Abstract
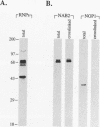
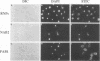
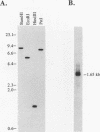
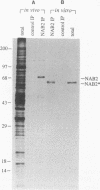
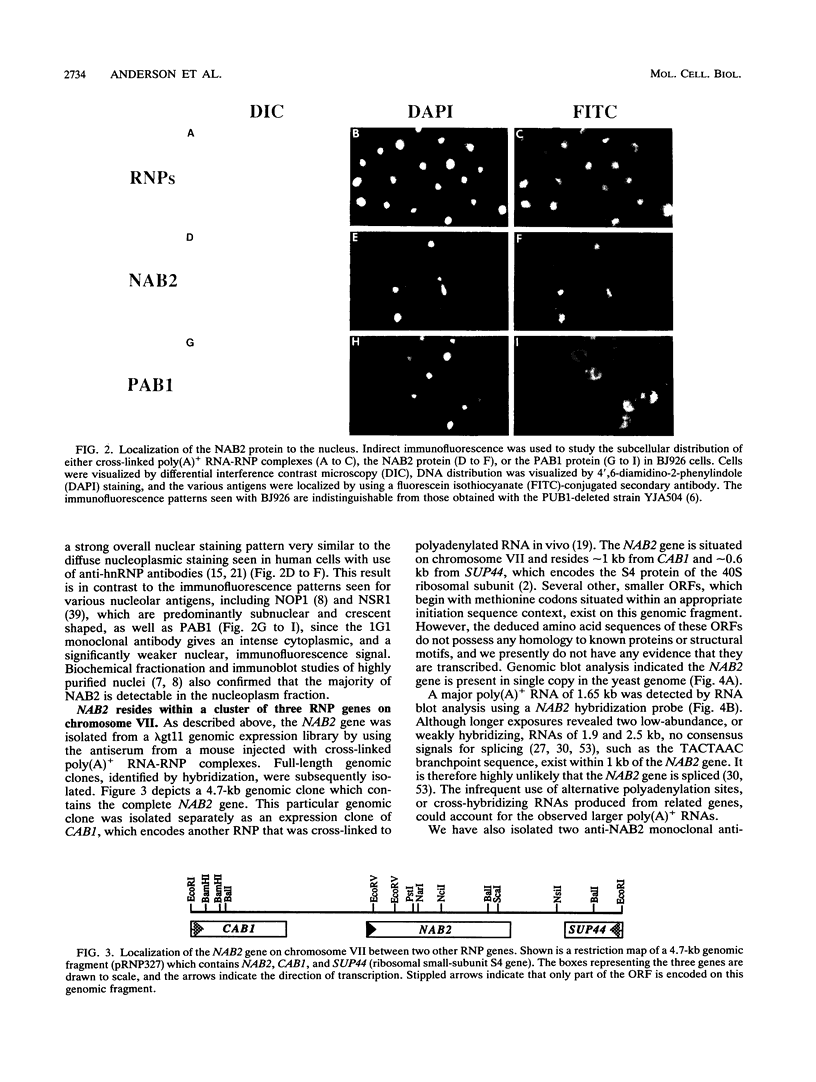
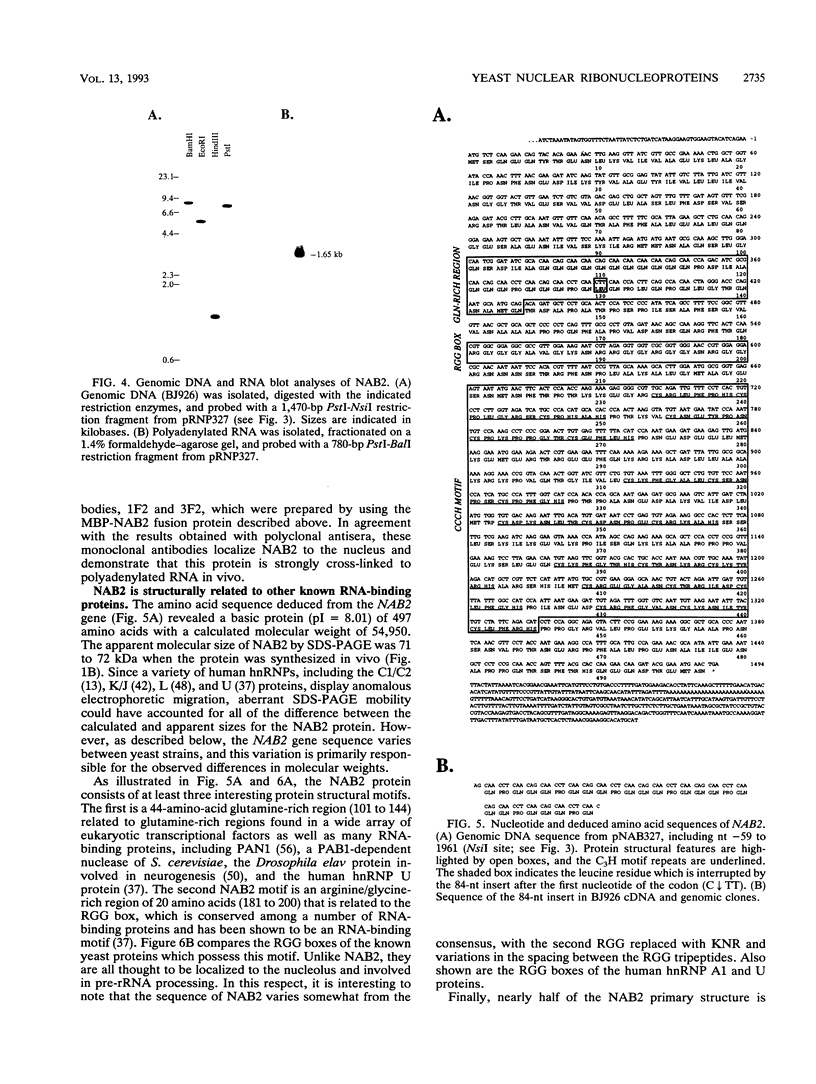
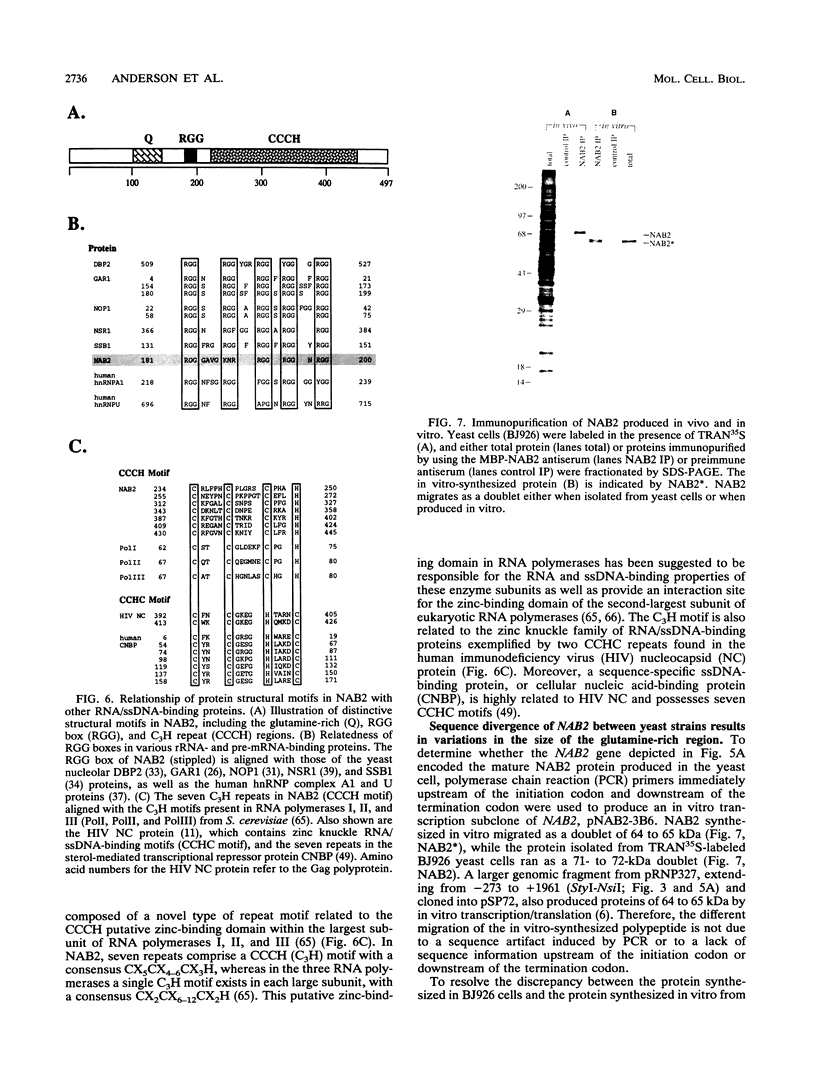
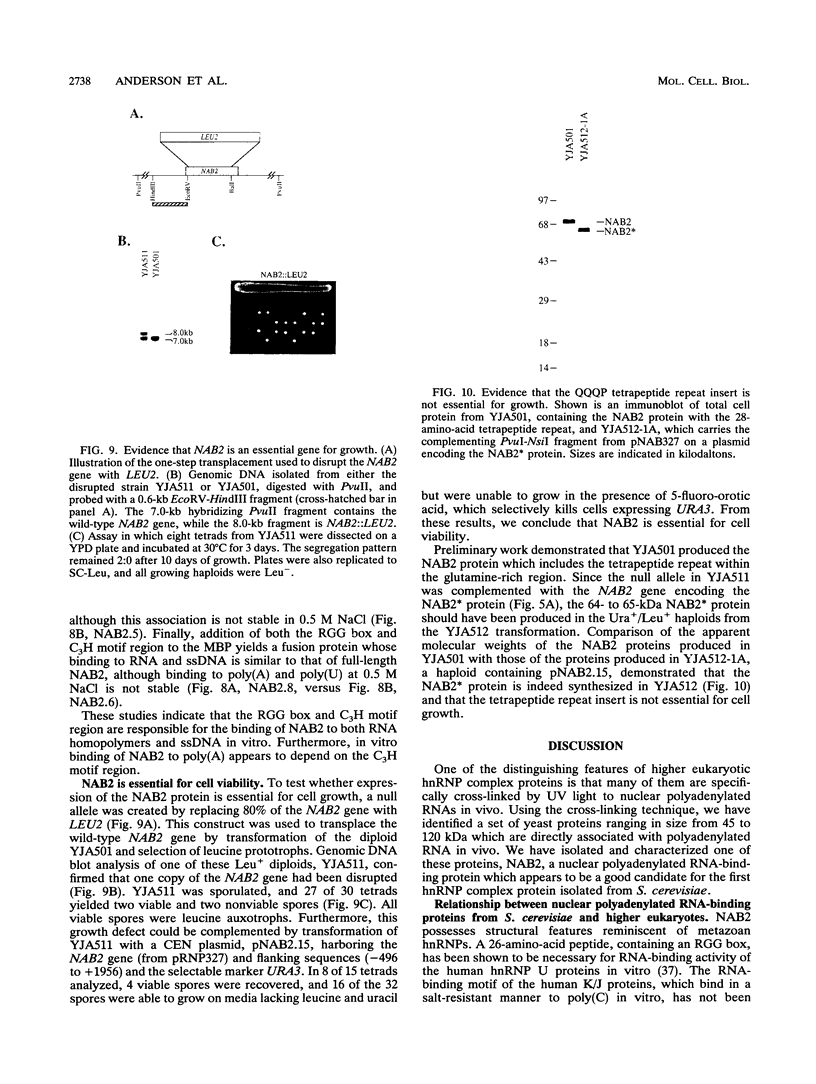
A variety of nuclear ribonucleoproteins are believed to associate directly with nascent RNA polymerase II transcripts and remain associated during subsequent nuclear RNA processing reactions, including pre-mRNA polyadenylation and splicing as well as nucleocytoplasmic mRNA transport. To investigate the functions of these proteins by using a combined biochemical and genetic approach, we have isolated nuclear polyadenylated RNA-binding (NAB) proteins from Saccharomyces cerevisiae. Living yeast cells were irradiated with UV light to covalently cross-link proteins intimately associated with RNA in vivo. Polyadenylated RNAs were then selectively purified, and the covalent RNA-protein complexes were used to elicit antibodies in mice. Both monoclonal and polyclonal antibodies which detect a variety of NAB proteins were prepared. Here we characterize one of these proteins, NAB2. NAB2 is one of the major proteins associated with nuclear polyadenylated RNA in vivo, as detected by UV light-induced cross-linking. Cellular immunofluorescence, using both monoclonal and polyclonal antibodies, demonstrates that the NAB2 protein is localized within the nucleus. The deduced primary structure of NAB2 indicates that it is composed of at least two distinct types of RNA-binding motifs: (i) an RGG box recently described in a variety of heterogeneous nuclear RNA-, pre-rRNA-, mRNA-, and small nucleolar RNA-binding proteins and (ii) CCCH motif repeats related to the zinc-binding motifs of the largest subunit of RNA polymerases I, II, and III. In vitro RNA homopolymer/single-stranded DNA binding studies indicate that although both the RGG box and CCCH motifs bind poly(G), poly(U), and single-stranded DNA, the CCCH motifs also bind to poly(A). NAB2 is located on chromosome VII within a cluster of ribonucleoprotein genes, and its expression is essential for cell growth.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986 Aug;6(8):2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- All-Robyn J. A., Brown N., Otaka E., Liebman S. W. Sequence and functional similarity between a yeast ribosomal protein and the Escherichia coli S5 ram protein. Mol Cell Biol. 1990 Dec;10(12):6544–6553. doi: 10.1128/mcb.10.12.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Goldstein A. L., Cole C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992 Jul;6(7):1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Amero S. A., Raychaudhuri G., Cass C. L., van Venrooij W. J., Habets W. J., Krainer A. R., Beyer A. L. Independent deposition of heterogeneous nuclear ribonucleoproteins and small nuclear ribonucleoprotein particles at sites of transcription. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8409–8413. doi: 10.1073/pnas.89.18.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988 Jul;107(1):17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Bennett M., Piñol-Roma S., Staknis D., Dreyfuss G., Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992 Jul;12(7):3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Burd C. G., Matunis E. L., Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991 Jul;11(7):3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Swanson M. S., Görlach M., Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Pepperkok R., Sproat B. S., Ansorge W., Swanson M. S., Lamond A. I. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 1991 Jul;10(7):1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R. Different types of messenger RNA editing. Annu Rev Genet. 1991;25:71–88. doi: 10.1146/annurev.ge.25.120191.000443. [DOI] [PubMed] [Google Scholar]
- Chung S. Y., Wooley J. Set of novel, conserved proteins fold pre-messenger RNA into ribonucleosomes. Proteins. 1986 Nov;1(3):195–210. doi: 10.1002/prot.340010302. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Yen T. J. Multiple determinants of eukaryotic mRNA stability. New Biol. 1989 Nov;1(2):121–126. [PubMed] [Google Scholar]
- Davis L. I., Fink G. R. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell. 1990 Jun 15;61(6):965–978. doi: 10.1016/0092-8674(90)90062-j. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Choi Y. D., Adam S. A. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984 Jun;4(6):1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M. S., Piñol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988 Mar;13(3):86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Elliott D. J., Bowman D. S., Abovich N., Fay F. S., Rosbash M. A yeast splicing factor is localized in discrete subnuclear domains. EMBO J. 1992 Oct;11(10):3731–3736. doi: 10.1002/j.1460-2075.1992.tb05458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W., Stutz F., Rosbash M., Wickens M. Defects in mRNA 3'-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992 Oct;6(10):1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Girard J. P., Lehtonen H., Caizergues-Ferrer M., Amalric F., Tollervey D., Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992 Feb;11(2):673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Groner B., Phillips S. L. Polyadenylate metabolism in the nuclei and cytoplasm of Saccharomyces cerevisiae. J Biol Chem. 1975 Jul 25;250(14):5640–5646. [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Henríquez R., Blobel G., Aris J. P. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J Biol Chem. 1990 Feb 5;265(4):2209–2215. [PubMed] [Google Scholar]
- Hopper A. K., Traglia H. M., Dunst R. W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990 Aug;111(2):309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo R. D., Jamieson D. J., MacNeill S. A., Southgate J., McPheat J., Lane D. P. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol. 1991 Mar;11(3):1326–1333. doi: 10.1128/mcb.11.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong A. Y., Clark M. W., Gilbert M., Oehm A., Campbell J. L. Saccharomyces cerevisiae SSB1 protein and its relationship to nucleolar RNA-binding proteins. Mol Cell Biol. 1987 Aug;7(8):2947–2955. doi: 10.1128/mcb.7.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Zhao Y., Tartakoff A. M. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992 Jul;11(7):2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H., Marselle L. M. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989 May 5;57(3):493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lee W. C., Xue Z. X., Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991 Apr;113(1):1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E. Nuclear mRNA export. Curr Opin Cell Biol. 1991 Dec;3(6):1004–1012. doi: 10.1016/0955-0674(91)90121-e. [DOI] [PubMed] [Google Scholar]
- Mattox W., Ryner L., Baker B. S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992 Sep 25;267(27):19023–19026. [PubMed] [Google Scholar]
- Matunis M. J., Michael W. M., Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992 Jan;12(1):164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Kataoka N., Shimura Y. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 1990 Dec 11;18(23):6989–6995. doi: 10.1093/nar/18.23.6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992 Feb 20;355(6362):730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S., Swanson M. S., Gall J. G., Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989 Dec;109(6 Pt 1):2575–2587. doi: 10.1083/jcb.109.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavashisth T. B., Taylor A. K., Andalibi A., Svenson K. L., Lusis A. J. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989 Aug 11;245(4918):640–643. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- Robinow S., Campos A. R., Yao K. M., White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988 Dec 16;242(4885):1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Rozen F., Sonenberg N. Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res. 1987 Aug 25;15(16):6489–6500. doi: 10.1093/nar/15.16.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991 Mar;7(3):79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986 Jun 20;45(6):827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Deardorff J. A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992 Sep 18;70(6):961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Snyder M., Elledge S., Sweetser D., Young R. A., Davis R. W. Lambda gt 11: gene isolation with antibody probes and other applications. Methods Enzymol. 1987;154:107–128. doi: 10.1016/0076-6879(87)54073-3. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988 May;8(5):2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Keller W. The biochemistry of 3'-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Yano R., Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991 Feb;11(2):754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Oakes M., Yamaghishi M., Dodd J. A., Nomura M. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Dec;12(12):5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]