the last wash was discarded, color development was initiated by adding 100 μl/well tetramethylbenzidine (TMB) peroxidase substrate solution (American Qualex) at room temperature. After 15 minutes of incubation at room temperature in the dark, 100 μl 1.0M phosphoric acid was added to each well to halt color development. Optical density was measured at 450 nm using a plate reader (Beckman Coulter, DTX 880 Multimode Detector). Values for each sample are reported as ‘mean pg ecdysteroid’, as the secreted ecdysteroid species could be either ecdysone or 20-HE. Each sample was assayed in triplicate and these triplicates were averaged for statistical analysis. EIA results from two independent cohorts of dsRNA-treated females are summarized (n=36).
2.8. Effect of RNAi on nutrient mobilization in Aedes aegypti females
We examined the role of specific genes in nutrient mobilization by quantifying nutrient levels in body carcasses and ovaries in RNAi-treated females. Whole body homogenates were processed from one subset of dsRNA-treated females prior to blood feeding. Another subset of dsRNA-treated females was analyzed 24 h PBM. For this subset, females were dissected to collect ovaries and remaining carcasses (without ovaries). Extraction of storage lipids (triacylglycerol) and soluble proteins from whole body or carcass homogenates entailed a procedure first described by Van Handel (1965) and modified for Ae. aegypti (Zhou et al., 2004). Ovaries from dsRNA-treated, 24 h PBM females underwent lipid and protein extraction following procedures outlined by Zhou et al. (2004). In all cases, extracts of lipid and protein were frozen until quantification by colorimetric-based assays. Total lipid levels were determined by a modified Vanillin reagent assay (Van Handel, 1985) and soluble protein content by the BCA protein assay reagent kit (Pierce). All nutrients are reported on a microgram per mg dry mass basis. Each sample for biochemical analysis was assayed in duplicate and these duplicates were averaged for inclusion in statistical analysis. Biochemical analyses from two independent cohorts of dsRNA-treated females are summarized (n=50).
2.9. qPCR analysis of RNAi-induced gene knockdown in Aedes aegypti females
At 48 h post blood meal (PBM), we dissected tissues from a subset of females to confirm dsRNA treatment reduced transcript abundance relative to dsEGFP controls. To quantify transcript knockdown, tissues (head, midgut, ovaries, and abdomen including fat body) were dissected from two sets of five dsRNA-injected females and total RNA isolation and cDNA synthesis proceeded as previously described (Subsection 2.4). Prepared cDNA served as template for qPCR using conditions described (Subsection 2.5) except we normalized transcript abundance in each tissue sample relative to the expression of actin. We chose dsEGFP as the control (calibrator) and abundance of OSBP and Apo transcripts were then reported relative to the calibrator using the Pfaffl (2001) method. The following formula was used to determine the expression ratio between sample and calibrator: Expression Ratio = (Etarget) ΔCT, target (calibrator-test) / (Eref) ΔCT, ref (calibrator-test), where E is PCR efficiency. Quantitative PCR results from two independent cohorts of dsRNA treated females are summarized (n=32).
2.10. Data Analysis
Transcript abundance in relation to autogenous egg development and reproductive strategy in Gc. atropalpus was analyzed by 1-way ANOVA for 3 different cohorts of animals. Comparisons of transcript levels between specific groups were made using linear contrasts. For Ae. aegypti, qPCR expression ratio in relation to dsRNA treatment was analyzed by 2-way ANOVA, with dsRNA treatment, tissue and an interaction term followed by the Tukey-Kramer HSD test to compare all means. Ovarian ecdysteroid production in relation to dsRNA treatment was analyzed by 1-way ANOVA followed by the Tukey-Kramer HSD test to compare all means. Levels of lipid and protein in carcasses of dsRNA treated females were analyzed separately using 2-way ANOVA, with dsRNA treatment, blood feeding time and an interaction term. Comparisons between specific groups were made using linear contrasts. Levels of lipid and protein in carcasses and ovaries of dsRNA-treated females were analyzed separately using 2-way ANOVA, with dsRNA treatment, tissue type and an interaction term. Comparisons between specific groups were made using linear contrasts. Data were statistically analyzed using JMP IN (version 6.0.2, SAS Institute Inc.). Least square means (with s.e.m.) were obtained from statistical models and illustrated using GraphPad Prism 4.0 (2006; GraphPad Software Inc., San Diego, CA).
3. Results and discussion
3.1 Construction and characterization of Gc. atropalpus ovary subtracted library
The SSH procedure relies on the hybridization of cDNA from a “tester” sample to an excess of cDNA from a “driver” sample, followed by PCR-enrichment of the unhybridized tester fraction representing differentially expressed transcripts. Since our goal was to isolate transcripts uniquely expressed in ovaries during autogenous oogenesis, this sample type served as the “tester” population in a forward experiment (i.e., a one-way SSH procedure). Ovaries from newly anautogenous females served as the excess “driver” population. A total of 249 clones produced usable sequence data of which 125 produced novel and significant alignments with sequences in database searches. These 125 ESTs were submitted to GenBank and assigned the accession numbers GT154619 – GT154743. A complete list of identified transcripts is presented in Table 1. Of the novel ESTs, our blastx search identified 105 ESTs with significant similarity to previously characterized mosquito sequences (M, Table 1), 2 ESTs with significant similarity to characterized sequences from other arthropods (A, Table 1), and 18 ESTs with significant similarity to uncharacterized sequences from either mosquitoes or other arthropods (hypothetical protein, Table 1). The remaining clones were either duplicate or did not yield significant matches to sequences in the database given our cutoff criteria.
Table 1.
ESTs from the subtracted Gc. atropalpus autogenous ovary library with significant BLAST matchesa
| Transcripts associated with cytoskeletal proteins
| |||
|---|---|---|---|
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
| AO_008 | GT154623 | XP_001652975.1 (M) | Moesin; epithelial integrity |
| AO_032a | GT154639 | XM_001648522.1 (M) | tubulin alpha chain |
| AO_075 | GT154661 | XM_001652925.1 (M) | cytoskeletal protein binding |
| AO_078a | GT154663 | XM_001663357.1 (M) | tubulin alpha chain |
| AO_109 | GT154681 | XM_001651313.1 (M) | microtubule associated protein |
| AO_119a | GT154685 | EU206967.1 (M) | Calponin-like actin-binding (signal transduction?) |
| AO_139 | GT154699 | XM_001650415.1 (M) | spectrin |
| AO_165a | GT154702 | XM_001655882.1 (M) | myosin light chain 1 |
| AO_170b | GT154704 | XM_001655511.1 (M) | lamin |
| AO_258 | GT154726 | XM_001842844.1 (M) | actin |
|
Transcripts associated with extracellular matrix
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_121a | GT154687 | XM_001846620.1 (M) | collagen - extracellular matrix structural constituent |
|
Transcripts associated with transport
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_015a* | GT154628 | XM_001655335.1 (M) | general substrate transporter |
| AO_026 | GT154634 | XM_001653416.1 (M) | ATP synthase |
| AO_033 | GT154641 | XM_001649152.1 (M) | cation transporter |
| AO_049a | GT154646 | XM_001843312.1 (M) | Importin, protein involved in nuclear import |
| AO_081a | GT154666 | XM_001650815.1 (M) | protein or nucleoside transporter |
| AO_084b | GT154670 | XM_001650080.1 (M) | Sec24B protein |
| AO_087 | GT154672 | XM_001662733.1 (M) | sugar transporter superfamily |
| AO_095a | GT154676 | XM_001658714.1 (M) | tyrosine transporter |
| AO_098 | GT154678 | XM_001663991.1 (M) | phosphatidylinositol-binding clathrin assembly protein |
| AO_137b | GT154697 | XM_001662995.1 (M) | Rab GDI protein |
| AO_177 | GT154707 | XM_001652466.1 (M) | UCP - mitochondria substrate/carrier protein |
| AO_257 | GT154725 | XM_001658180.1 (M) | Amino acid transport (maybe metabolism) |
|
Transcripts associated with signal transduction
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_006 | GT154622 | XM_001850285.1 (M) | GTP-binding nuclear protein |
| AO_014a** | GT154627 | XM_001651481.1 (M) | Oxysterol Binding Protein - steroid metabolic process |
| AO_031b | GT154638 | XM_001650876.1 (M) | Calpains |
| AO_050 | GT154648 | XM_001658794.1 (M) | Bleomycin hydrolases |
| AO_067b | GT154659 | XM_001870624.1 (M) | G-protein signaling |
| AO_078b | GT154664 | XM_001659729.1 (M) | Peptidase S9, prolyl oligopeptidase |
| AO_094b | GT154675 | XM_001848979.1 (M) | serine-type peptidase activity |
| AO_100 | GT154679 | XM_001849410.1 (M) | Serine/threonine-specific protein phosphatase |
| AO_110 | GT154682 | XM_001865810.1 (M) | serine protease |
| AO_122 | GT154689 | XM_001661301.1 (M) | plexin a - tissue development guidance |
| AO_126 | GT154690 | XM_001655818.1 (M) | acetylcholinesterase |
| AO_233a | GT154721 | XP_001650933.1 (M) | armadillo (intracellular signalling) |
| AO_295 | GT154733 | XM_001649024.1 (M) | polo kinase kinase |
| AO_303 | GT154734 | XM_001654456.1 (M) | dullard (phosphatase activity) |
| AO_317 | GT154739 | XM_001655061.1 (M) | 14-3-3 protein |
| AO_319 | GT154740 | XM_001660391.1 (M) | Arf-like: GTP-binding proteins |
|
Transcripts associated with nuclear regulation
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_001b | GT154620 | XM_001845856.1 (M) | DNA repair endonuclease |
| AO_049b | GT154647 | AF411297.1 (M) | Ribonucleotide reductase 2 |
| AO_057 | GT154653 | XM_001648649.1 (M) | DNA replication licensing factor MCM8 |
| AO_066 | GT154657 | AB019403.1 (A) | Histone 3 |
| AO_093b | GT154674 | XM_001660927.1 (M) | ribonucleotide reductase |
| AO_201 | GT154715 | XM_001660578.1 (M) | cyclin b (cell cycle regulator) |
| AO_307a | GT154736 | XM_001868813.1 (M) | cell division cycle and apoptosis regulator protein 1 |
|
Transcripts associated with transcriptional machinery
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_012** | GT154626 | XM_001654797.1 (M) | gonadotropin inducible transcription factor (Zinc finger?) |
| AO_024 | GT154632 | XM_001658298.1 (M) | zinc finger domain (C2H2-type) |
| AO_037 | GT154643 | XM_001659237.1 (M) | RNA helicase |
| AO_051 | GT154649 | XM_001654618.1 (M) | zinc finger domain (BTB family) |
| AO_064 | GT154655 | XM_001657187.1 (M) | RNA binding motif protein 4, lark |
| AO_083 | GT154668 | XM_001654567.1 (M) | transcriptional regulator ATRX (X-linked helicase II) |
| AO_085 | GT154671 | XM_001864820.1 (M) | vigilin - RNA binding |
| AO_116 | GT154684 | XM_001650994.1 (M) | heterogeneous nuclear ribonucleoprotein |
| AO_145a | GT154701 | XM_001851227.1 (M) | quaking - RNA binding protein |
| AO_183 | GT154710 | XM_001658569.1 (M) | PUA - RNA binding motif |
| AO_187b | GT154711 | XM_001658657.1 (M) | La protein (RNA binding protein?) |
| AO_198 | GT154713 | XP_001663221.1 (M) | zinc finger protein (perhaps protein modification?) |
| AO_204 | GT154717 | XP_001656937.1 (M) | RNA pol II accessory factor, Cdc73 |
| AO_232 | GT154720 | XM_001654559.1 (M) | zinc finger protein |
| AO_237b | GT154723 | XP_001653007.1 (M) | E2F transcription factor dimerization partner |
|
Transcripts associated with protein synthesis and modification
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_001a | GT154619 | XM_001652519.1 (M) | Nucleolar NOP-1; Generation of 28S rRNA |
| AO_003b | GT154621 | XM_001663233.1 (M) | glycotransferase activity |
| AO_009 | GT154624 | XM_001655341.1 (M) | ribosomal protein |
| AO_027a | GT154635 | XM_001656021.1 (M) | isoleucyl tRNA synthetase |
| AO_034 | GT154642 | XM_001650943.1 (M) | ubiquitin-activating enzyme E1 |
| AO_040 | GT154644 | XM_001657290.1 (M) | Nop14 (ribosome production and maturation) |
| AO_053a | GT154651 | XM_001842705.1 (M) | mahogunin; E3 ubiquitin ligase |
| AO_065b | GT154656 | XM_001655602.1 (M) | Nascent polypeptide-associated complex (NAC) |
| AO_067a | GT154658 | XM_001663392.1 (M) | Peptidyl-prolyl cis-trans isomerase |
| AO_102a | GT154680 | XM_001649725.1 (M) | protein disulfide isomerase |
| AO_114 | GT154683 | EU528307.1 (A) | Elongation Factor-1 alpha |
| AO_119b | GT154686 | XM_001660482.1 (M) | chaperone atp11p |
| AO_136b | GT154695 | XM_001653574.1 (M) | sentrin/sumo-specific protease |
| AO_137a | GT154696 | XM_001656838.1 (M) | 40S ribosomal protein S5 |
| AO_166 | GT154703 | DQ440225.1 (M) | Heat shock protein Hsp70 |
| AO_178 | GT154708 | XM_001659146.1 (M) | protein disulfide isomerase |
| AO_251 | GT154724 | XM_001654966.1 (M) | Ribosomal protein 60S |
| AO_266 | GT154728 | XM_001660545.1 (M) | chaperonin |
| AO_304 | GT154735 | EU206064.1 (M) | proteasome complex |
| AO_328 | GT154742 | XM_001656813.1 (M) | cullin - scaffolds for ubiquitin ligases |
|
Transcripts associated with intermediate metabolism
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_010 | GT154625 | XM_001656036.1 (M) | saccharopine dehydrogenase (lysine metabolism) |
| AO_021a* | GT154630 | XM_001653891.1 (M) | pyruvate carboxylase activity - gluconeogenesis |
| AO_023* | GT154631 | XM_001658008.1 (M) | Perilipin, lipid metabolism |
| AO_029 | GT154636 | XM_001653700.1 (M) | phosphopyruvate hydratase activity, glycolysis |
| AO_054a** | GT154652 | XM_001659474.1 (M) | apolipophorin3 |
| AO_070 | GT154660 | XM_001660771.1 (M) | sulfuric ester hydrolase activity |
| AO_096 | GT154677 | XM_316061.3 (M) | aromatic amino acid hydroxylase |
| AO_128a | GT154691 | EU210262.1 (M) | testosterone 17-beta-dehydrogenase (steroid metabolism) |
| AO_134b | GT154694 | XM_001648221.1 (M) | Cytochrome P450 |
| AO_141b | GT154700 | XM_001851193.1 (M) | ethanolamine-phosphate cytidylyltransferase |
| AO_171a | GT154705 | AF311261.1 (M) | cytochrome b |
| AO_174 | GT154706 | XM_001648169.1 (M) | Enoyl-CoA hydratase |
| AO_181b | GT154709 | XM_001651897.1 (M) | ribose-phosphate pyrophosphokinase |
| AO_196 | GT154712 | XP_001654813.1 (M) | ferritin-like diiron-carboxylate protein (metal ion binding?) |
| AO_228b | GT154719 | EU206957.1 (M) | Glyceraldehyde 3 phosphate dehydrogenase |
| AO_268 | GT154729 | XM_001653376.1 (M) | glycoprotein sc2 (lipid (steroid) metabolic process) |
| AO_288 | GT154731 | XM_001649983.1 (M) | cytochrome B5 |
| AO_294 | GT154732 | XM_001660189.1 (M) | elongase (long chain fatty acid elongation) |
| AO_310 | GT154738 | XM_001648782.1 (M) | dihydrolipoamide S-acetyltransferase (part of PDH complex) |
|
Transcripts associated with oogenesis
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_019 | GT154629 | S54555.1 (M) | vitelline membrane protein |
| AO_084a | GT154669 | XM_001657708.1 (M) | cathepsin L |
| AO_092 | GT154673 | EU207483.1 (M) | dopa decarboxylase |
| AO_128b | GT154692 | DQ314204.1 (M) | maternal oskar protein |
| AO_199 | GT154714 | DQ459306.1 (M) | cathepsin L |
| AO_203a_ | GT154716 | XP_001858485.1 (M) | maternal exuperantia protein |
| AO_207 | GT154718 | XP_001660748.1 (M) | imaginal disc growth factor |
|
Transcripts associated with unknown function
| |||
| EST# | GenBank accession# | Best blastx match (organism) | comments (similar to/putative function) |
|
| |||
| AO_025 | GT154633 | XM_001651466.1 (M) | Hypothetical protein |
| AO_031a | GT154637 | XM_001657440.1 (M) | Hypothetical protein |
| AO_032b | GT154640 | XM_001649116.1 (M) | Hypothetical protein |
| AO_045 | GT154645 | XM_001649556.1 (M) | Hypothetical protein |
| AO_052* | GT154650 | XM_001661476.1 (M/A) | Hypothetical protein (ecdysone-inducible Vulcan?) |
| AO_058 | GT154654 | XM_001652317.1 (M) | Hypothetical protein |
| AO_077 | GT154662 | XM_001655064.1 (M) | Hypothetical protein |
| AO_079 | GT154665 | XM_001655345.1 (M) | Hypothetical protein |
| AO_081b | GT154667 | XM_001652776.1 (M) | Hypothetical protein |
| AO_121b | GT154688 | XM_001650232.1 (M/A) | Hypothetical protein (Dmel gene Arrest or Bruno?) |
| AO_132 | GT154693 | XM_001851648.1 (M) | Hypothetical protein |
| AO_138 | GT154698 | XM_001654743.1 (M) | Hypothetical protein |
| AO_233b | GT154722 | XP_001648321.1 (M/A) | Hypothetical protein (BWK-1, protein scaffolding?) |
| AO_264 | GT154727 | XM_001659206.1 (M) | Hypothetical protein |
| AO_284 | GT154730 | XM_001662296.1 (M) | Hypothetical protein |
| AO_309 | GT154737 | XM_001864543.1 (M/A) | Hypothetical protein (tyrosine hydroxylase?) |
| AO_326 | GT154741 | XM_001657060.1 (M) | Hypothetical protein |
| AO_339 | GT154743 | XM_001658229.1 (M) | Hypothetical protein |
ESTs were tabulated according to putative functional categories. A cutoff for assigning significant amino acid sequence homology was set at an E value of 10-4.
ESTs that were analyzed by RT-PCR are indicated by an “*” and by both RT-PCR and quantitative real-time PCR by an “**”.
3.2 Sequence identification of cDNA clones from the Gc. atropalpus ovary subtracted library
Supporting evidence for the successful preparation of a subtracted Gc. atropalpus ovary library (hereafter referred to as the GcaOv library) comes from a microarray analysis of genes expressed at the whole body level in An. gambiae 24 h after blood feeding (Marinotti et al., 2005). Their analysis showed major changes in transcript abundance in a multitude of genes presumably involved in processes associated with egg maturation. Many of these same genes were identified in our study of the ovaries of Gc. atropalpus during their autogenous phase of egg development. Further support for the success of the subtraction was obtained following RT-PCR and qPCR analyses of the abundance and distribution of certain GcaOv library transcripts in females in the midst of either the autogenous or anautogenous phases of egg production (see Sections 3.3 and 3.4 below). ESTs from the GcaOv library with significant blastx scores were classified into nine groups using the classification scheme of Marinotti et al. (2005) with modification.
3.2.1 Transcripts associated with cytoskeleton proteins and extracellular matrix
Immature egg chambers are more or less spherical in shape but elongate as they mature and increase in volume during vitellogenesis and yolk uptake. Morphogenesis of the follicular epithelium that surrounds oocytes and nurse cells requires a great deal of cytoskeletal reorganization. A total of eleven ESTs matched significantly to genes putatively coding for various cytoskeleton components (Table 1).
3.2.2 Transcripts associated with transport
A major undertaking during oogenesis is the mobilization of nutrients and substrates to fuel ovarian development and provision eggs. We found a total of eleven ESTs that matched significantly to genes encoding transporters of diverse substrate molecules (Table 1).
3.2.3 Transcripts associated with signal transduction
Considering the extensive inter- and intracellular communication mandated by oogenesis, it was not surprising to see numerous signal transduction molecules represented in the GcaOv EST library (Table 1). Among these, a number of ESTs (AO_031b, _078b, _094b, _110) encode serine proteases involved in protease cascades that play a large role in signaling processes (LeMosy et al., 1999; Schüpbach, 2009). We also found several ESTs (AO_006, _067b, _319) that encode putative small GTPases Ran and Arf involved in G-protein signaling.
During the course of construction of the subtracted GcaOv library, our lab had initiated construction of a second subtracted library to isolate transcripts uniquely expressed in brains of Gc. atropalpus undergoing autogenous oogenesis (unpublished data). We applied similar procedures for cDNA preparation and subtractive hybridization as those described for construction of the GcaOv library and used heads (brains) during autogenous oogenesis as the “tester” population and heads from newly anautogenous females as the “driver” population. BLAST analysis of one particular cDNA clone (AHd_03, accession number GT154744) revealed only one significant hit within arthropods. This clone has been putatively identified as a homolog of Ae. aegypti small GTPase of the Ras-like family. This EST warranted consideration as part of this study as it surprisingly showed greater expression in ovaries from autogenous females compared to head tissues and was preferentially expressed in ovaries of females derived from well-fed larvae during preliminary RT-PCR analyses (data not shown). A Drosophila homolog of the small GTPase of the Ras-like family was recently identified as playing a key regulatory role in determining the fate of polar cells during oogenesis (Ghiglione et al., 2008). Quantitative data on EST AHd_03 expression is given in Section 3.4.
A number of ESTs (_100, _295, _303) encode cellular protein kinases and phosphatases that govern reversible phosphorylation events, and other ESTs in this category (AO_014a, _122, _126, _233a, _317) encode products playing diverse roles in signal cascades. For example, EST AO_014a (accession number GT154627) encodes a putative oxysterol binding protein (OSBP) that binds cholesterol and plays a large role in mediating cholesterol homeostasis in mammals. Oxysterols are oxygenated derivatives of cholesterol that repress synthesis and uptake of cholesterol. Members of the large family of OSBP-related proteins (ORPs) play critical roles in cellular lipid metabolism, vesicle transport and cell signaling (Lehto and Olkkonen, 2003) and are vital for embryonic development in mice (Im et al., 2005). Recent structural analyses suggest OSBP may figure significantly in sterol transfer and signaling (Im et al., 2005). The method by which mosquito ovaries acquire cholesterol has not been determined but it seems clear that cholesterol trafficking would be important during oogenesis for membrane support and synthesis but also for steroid biosynthesis. In insects, homologs of mammalian OSBP have been identified in Drosophila (Alphey et al., 1998; Ma et al., 2010), Ae. aegypti (Fu et al., 2011) and now in Gc. atropalpus. Our RT-PCR analysis found OSBP products in both midgut and ovaries (Fig 1), further supporting its role in cholesterol trafficking. Quantitative details for OSBP are given in Results section 3.4.
Figure 1.
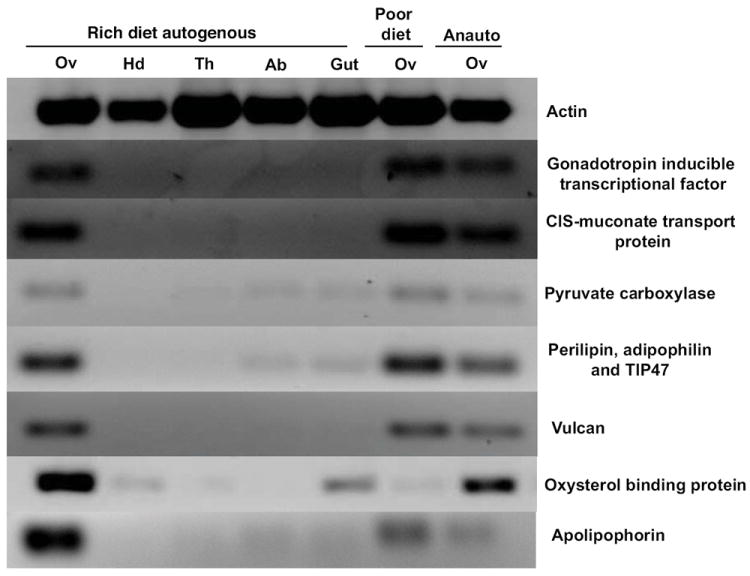
RT-PCR analysis of the tissue distribution of six gene transcripts in relation to female nutritional condition and reproductive stage along with tissue-specific and loading controls (Gc. atropalpus actin). Rich diet autogenous = tissues collected from autogenous females derived from larvae fed ad libitum. Poor diet = ovaries only were collected from autogenous females derived from larvae fed low amounts of food. Anauto = ovaries from newly anautogenous females that are developmentally arrested at Christopher’s stage 1. Ov = ovary pairs, Hd = head, Th = thorax, Ab = abdominal wall, Gu = gut.
3.2.4 Transcripts associated with nuclear regulation
ESTs placed in this category matched significantly to genes that play a role in maintaining the structure and function of DNA and chromatin (Table 1). For example, we found two ESTs (AO_049b, _093b) that showed high similarity to a gene for ribonucleotide reductase (RNR), an enzyme that catalyzes de novo synthesis of deoxyribonucleotides needed to support cell division. EST AO_049b matched specifically to Ae. aegypti Class I R2 subunit of the RNR holoenzyme. Earlier reports indicate RNR enzyme activity as a rate-limiting step in DNA synthesis and that these RNR subunits are significantly induced in Ae. aegypti after blood feeding, with specific induction of R2 in ovaries (Pham et al., 2002; Pham et al., 2006). The presence of transcripts for an RNR-R2 homolog in our GcaOv library suggests this protein is similarly upregulated during autogenous oogenesis.
3.2.5 Transcripts associated with transcriptional machinery
Using larval-derived nutrient reserves, Gc. atropalpus females initiate egg development shortly after emergence and typically oviposit eggs 48 – 72 hrs post-eclosion. Production of proteins needed to support egg development requires hormonal activation of the transcriptional machinery. Oocyte-specific transcriptional control is required to coordinate expression of genes involved in both oocyte and early embryonic development. A total of fifteen sequences were associated with transcriptional machinery elements (Table 1). BLAST analysis identified EST AO_012 as a putative gonadotropin inducible transcriptional factor (GITF) (accession number GT154626). No putative orthologs were found in vertebrates, suggesting GITF is a novel transcription factor among arthropods. Earlier studies using Ae. aegypti showed that the neuropeptide ovarian ecdysteroidogenic hormone (OEH) (Matsumoto et al., 1989; Brown et al., 1998) is released from the medial neurosecretory cells (MNC) in the female brain at a critical time after blood feeding and is crucial for oogenesis to resume from its arrested state (Klowden, 1997). More recently, OEH was also found to regulate egg maturation in Gc. atropalpus (Gulia-Nuss et al. 2012). Transcription factor GITF has not been studied in mosquitoes but it is tempting to speculate on its potential role as a regulatory link between the gonadotropin OEH and genes involved in ecdysteroid production. In D. melanogaster, the best BLAST match to the putative GITF homolog in the GcaOv library was the gene suppressor of Hairy wing (Dmel\su(Hw), Flybase ID FBgn0003567). Su(Hw) protein seems to play an essential role in D. melanogaster oogenesis as mutations at this locus result in larvae developing into sterile but otherwise normal flies (Müller, 2000). Quantitative data on EST GITF expression is given in Section 3.4.
3.2.6 Transcripts associated with protein synthesis and modification
The ovaries of Gc. atropalpus females 18 h post-emergence, the time point used for autogenous transcript collection in this study, are at late second or early third Christopher’s stage. Ovaries at these stages are actively engaged in yolk uptake and are also close to the time when ECD biosynthesis and secretion peaks (Telang et al., 2006). Mobilization of teneral nutrient reserves to support vitellogenesis as well as maintenance of metabolic needs would be taking place at this time. It is not surprising then that we found a total of twenty ESTs that showed significant similarity to products involved in synthesis and modification of proteins supporting these processes (Table 1).
3.2.7 Transcripts associated with intermediary metabolism
Intermediary metabolism refers to the production and use of energy and is represented by the total of catabolic, anabolic, and amphibolic reactions that work together to synthesize and degrade proteins, carbohydrates, and lipids. All cellular processes, including oogenesis, depend on this energy. In other studies regarding mosquito reproduction, transcripts related to products involved in intermediary metabolism have been isolated. In these studies, either whole abdomen or whole body carcasses were used as starting material in order to examine gene expression in multiple tissues involved in egg development. As such, transcripts encoding products functioning as metabolic machinery were often attributed to tissues other than ovaries. A number of ESTs derived from either abdomen tissues (Dana et al., 2005) or whole-body carcasses (Marinotti et al., 2005) were catalogued in the role of intermediary metabolism and attributed to the midgut given the involvement of this organ in blood meal digestion and absorption of amino acid products. Likewise, Raikhel and colleagues have identified specific genes related to amino acid metabolism in the mosquito fat body (Hansen et al., 2004; Attardo et al., 2006). Our transcriptome study found nineteen ESTs that significantly matched products involved in intermediary metabolism (Table 1). These findings suggest that the ovaries may also serve as a tractable system to study this aspect of cell metabolism.
We also found a large number of ESTs involved in lipid metabolism (AO_023, _54a, _141b, _268, _294). EST AO_023 encodes a putative perilipin, a lipid droplet-associated protein and modulator of lipid metabolism (Miura et al., 2002). Lipids make up a large part of insect yolk, and studies of the D. melanogaster perilipin homolog Lsd2 showed it was predominately expressed in the fat body and female germ line, tissues exhibiting high levels of lipid metabolism. Further, ovaries of Lsd2 mutant flies showed an abnormal pattern of lipid accumulation in oocytes that resulted in reduced lipid deposition in eggs (Teixeira et al., 2003). While Drosophila studies show that perilipin plays a lipid storage role in oocytes, studies in mammals have found perilipin transcript and protein expression in adipocytes and steroidogenic cells (Kimmel et al., 2010). It is likewise interesting that EST AO_268 encodes a putative glycoprotein sc2 that bears sequence similarity to steroid 5-alpha-reductase, an enzyme involved in steroid metabolism in mammals (Johnston et al., 1992). Whether the perilipin and glycoprotein sc2 homologs identified in this study play a dual role in lipid metabolism and hormone biosynthesis in mosquito ovaries awaits further study. EST AO_054 from the GcaOv library displayed significant homology to the lipid transfer protein apolipophorin III (Apo III; accession number GT154652). Previous work using Ae. aegypti determined that lipophorin was synthesized in fat body and its transcript expression was induced by blood feeding (van Heusden et al., 1998). A gene encoding a lipophorin receptor in Ae. aegypti ovaries has been characterized and it shows peak expression after a blood meal (Seo et al., 2003). Details of our work with Apo III will be further discussed in Section 3.4.
Given the ecdysteroidogenic activity of mosquito ovaries, we were pleased that several ESTs isolated in this study appear to encode proteins with steroidogenic roles (Table 1). One EST (AO_134b) matched significantly to a cytochrome P450 enzyme. While these enzymes contribute to a diversity of processes such as carbon source assimilation, metabolism of xenobiotics, cholesterol metabolism in addition to the role of hormone biosynthesis (Werck-Reichhart and Feyereisen, 2000), the location of this EST in ovaries warrants investigation for its potential role in steroidogenesis. EST AO_288 matched significantly to a gene putatively coding for cytochrome b5, a small membrane-bound electron carrier hemoprotein that is now thought to influence properties of cytochrome P450 functions (Murataliev et al., 2008).
3.2.8 Transcripts associated with oogenesis and embryogenesis
Transcripts placed in the category of cytoskeletal proteins and extracellular matrix (see Section 3.2.1 above) likely support the rapid overall structural development of the ovaries during oogenesis as well and so could also be described here. A total of seven transcripts were placed in this category to reflect their roles in processes supporting embryogenesis. EST AO_019 codes for an endochorion protein (previously referred to as a vitelline membrane or envelope protein). Endochorion proteins are secreted by the follicular epithelium during the ECD peak following a blood meal in anautogenous Ae. aegypti and coalesce to form the innermost layer of eggs (Edwards et al., 1998). We also found ESTs (AO_128b, _203a) that putatively encode oskar and exuperantia, proteins derived from maternal transcripts stored in the oocyte to facilitate embryo development (Riechmann and Ephrussi, 2004; Juhn and James, 2006).
3.2.9 Transcripts with unknown function
Among the large number of predicted protein-encoding sequences that emerged from the Ae. aegypti genome project, some were designated as coding for either “hypothetical proteins” or “conserved hypothetical proteins” during annotation if criteria necessary for more specific associations were not met (Nene et al., 2007). We found a total of eighteen ESTs in the GcaOv library that code for products with currently unknown functions that were upregulated during autogenous egg production. One of the more interesting ESTs in this category is AO_052 (accession # GT154650, Table 1). In D. melanogaster, the best BLAST match to this EST was the gene vulcan (Flybase ID FBgn0259978). Vulcan functions in imaginal disc-derived leg morphogenesis during fly metamorphosis, a process that is driven by epithelial morphogenesis. During fly metamorphosis this process is dependent on pulses of ecdysone, so vulcan may serve as a link between the hormone signal and epithelial morphogenesis (Gates and Thummel, 2000). Whether any of these ESTs with currently unknown functions also represent novel proteins in this autogenous mosquito awaits further investigation.
3.3 Tissue distribution of transcripts of interest in Gc. atropalpus
We used RT-PCR to localize tissue expression of seven of the genes represented by the ESTs in Table 1. In order to be selected for RT-PCR analysis, ESTs were required to have a best-match E-value lower than e-30 and be at least 75% identical to the Ae. aegypti sequences to which they were most similar. Candidate genes were chosen according to putative functions in nutrient transport, ecdysteroid biosynthesis, ecdysone responsiveness, or egg development. Expression patterns for these genes are shown (Fig 1). With the exception of oxysterol binding protein (OSBP), all of the genes analyzed were more abundant in ovaries than the head, thorax or abdomen regions. Further, transcripts putatively encoding the proteins Vulcan (AO_052), OSBP and APO III were found to be more abundant in the ovaries of females derived from well-fed larvae compared to the ovaries of autogenous females fed a poor diet as larvae. The actin product was amplified in all samples, thus confirming the integrity and consistency of the RNA and cDNA preparations.
3.4 Quantitative PCR analysis of transcripts of interest in Gc. atropalpus ovaries
A number of the genes identified in the GcaOv library appeared to be good candidates for oogenic markers cued to female nutritional status, so we analyzed these templates further using qPCR. We focused our attention on the OSBP (AO_014a, accession number GT154627), Apo III (AO_054a, accession number GT154652), GITF (AO_012, accession number GT154626) genes analyzed previously by RT-PCR (see expression profiles in Fig 1) and on a small GTPase identified in a mosquito head cDNA library (AHd_03, accession number GT154744). As Figure 2A-D shows, all four genes displayed a rise and subsequent fall in transcript abundance in Gc. atropalpus ovaries over the course of autogenous egg developmental stages 1-4 (ANOVA p<0.0001 for each of the transcripts). Expression levels were greatest in Stage 2/3 ovaries in which peak ecdysteroid production and, presumably, nutrient mobilization takes place (p<0.0001, from a linear contrast of stage 2/3 vs stages 1 + 4). In contrast, lower transcript numbers were associated with ovaries from females entering their anautogenous reproductive cycle prior to blood feeding compared to ovaries underway in autogenous egg development (p<0.0001, from a linear contrast). For OSBP, its expression pattern in Gc. atropalpus is supported by a recent study reporting up-regulation of OSBP in ovaries of blood-fed Ae. aegypti (Fu et al., 2011). Interestingly, transcripts from all four genes were also most abundant in autogenous ovaries from females derived from well-nourished larvae compared to ovaries from females poorly nourished as larvae (p<0.0001, from a linear contrast), suggesting these transcripts may indeed be cued to female nutritional condition (Figure 2A-D).
Figure 2.
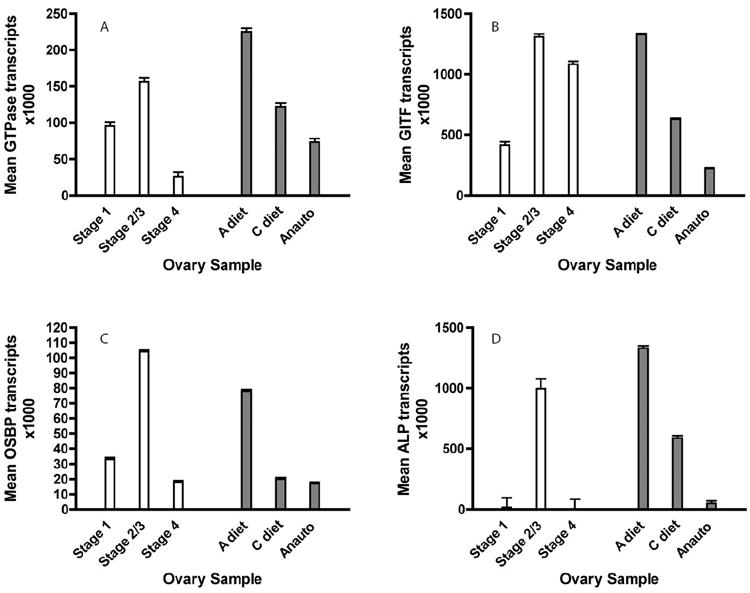
Real-time quantitative PCR (qPCR) analysis of gene transcripts in ovaries at discrete stages of Gc. atropalpus autogenous follicle development (Stages 1 – 4, white bars), in relation to rich- or poor- female nutritional condition during autogenous oogenesis (A -, C-diet, gray bars), and reproductive state (anautogenous, follicles arrested at Christophers stage I, gray bars). Graphs display mean estimated transcript copies X 1000 ± s.e.m for (A) GcaGTPase (Ras-small GTPase), (B) GcaGITF (gonadotropin inducible transcription factor), (C) GcaOSBP (oxysterol binding protein), and (D) GcaALP (apolipophorin III). Error bars represent s.e.m.
3.5. RNAi bioassays in Ae. aegypti females
Quantitative PCR analysis confirmed dsRNA treatment led to lower transcript levels. Injection of dsAPO led to about 28-95% reduction of APO transcripts among tissues examined (ANOVA p<0.0001), with the greatest reduction in the FB/Abdomen (p<0.05, Tukey) (Fig 3A). Injection of dsOSBP led to a 24-40% reduction of OSBP transcripts among tissues (ANOVA p<0.0001) but a 7-fold increase in head tissue (p<0.05, Tukey) (Fig 3B). We used relative quantitation with actin as our normalizer after confirming that actin transcript was not affected by dsRNA treatment (ANOVA p=0.120). We utilized an ecdysteroid EIA to quantify ovarian ecdysteroid (Ecd) synthesis in response to knockdown of APO or OSBP transcripts in order to determine a role for APO or OSBP products in Ecd production. We found suggestive evidence that Ecd levels were reduced in dsOSBP injected females compared to control (p=0.05) (Fig 4).
Figure 3.
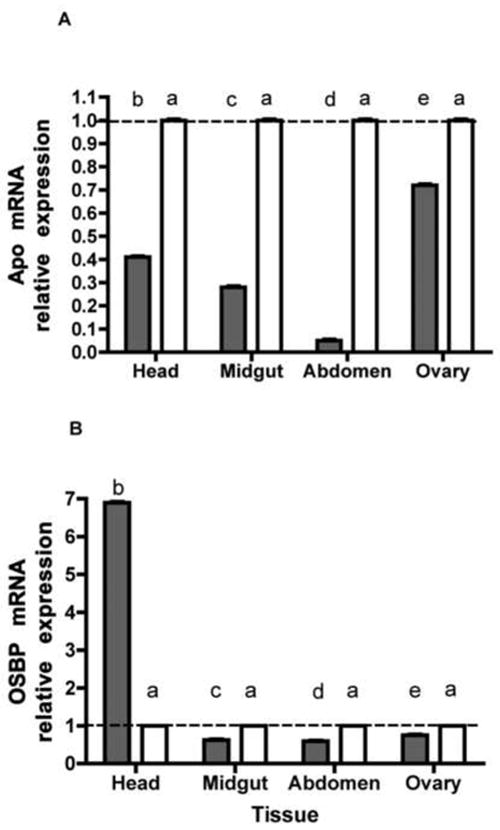
qPCR analysis of (A) APO and (B) OSBP transcript abundance in heads, guts (without blood), abdominal walls (including fat body), and ovaries of Aedes aegypti females in response to gene of interest (gray bars) or EGFP control (white bars) dsRNA treatment. Bars represent normalized levels of transcripts in each tissue with tissues from females treated with EGFP dsRNA chosen as calibrator. Calibrator ratio of 1.0 indicated by dashed line. Within each figure, columns with different letters are significantly different (from a Tukey-Kramer HSD P<0.05). Error bars represent s.e.m.
Figure 4.
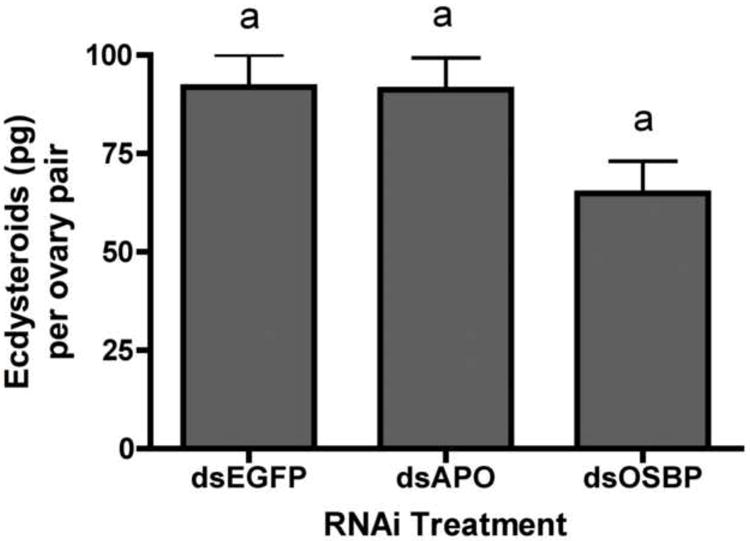
In vitro ecdysteroid secretion by ovary pairs dissected from Aedes aegypti females in response to dsRNA treatment. Columns with different letters are significantly different (from a Tukey-Kramer HSD P<0.05). Error bars represent s.e.m.
Lastly, we hypothesized that OSBP and APO III may play a role in lipid mobilization. We used biochemical assays to determine whole body lipid mobilization from reserves (fatbody, using carcass without ovaries) to ovaries in females treated with dsAPO and dsOSBP. We used biochemical assays to confirm that protein levels were not altered in response to dsRNA treatments. As predicted, no change in protein reserves occurred in non-blood fed females when injected with dsAPO, dsOSBP or dsEGFP control (p=0.52) (Fig 5A). At 24 PBM, injection of dsAPO or dsOSBP had affected neither carcass nor ovarian protein levels (Fig 5A). In contrast, carcass lipid levels were significantly reduced in dsAPO-treated females compared to dsEGFP control prior to blood feeding and remained low at 24 PBM (p<0.0001, from linear contrasts in regards to both time points) (Fig 5B). On the other hand, carcass lipid level was significantly higher in dsOSBP-treated females compared to dsEGFP control both before blood feeding and 24 hr PBM (p<0.0001, from linear contrasts in regards to both time points) but no change was seen in ovarian lipid level in dsOSBP-treated females (p=0.13). We did, however, observe reduced lipid level in ovaries from dsAPO-treated females compared to control (p<0.0001, from linear contrast) (Fig 5B). This was supported by our observation of a reduced number of primary follicles and delayed development of follicles in dsAPO-treated females compared to controls (data not shown).
Figure 5.
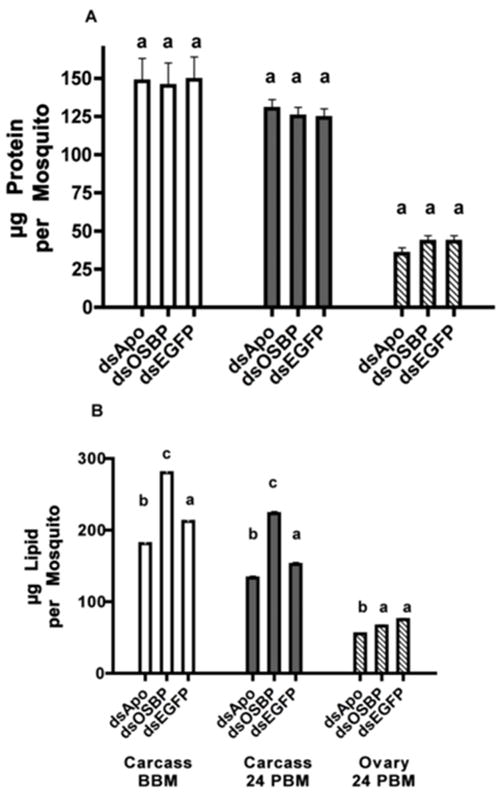
Effect of dsRNA treatment on amount of nutrients in Aedes aegypti females. (A) Protein and (B) lipid amounts in carcasses from females treated with dsRNA before blood feeding (white bars), in carcasses (without ovaries) from females treated with dsRNA, 24 h after blood feeding (dark gray bars), and in ovaries from females treated with dsRNA, 24 h after blood feeding (striped bars). Within grouped bars for each figure, columns with different letters are significantly different (from linear contrast test P<0.001). Error bars represent s.e.m.
4. Concluding remarks
Anautogenous mosquitoes are obligatory blood feeders that use the nutrients in blood to provision their eggs. Ovaries of newly emerged anautogenous species are halted at an early stage of development. Upon ingestion of a blood meal, a complex hormonal cascade ensues leading to the resumption of ovarian development and culminating in the maturation of a batch of eggs. Autogenous mosquitoes carry over nutrient reserves from their larval stage for both metamorphosis and oogenesis (Telang and Wells, 2004). Unlike those of anautogenous females, the ovaries of newly emerged autogenous females of the species Georgecraigius atropalpus appear to develop without a period of arrest, producing the ecdysteroids associated with egg maturation in the absence of a blood meal (Telang et al., 2006). Research on mosquito reproduction has relied heavily on the much-used anautogenous Aedes aegypti. However, a critical barrier to our understanding of how both nutrients and hormones regulate oogenesis has been the complex physiological background of blood feeding itself. Our utilization of the autogenous mosquito, Gc. atropalpus, to initiate our transcriptome study is a novel approach to this research area and has already contributed a suite of candidate genes for further analysis in Ae. aegypti.
Despite the small number of ESTs sequenced in this study, our focused transcriptome analysis of autogenous ovaries identified a suite of genes directly relevant to processes supporting egg maturation. For example, the clone AO_054a showed highest homology to Aea apolipophorin-III (Apo). APO transcript knockdown at the whole-animal level in Ae. aegypti resulted in reduced lipid levels in both carcass and ovary. It is possible that the encoded APO product is needed to “lock-up” lipids for egg development, and, in the face of reduced APO protein, the animal uses lipid for somatic maintenance instead. Alternatively, APO may be used both to transport lipid from the fatbody to the ovaries and to transport lipid from a blood meal to the fat body. Thus, knockdown of APO could mean less that fat body lipid is being mobilized for somatic maintenance and more that the lipid is not being properly taken up from the bloodmeal to begin with.
Oxysterols are oxygenated derivatives of cholesterol and are thought to repress the synthesis and uptake of cholesterol in addition to performing a number of other important functions. Oxysterol binding protein (OSBP) binds a range of oxysterols to regulate sterol metabolism and regulatory actions of oxysterols. We observed that OSBP transcript knockdown at the whole-animal level in Ae. aegypti resulted in higher lipid levels in carcass and possibly leads to a reduction in ecdysteroid production. This suggests OSBP may play a role in overall lipid and sterol homeostasis. Our gene functional assays are supported by a recent study in which transfected Ae. aegypti Aag-2 cells increased cholesterol uptake when over-expressing AeOSBP (Fu et al., 2011). Interestingly, our finding that OSBP transcript levels increased in the head (brains) in dsOSBP-treated females, even as transcript levels decreased elsewhere, suggests that the head may be the primary site of OSBP synthesis. If this were the case, we would expect this site to be responsive to a drop in OSBP transcript levels elsewhere in the body through a feedback mechanism involving OSBP directly or perhaps via some intermediary effector molecule originating in the body.
The majority of ESTs in our study matched significantly with proteins of known function, but we also found nineteen ESTs that did not match significantly to characterized proteins at the time of our analysis. Identification and characterization of these genes may expand our understanding of autogeny and hematophagy as well as provide new potential intervention points for the regulation of mosquito egg production. We will continue to use this comparative approach between Gc. atropalpus and Ae. aegypti to elucidate functions of key genes involved in egg production.
Supplementary Material
Research Highlights.
Autogenous mosquito Gc. atropalpus offers a study of oogenesis sans bloodmeal.
SSH of transcripts from autogenous ovaries identified ESTs relevant to oogenesis.
RNAi studies in Ae. aegypti revealed potential functions of specific genes.
Knockdown of APO transcripts lead to reduced lipid levels in tissues.
Knockdown of OSBP transcripts suggests a role in ecdysteroid production.
Acknowledgments
We thank John Frey for assistance in rearing mosquitoes used in this study. This work was funded by NIH/NIAID grant (K22AI070644) to A.T. The conjugated ecdysone and primary antiserum were purchased from Dr. Tim Kingan. The authors thank Dr Tim Kingan and Dr Dale Gelman for their assistance in helping to establish the EIA in our laboratory.
Footnotes
Appendix A. Supplementary data
Table 2. Primers used for quantitative Real-Time PCR (qPCR) analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alphey L, Jimenez J, Glover D. A Drosophila homologue of oxysterol binding protein (OSBP) - implications for the role of OSBP. Biochimica et Biophysica Acta. 1998;1395:159–164. doi: 10.1016/s0167-4781(97)00159-0. [DOI] [PubMed] [Google Scholar]
- Attardo G, Hansen IA, Shiao S-H, Raikhel AS. Identification of two cationic amino acid transporters required for nutritional signaling during mosquito reproduction. Journal of Experimental Biology. 2006;209:3071–3078. doi: 10.1242/jeb.02349. [DOI] [PubMed] [Google Scholar]
- Brown MR, Graf R, Swiderek KM, Fendley D, Stracker TH, Champagne DE, Lea AO. Identification of a steroidogenic neurohormone in female mosquitoes. The Journal of Biological Chemistry. 1998;273:3967–3971. doi: 10.1074/jbc.273.7.3967. [DOI] [PubMed] [Google Scholar]
- Christophers SR. The development of the egg follicle in Anophelines. Paludism. 1911;2:73–87. [Google Scholar]
- Clements AN. The Biology of Mosquitoes. Vol. 1. Chapman & Hall; London: 1992. [Google Scholar]
- Dana AN, Hong YS, Kern MK, Hillenmeyer ME, Harker BW, Lobo NF, Hogan JR, Romans P, Collins FH. Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genomics. 2005 doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YFC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MJ, Severson DW, Hagedorn HH. Vitelline envelope genes of the yellow fever mosquito, Aedes aegypti. Insect Biochemistry and Molecular Biology. 1998;28:915–925. doi: 10.1016/s0965-1748(98)00083-6. [DOI] [PubMed] [Google Scholar]
- Fu Q, Lynn-Miller A, Lan Q. Characterization of the oxysterol-binding protein gene family in the yellow fever mosquito, Aedes aegypti. Insect Molecular Biology. 2011;20:541–552. doi: 10.1111/j.1365-2583.2011.01087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J, Thummel CS. An enhancer trap screen for ecdysone-inducible genes required for Drosophila adult leg morphogenesis. Genetics. 2000;156:1765–1776. doi: 10.1093/genetics/156.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Cerezo D, Noselli S. Drosophila RalA is essential for the maintenance of Jak/Stat signalling in ovarian follicles. EMBO Reports. 2008;9:676–682. doi: 10.1038/embor.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M, Jai-Hoon E, Strand MR, Brown MR. Ovary ecdysteroidogenic hormone activates egg maturation in the mosquito Georgecraigius atropalpus after adult eclosion or a blood meal. The Journal of Experimental Biology. 2012;215:3758–3767. doi: 10.1242/jeb.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen IA, Attardo G, Park J, Peng Q, Raikhel AS. Target of rapamysin-mediated amino acid signaling in mosquito anautogeny. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10626–10631. doi: 10.1073/pnas.0403460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JY, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston IG, Rush SJ, Gurd JW, Brown IR. Molecular cloning of a novel mRNA using an antibody directed against synaptic glycoproteins. Journal of Neuroscience Research. 1992;32:159–166. doi: 10.1002/jnr.490320205. [DOI] [PubMed] [Google Scholar]
- Juhn J, James AA. oskar gene expression in the vector mosquitoes, Anopheles gambiae and Aedes aegypti. Insect Molecular Biology. 2006;15:363–372. doi: 10.1111/j.1365-2583.2006.00655.x. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of perilipin as a unifying nomenclature for the mammalian PAT-family of intracellular, lipid storage droplet proteins. Journal of Lipid Research. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingan TG. A competitive enzyme-linked immunosorbent assay: applications in the assay of peptides, steroids, and cyclic nucleotides. Analytical Biochemistry. 1989;183:283–289. doi: 10.1016/0003-2697(89)90481-8. [DOI] [PubMed] [Google Scholar]
- Klowden MJ. Endocrine aspects of mosquito reproduction. Archives of Insect Biochemistry and Physiology. 1997;35:491–512. [Google Scholar]
- Lehto M, Olkkonen VM. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochimica et Biophysica Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- LeMosy EK, Hong CC, Hashimoto C. Signal transduction by a protease cascade. Trends in Cell Biology. 1999;9:102–107. doi: 10.1016/s0962-8924(98)01494-9. [DOI] [PubMed] [Google Scholar]
- Ma Z, Lie Z, Huang X. OSBP- and FAN-mediated sterol requirement for spermatogenesis in Drosophila. Development. 2010;137:3775–3784. doi: 10.1242/dev.049312. [DOI] [PubMed] [Google Scholar]
- Margam VM, Gelman DB, Palli SR. Ecdysteroid titers and developmental expression of ecdysteroid-regulated genes during metamorphosis of the yellow fever mosquito, Aedes aegypti (Diptera: Culicidae) Journal of Insect Physiology. 2006;52:1–11. doi: 10.1016/j.jinsphys.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JMC. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Molecular Biology. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Brown MR, Suzuki A, Lea AO. Isolation and characterization of ovarian ecdysteroidogenic hormones from the mosquito, Aedes aegypti. Insect Biochemistry. 1989;19:651–656. [Google Scholar]
- Miura S, Gan J-W, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. Journal of Biological Chemistry. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- Müller J. Transcriptional control: The benefits of selective insulation. Current Biology. 2000;10:R241–R244. doi: 10.1016/s0960-9822(00)00374-2. [DOI] [PubMed] [Google Scholar]
- Murataliev MB, Guzov VM, Walker FA, Feyereisen R. P450 reductase and cytochrome b5 interactions with cytochrome P450: Effects on house fly CYP6A1 catalysis. Insect Biochemistry and Molecular Biology. 2008;38:1008–1015. doi: 10.1016/j.ibmb.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara GF. Variable expressions of autogeny in three mosquito species. International Journal of Invertebrate Reproduction. 1979;1:253–261. [Google Scholar]
- O’Meara GF. Gonotrophic interactions in mosquitos - kicking the blood-feeding habit. Florida Entomologist. 1985;68:122–133. [Google Scholar]
- Paton MG, Karunaratne SH, Giakoumaki E, Roberts N, Hemingway J. Quantitative analysis of gene amplification in insecticide-resistant Culex mosquitoes. Biochemical Journal. 2000;346:17–24. [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham DQD, Blachuta BJ, Nichol H, Winzerling JJ. Ribonucleotide reductase subunits from the yellow fever mosquito, Aedes aegypti: cloning and expression. Insect Biochemistry and Molecular Biology. 2002;32:1037–1044. doi: 10.1016/s0965-1748(02)00041-3. [DOI] [PubMed] [Google Scholar]
- Pham DQD, Kos PJ, Mayo JJ, Winzerling JJ. Regulation of the ribonucleotide reductase small subunit (R2) in the yellow fever mosquito, Aedes aegypti. Gene. 2006;372:182–190. doi: 10.1016/j.gene.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Riechmann V, Ephrussi A. Par-1 regulates bicoid mRNA localisation by phosphorylating Exuperantia. Development. 2004;131:5897–5907. doi: 10.1242/dev.01515. [DOI] [PubMed] [Google Scholar]
- Schüpbach T. Developmental biology: Pipe’s smoking guns. Current Biology. 2009;19:R548–R550. doi: 10.1016/j.cub.2009.05.053. [DOI] [PubMed] [Google Scholar]
- Seo S-J, Cheon H-M, Sun J, Sappington TW, Raikhel AS. Tissue- and stage-specific expression of two lipophorin receptor variants with seven and eight ligand-binding repeats in the adult mosquito. The Journal of Biological Chemistry. 2003;278:41954–41962. doi: 10.1074/jbc.M308200200. [DOI] [PubMed] [Google Scholar]
- Sieglaff DH, Duncan KA, Brown MR. Expression of genes encoding proteins involved in ecdysteroidogenesis in the female mosquito, Aedes aegypti. Insect Biochemistry and Molecular Biology. 2005;35:471–490. doi: 10.1016/j.ibmb.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Rabouille C, Rørth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mechanisms of Development. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Telang A, Li Y, Noriega FG, Brown MR. Effects of larval nutrition on the endocrinology of mosquito egg development. Journal of Experimental Biology. 2006;209:645–655. doi: 10.1242/jeb.02026. [DOI] [PubMed] [Google Scholar]
- Telang A, Wells MA. The effect of larval and adult nutrition on successful autogenous egg production by a mosquito. Journal of Insect Physiology. 2004;50:677–685. doi: 10.1016/j.jinsphys.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Van Handel E. Microseparation of glycogen, sugars, and lipids. Analytical Biochemistry. 1965;11:266–271. doi: 10.1016/0003-2697(65)90014-x. [DOI] [PubMed] [Google Scholar]
- Van Handel E. Rapid determination of total lipids in mosquitoes. Journal of the American Mosquito Control Association. 1985;1:302–304. [PubMed] [Google Scholar]
- Van Heusden MC, Thompson F, Dennis J. Biosynthesis of Aedes aegypti lipophorin and gene expression of its apolipoproteins. Insect Biochemistry and Molecular Biology. 1998;28:733–738. doi: 10.1016/s0965-1748(98)00068-x. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biology. 2000;1:3003.1–3003.9. doi: 10.1186/gb-2000-1-6-reviews3003. http://genomebiology.com/2000/1/6/reviews/3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Flowers M, Friedrich K, Horton J, Pennington J, Wells MA. Metabolic fate of [14C]-labeled meal protein amino acids in Aedes aegypti mosquitoes. Journal of Insect Physiology. 2004;50:337–349. doi: 10.1016/j.jinsphys.2004.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


