Abstract
Cytochrome P4501B1 (Cyp1b1) is expressed specifically in certain neural crest (NC) cells during embryogenesis. Mesenchymal progenitor cells that develop from NC cells are modeled here by mouse C3H10T1/2 and 3T3-L1 cells. Dexamethasone in combination with methylisobutylxanthine (DM) induces Cyp1b1 and a 6.7 kb mouse Cyp1b1 promoter-luciferase reporter in each cell type prior to adipogenesis. An 18 base sequence (at −6.11 kb) (PaxE) which was essential for this reporter stimulation in 3T3-L1 cells bound the transcription factor Pax6. This is shown by gel mobility shifts and sequence mutations. Heterologous vector expression of Pax6 in 3T3-L1 cells enhanced DM stimulated Cyp1b1 promoter activity through cooperation with two Sp1 sites in the proximal promoter region. Chromatin immunoprecipitation showed that DM stimulated binding of Pax6 adjacent to Sp1 in the proximal promoter more than in the PaxE region. The Cyp1b1 induction by DM in C3H10T1/2 cells was more rapid but independent of Pax6. The far upstream enhancer region (FUER) found in rat Cyp1b1 responded to DM but was inactive in the mouse promoter due to key sequence changes. The expression patterns of Pax6 and Cyp1b1 frequently overlap during mouse embryogenesis. The relationship between Pax6 and Cyp1b1 expression warrants further investigation, particularly in the NC.
Keywords: Cyp1b1, Adipogenesis, Pax6, Sp1, PPARγ, Embryo fibroblast
Introduction
Cytochrome P4501B1 (Cyp1b1) is well known for a key role in the metabolic activation of carcinogenic polycyclic aromatic hydrocarbons [1]. However, accumulating data reveal that Cyp1b1 also plays major roles in multiple endogenous cell signaling pathways and in developmental processes. Thus Cyp1b1 is specifically expressed in rhombomere 4 in the developing neural tube [2]. The function remains elusive. There is however evidence for a role in retinoic acid synthesis even though this process is more commonly directed by Aldh1a forms [2]. In endothelia, Cyp1b1 promotes vasculogenesis by detoxifying reactive oxygen species that affect cell morphology [3]. Cyp1b1 was first identified as the predominant cytochrome P450 isoform in mouse embryo fibroblasts (MEFs)2 [4], as an AhR-inducible isoform in human keratinocytes [5] and as a cAMP-induced isoform in rat adrenals [6]. Cyp1b1 is also substantially expressed in epithelial cells located in skin, uterus, mammary and the prostate [7, 8]. Epigenetic effects provided by promoter methylation have a major impact on the cell selectivity of Cyp1b1 expression [9], providing another mechanism of cell specific control and again suggesting important physiologic roles for Cyp1b1.
Cyp1b1 plays an important role in human and mouse eye development [10–12]. The unique temporal and positional expression of Cyp1b1 in developing mouse embryos [13] and zebra fish [14] as well as the selective hormonal regulation of Cyp1b1 in rat steroidogenic tissues [6, 15–17] strongly suggest that there are important physiological substrates. Cyp1b1 is expressed early during the process of adipogenesis [18], suggesting a possible important function for Cyp1b1 substrates in this process. Multi-potential mouse C3H10T1/2 (10T1/2) embryo fibroblasts (MEF) cells and 3T3-L1 pre-adipocytes each undergo adipogenesis following stimulation by a standard IDM cocktail consisting of insulin (I), dexamethasone (D) and methylisobutylxanthine (M), a cAMP phosphodiesterase inhibitor [19, 20]. Cyp1b1 expression is stimulated in 10T1/2 cells in parallel with PPARγ (peroxisome proliferator-activated receptor gamma), a critical mediator of this differentiation process [21, 22]. During adipogenesis one or more endogenous ligands are likely to mediate PPARγ activation [23]. Natural activators including fatty acid oxidation products found in lipoproteins such as 13-HODE (13-hydroxy octadecadienoic acid), as well as arachidonic acid metabolites such as prostaglandin J2-derivatives [24, 25] can function as potential CYP substrates. Such substrates are often ligands for the aryl hydrocarbon receptor (AhR) which activates Cyp1b1 expression [26, 27]. Cyp1b1 deletion in endothelial cells enhances oxidative stress again suggesting an important role for Cyp1b1 in the lipid oxidation [3, 28].
Although constitutive expression of Cyp1b1 in MEFs depends in part on AhR [29], expression of this receptor declines substantially during adipogenesis as Cyp1b1 increases [18, 30–32]. This suggests that AhR is not the predominant stimulant for Cyp1b1 during adipogenesis. Although this increase occurs concomitantly with the stimulation of PPARγ [22], we have developed clonal 10T1/2 lines that distinguish Cyp1b1 and PPARγ increases. However, only the latter increase is coupled to DNA synthesis, a requirement for the adipogenesis [22, 33]. In previous reports we have demonstrated that Cyp1b1 expression in rat steroidogenic tissues is highly stimulated by cAMP/Protein kinase A through a far upstream enhancer region (FUER). This 189 bp sequence which is located 5.1 kb upstream from the transcription start site functions through the cooperation of one CREB site and three SF-1 sites [16, 17, 34].
The central hypothesis for the present experiments is that Cyp1b1 is stimulated during differentiation processes by factors other than AhR that may function at distal sites like the FUER. The expression of Cyp1b1 in neural crest cells of the early embryo is particularly intriguing in this regard [35]. Cyp1b1 converts retinol to retinoic acid [36] which has been linked to effects of Cyp1b1 deletion in these cells [2]. Neural crest cells differentiate during embryogenesis to both neurons and mesenchymal progenitor cells that then give rise to bone, cartilage and adipose cells [37]. 10T1/2 cells provide an excellent model for these mesenchymal progenitor cells [33] while 3T3-L1 cells represent a further stage of commitment as precursors for adipogenesis [33]. In this manuscript we utilize a series of rat and mouse Cyp1b1 promoter-luciferase reporters to demonstrate that stimulation of Cyp1b1 during adipogenesis in 10T1/2 and 3T3-L1 cells is indeed enhanced by far upstream sequences and that these cells differ appreciably in their Cyp1b1 response mechanisms.
Here we provide evidence that the rat FUER alone responds rapidly to adipogenic stimulation in these cells but nevertheless can be deleted from these reporter constructs without affecting their substantial stimulation during adipogenesis. We show that this stimulation of Cyp1b1 by DM in mouse 3T3-L1 cells depends on a distal element which is also completely conserved in the rat, and that DM treatment increases binding of the key developmental transcription factor Pax6 to this regulatory element. We refer to this conserved sequence as the Cyp1b1 Pax6 element (PaxE). We subsequently found that the Cyp1b1 proximal promoter contains an alternative site for Pax6 activity that is linked to stimulation of Sp1 activity. We show that adipogenic stimulation of Cyp1b1 in 10T1/2 which occurs much faster than in 3T3-L1 cells can also depend on Sp1 but is less dependent on Pax6.
Pax6 is a highly conserved transcription factor which regulates important events during embryonic development particularly in the hind brain and the eye [38]. Pax6 is expressed coordinately in the mouse embryo with Cyp1b1 [35, 38]. Pax6 also plays a key role in development of the central nervous system and pancreas [39–41] including the control of insulin and glucagon. Pax family proteins primarily function through a paired domain (PD) but also contain a homeobox domain (HD) which interacts with distinct DNA motifs [42, 43]. The PaxE sequence corresponds to elements in many genes that respond to Pax6 through interaction with the PD domain. We also report that PaxE is part of a 194 base sequence that is over 90 percent conserved between rats and mice and which includes two core homeobox elements. The significance of these findings in relation to the expression of Cyp1b1 during development is discussed.
Material and methods
Chemicals
Insulin, dexamethasone, and methylisobutylxanthine were purchased from Sigma-Aldrich (St Louis, MO).
Antibodies
Rabbit anti-mouse PPARγ monoclonal antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-mouse Pax6 polyclonal antibody was purchased from Chemicon International, Inc. (Temecula, California). Rabbit anti-mouse Cyp1b1 antibody was generated in this laboratory as previously described [4]. Rabbit anti-mouse Sp1 antibody was purchased from Biotechnology (Charlottesville, VA)
Cell culture
3T3-L1 preadipocytes and 10T1/2 cells were obtained from ATCC (Bethesda, MD) and were maintained in DMEM/F12 medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 µg/ml streptomycin. Cells were cultured with initial plating density of 3×105 cells/well in 6-well plates, or 1×105 cells/well in 12-well plates. At 100% confluence, cells received fresh medium supplemented with 10% FBS. After being maintained at confluence for 2 days, cells were treated with 1.74 µM insulin, 1 µM dexamethasone, 0.5 µM methylisobutylxanthine, or combinations of these agents. For the time course experiments shown, the initiation of treatment is designated as zero hour (0 h) and continues for up to 48 h. Adipogenic differentiation of these cells following treatment with insulin has previously been described by this laboratory and many others [8, 21, 22, 31].
Plasmid constructions and cell transfection
Mouse Pax6 expression vector (pmPax6) was generously provided by Dr. Masaharu Sakai (Hokkaido University School of Medicine, Japan) [44].
A luciferase reporter vector pGL3-basicPlus was created by modifying the pGL3-basic vector (Promega, Madison, WI) by introduction of a PstI-SmaI-KpnI-EcoRI-XhoI-NheI-BglII-HindIII-cloning-site sequence in place of a multi-cloning-site sequence in pGL3-basic. PCR fragments of mouse Cyp1b1 5'-flanking sequence were subcloned into pGL3-basicPlus or pGL3-basic according to the primers used. Primers with a KpnI site, m1B1(−6712)KpnIF, m1B1(−6110)KpnIF, and m1B1(−6091)KpnIF (Table 1) were used to generate fragments starting at sites –6712, −6110, and −6091 on 5’-flanking sequence of mouse Cyp1b1 gene, respectively. Primer m1B1(−203)EcoRIF (with an EcoRI site) was used to generate a fragment starting at site −203. Primers with an EcoRI site, m1B1(−5966) EcoRIR, m1B1(−5706)EcoRIR, m1B1(−5186)EcoRIR and a primer with an NheI site, m1B1(+130)NheIR, were used to generate fragments ending at sites −5966, −5706, −5186, and +130, respectively. Mutant Pax6, Sp1a, and Sp1b sites were made with the ExSite PCR-Based Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's standard protocol. A luciferase vector primer m1B1Pax6R was used to mutate the Pax6 site in 5-flanking sequence of mouse Cyp1b1 with one of primers m1B1Pax6Mut1F, m1B1Pax6Mut2F, m1B1Pax6Mut3F, and m1B1Pax6Mut4F. The following sets of primers were used to mutate Sp1a and Sp1b sites in 5-flanking sequence of mouse Cyp1b1: m1B1MutSp1aF and m1B1MutSp1aR and m1B1MutSp1bF and m1B1MutSp1bR. The small letters in the sequences indicate the mutant bases.
Table 1.
The oligonucleotides used in subcloning of luciferase reporters and EMSA.
| Oligonucleotides | Sequences |
|---|---|
| Subcloning | |
| m1B1(-6712)KpnIF | 5’ -AAAGGTACCGAGCTCCTTGATCACCTTCTT |
| m1B1(-6110)KpnIF | 5’ -AAAGGTACCATAACTCAGGCATGAATAC |
| m1B1(-6091)KpnIF | 5’ -AAAGGTACCAGCTTTCCTCCCCGCCCCCA |
| m1B1(-203)EcoRIF | 5’ -AAAGAATTCTATTGTTTCAGGAGGGTCC |
| m1B1(-5966)EcoRIR | 5’ -AAAGAATTCATTCTTCTGATCCTTTGACC |
| m1B1(-5706)EcoRIR | 5’ -AAAGAATTCGAGTAGACTTGGAGGGGGGC |
| m1B1(-5186)EcoRIR | 5’ -AAAGAATTCAGCCGCAACACTGCCTTTG |
| m1B1(+130)NheIR | 5’ -AAAGCTAGCGCTTTCTCACGGAGTTGGGG |
| m1B1Pax6R | 5’ -GTACCCGGGCTGCAGTACCT |
| m1B1Pax6Mut1F | 5’ -CATAAtaCAGGCATGAATACAGCTTTCCTCCCCGC |
| m1B1Pax6Mut2F | 5’ -CATAACTatGGCATGAATACAGCTTTCCTCCCCGC |
| m1B1Pax6Mut3F | 5’ -CATAACTCAttCATGAATACAGCTTTCCTCCCCGC |
| m1B1Pax6Mut4F | 5’ -CATAACTCAGGtcTGAATACAGCTTTCCTCCCCGC |
| m1B1MutSp1aF | 5’ -CGCCCCACCCCCTAAGGTCC |
| m1B1MutSp1aR | 5’ -TGAGGttgGGGACGCACTGCGAAAATCC |
| m1B1MutSp1bF | 5’ -CGCCCCcaaCCCTAAGGTCCCGCTTCCTC |
| m1B1MutSp1bR | 5’ -TGAGGGGCGGGACGCACTGC |
| EMSA | |
| m1B1Pax6 (Cyp1b1 Pax6) | 5’ -TTTTCATAACTCAGGCATGAATACAGCTTTAAAAGTATTGAGTCCGTACTTATGTCGAAA-5’ |
| Mut2Pax6 (Cyp1b1 Pax6 mutation 2) | 5’ -TTTTCATAACTatGGCATGAATACAGCTTTAAAAGTATTGAtaCCGTACTTATGTCGAAA-5’ |
| Consensus Sp1 | 5’ -CCCTTGGTGGGGGCGGGGCCTAAGCTGCGGGGAACCACCCCCGCCCCGGATTCGACGC-5’ |
| Mut Sp1 (mutant Sp1) | 5’ -CCCTTGGTGGGttgGGGGCCTAAGCTGCGGGGAACCACCCaacCCCCGGATTCGACGC-5’ |
| Sp1a | 5’ -TCGCAGTGCGTCCCGCCCCTCACAGCGTCACGCAGGGCGGGGAGTG-5’ |
| Sp1b | 5’ -CCTCACGCCCCACCCCCTAAGGTGGAGTGCGGGGTGGGGGATTCCA-5’ |
| Mut Sp1a (mutant Sp1a) | 5’ -TCGCAGTGCGTCCCcaaCCTCACAGCGTCACGCAGGGgttGGAGTG-5’ |
| Mut Sp1b (mutant Sp1b) | 5’ -CCTCACGCCCCcaaCCCTAAGGTGGAGTGCGGGGgttGGGATTCCA-5’ |
The small letters in the sequences shown indicate mutant bases.
Transient transfection of these Cyp1b1 reporters into mouse 10T1/2 and 3T3-L1 cells were performed with TransIT-LT1 (Mirus Bio Corporation, Madison, WI) following the manufacturer's recommended protocol. Approximately 24 h prior to transfection, cells were plated at a cell density of 1×105 cells in complete growth medium per well of a 12-well plate. Each transfection for a well requires 0.83 µg of DNA and 2.5 µl of TransIt-LT1. Cells were incubated with TransIT-LT1-DNA mixture for 48 h. Cotransfection of mouse Pax6 expression vector pmPax6 with Cyp1b1 reporters was carried out at the ratio 0.38 µg : 0.44 µg (molar ratio at 1 : 1, Pax6 vector : reporter). Luciferase activity was normalized by Dual-luciferase Reporter Assay System (Promega, Madison, WI). The statistical analysis of significance was performed by Student’s t test.
Western Immunoblotting
Following treatments, the cells were washed three times with ice-cold PBS. Total cellular protein was extracted using RIPA buffer (50 mM Tris pH 7.4, 0.25% sodium deoxycholate, 1% NP-40, 1 mM EGTA, 1 mM EDTA, 150 mM NaCl, 0.05% SDS, 1 mM PMSF, 1 µg/ml leupeptin, 1 µg/ml aprotinin). Protein concentrations were determined by using the BCA protein assay kit (Pierce Biotechnology, Rockford, IL). For analysis, 30 µg/lane of total cellular protein was heated at 95 °C for 5 min before being loaded on 10% SDS-polyacrylamide gels. Total cellular protein was separated by electrophoresis, and transferred to a nitrocellulose membrane. After transferring, the membrane was incubated with the primary antibody of interest at 4 °C over night, and further with horse radish peroxidase-conjugated secondary antibodies. Immune complexes were visualized using the enhanced chemiluminescence (ECL) detection method per manufacturer’s protocol (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Band densities in the immunoblots were quantified using the ImageQuant 5.2 Software (Amersham Biosciences, Pittsburgh, PA). Expression of numerous genes, including many housekeeping genes, changes during adipogenesis (including β-actins). Therefore, gels were loaded based upon total sample protein content as the most reliable basis for comparison. Western blots were repeated at least twice with similar results.
Electrophoretic Mobility Shift Assay (EMSA)
3T3-L1 cell and 10T1/2 nuclear extracts were prepared from either untreated or DM treated cells. Oligonucleotide probes were end-labeled by T4 polynucleotide kinase. Pax6 Binding reactions were performed in 10 µl total volume with 10 µg 3T3-L1 nuclear extract and 20,000 cpm of labeled probe in specific binding buffer. Pax6 binding buffer contained 20 mM HEPES (pH 7.9), 0.2 µg/µl Poly(dI-dC), 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.1% NP-40, 100 mM KCl, 0.1 mM MgCl2, and 5% (v/v) glycerol. The binding reactions were resolved on a 6% non-denaturing polyacrylamide gel at 220 volts for 2.5h at 4°C. Following electrophoresis, DNA-protein complexes were analyzed with a PhosphoImager. For competition experiments a 100-fold molar excess of unlabeled oligonucleotides or 3 µl of Pax6 antibodies was added to the nuclear extracts before addition of radio labeled probe. Sp1 EMSA was carried out as described previously [17]. Sequences of oligonucleotides used in EMSA were shown in Table 1.
Chromatin Immunoprecipitation (ChIP) Assay
To cross-link protein to DNA, formaldehyde was directly added to the dishes at a final concentration of 1% at room temperature with gentle agitation for 10 min. Glycine was added to quench the reaction. Cells were then washed with ice-cold PBS twice and collected by centrifugation. Nuclei were isolated by incubation in cell lysis buffer (10 mM Tris, 10 mM NaCl, 0.2% Nonidet P-40, 1x phosphatase inhibitor cocktail (6.25 mM NaF, 12.5 mM β-glycerophosphate, 12.5 mM para-nitrophenyl phosphate, 1.25 mM NaVO3), 1x protease inhibitor cocktail (Sigma-Aldrich, St Louise, MO)) for 10 min. Resuspended nuclei were lysed in nuclei lysis buffer (50 mM Tris, 10 mM EDTA, 1% SDS, 1x phosphatase inhibitor cocktail, and 1x protease inhibitor cocktail) for 10 min. The lysate was sonicated with 8 pulses of 15 seconds each at output level 60%. Sheared chromatin was cleared by centrifugation in desktop centrifuge at 14, 000 rpm for 10 min. The chromatin was diluted with IP dilution buffer (20 mM Tris, 150 mM NaCl, 2 mM EDTA, 0.01% SDS, 1% Triton X-100, 1x phosphatase and protease inhibitor cocktail) to reduce the SDS concentration to below 0.2%, and incubated with the primary antibody, which was pre-bound to Dynabeads (Invitrogen Corporation, Carlsbad, CA) for 2 h at 4 °C. The tube was placed on the magnet for 1 min and the supernatant was removed. The Dynabeads were washed twice with 500-µl aliquots of IP wash buffer 1 (20 mM Tris, 50 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, pH 8.0), once with IP wash buffer 2 (10 mM Tris, 0.25 M LiCl, 1 mM EDTA, 1% Nonidet P-40, 1% deoxycholate, pH 8.1), and twice with TE (10 mM Tris, 1 mM EDTA, pH 8.0). 10% of Chelex-100 was added to the washed beads, and the mix was vortexed and followed by boiling for 10 min. Proteinase K was added to beads and the mix was incubated at 55°C for 30 min, followed by another boiling for 10 min. The eluate was used as a template in quantitative real-time PCR (qPCR). For the qPCR evaluation of Cyp1b1 proximal region, forward primer, 5'-CTAAGGTCCCGCTTCCTCCA-3'; and reverse primer, 5'-ATGCGCCATCCCTCTACTCC-3' were used. For the qPCR evaluation of PaxE region, forward primer, 5'-TCAGCTCTTGTGTTCCTTTCG-3'; and reverse primer, 5'-GGGGAGGAAAGCTGTATTCAT-3' were used. ChIP-qPCR results were calculated based on the ΔΔCt method using the SuperArray ChIP-qPCR Data Analysis Template (Frederick, MD) according to the SuperArray manual. ChIP DNA fractions were normalized to input DNA (ΔCt) to account for any differences due to chromatin sample preparation.
Statistical Analyses
Statistical analysis was done using Excel ((Microsoft, Redmond, WA). Statistical significance was determined by two-tailed Student's t test for unpaired or paired observations (P < 0.05).
Results
We have previously reported that combinations of insulin (I), dexamethasone (D) and methyl isobutyl xanthine (M) (IDM or DM) which induce adipogenesis in the mouse mesenchymal progenitor 10T1/2 line also extensively stimulate the expression of Cyp1b1 [22, 33]. We have also shown that sequences more than 5 kb upstream from the transcription start site regulate Cyp1b1 during hormonal stimulation [15]. Here we use a variety of methods, including immunoblotting, chromatin immunoprecipitation and Cyp1b1-promoter luciferase reporters, to test whether far upstream sequences also control the substantial stimulation of Cyp1b1 during the early stages of adipogenesis in 3T3-L1 cells [21]. In these cells, subsequent addition of insulin to the cell culture media induces formation of intracellular lipid droplets over the course of an additional 4 days of treatment. This is a similar yet slower response than observed for 10T1/2 cells [18]. We then test whether the same mechanism is conserved in 10T1/2 cells. 3T3-L1 cells share with 10T1/2 cells a common MEF progenitor source but represent a further stage of commitment towards adipocytes. The experimental designs for immunoblotting, chromatin immunoprecipitation, and comparisons of Cyp1b1 promoter-activity using luciferase reporters are shown schematically in Figure 1A.
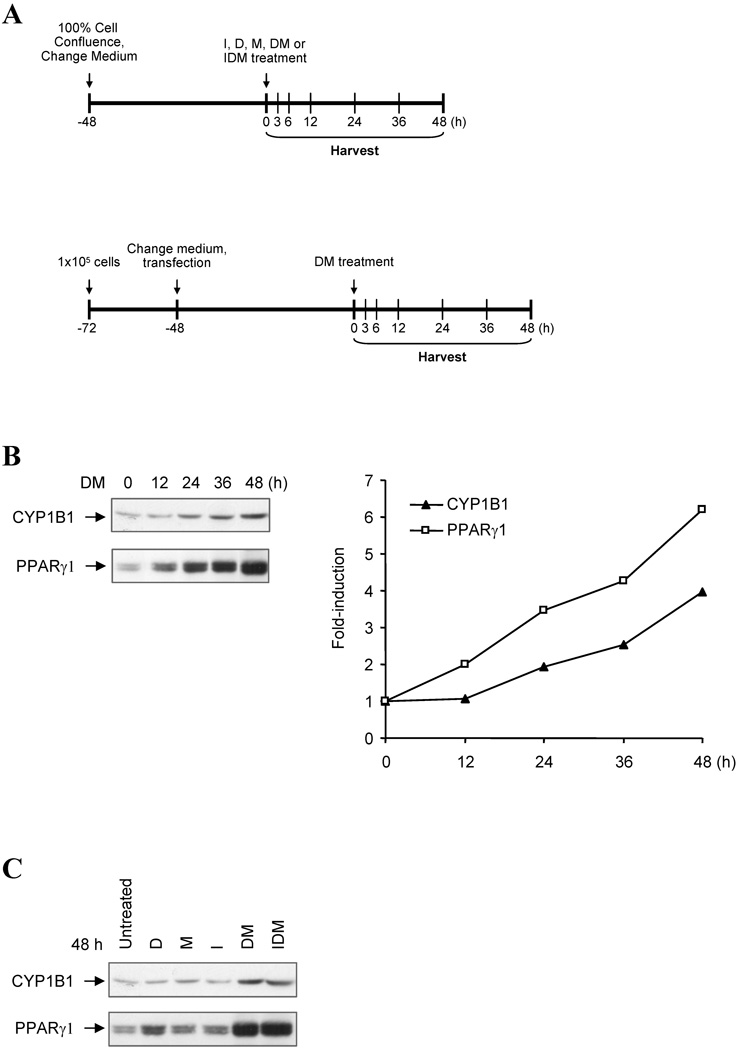
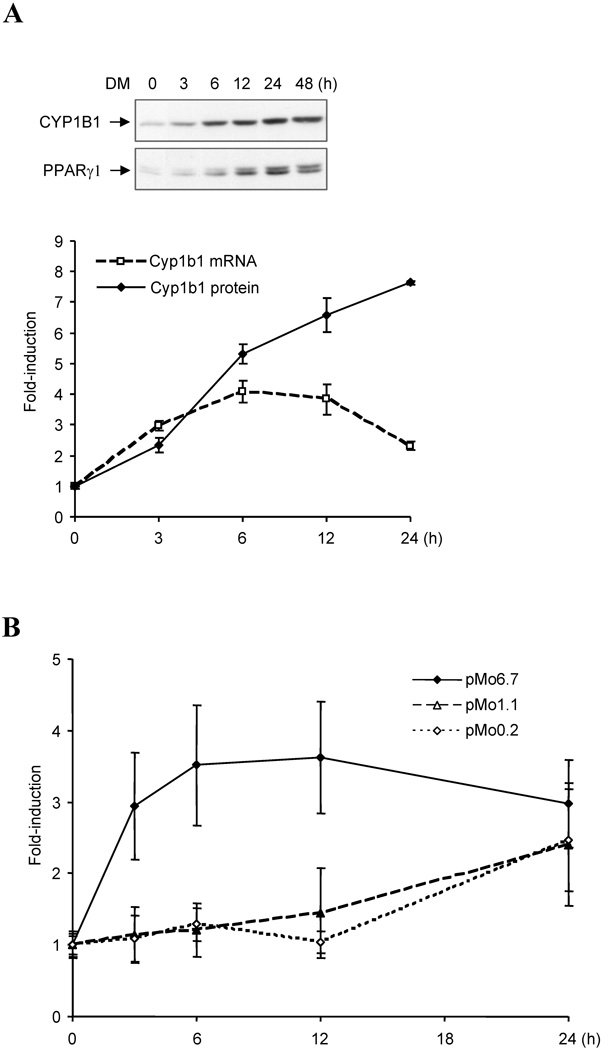
Figure 1. Cyp1b1 and PPARγ are similarly expressed in 3T3-L1 preadipocytes.
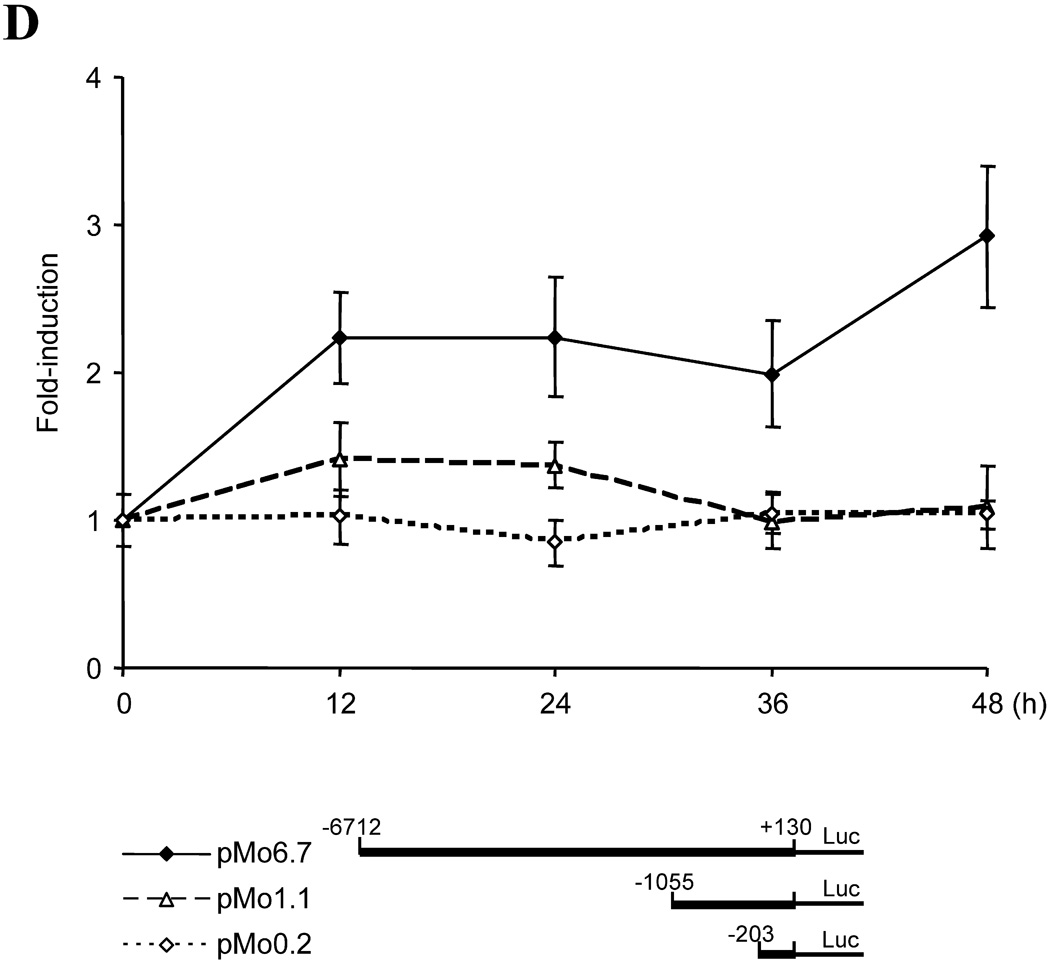
(A) Experimental designs for analysis of Cyp1b1 responses to adipogenic stimuli. Top panel, experimental designs for Western-blotting and ChIP assay. 3T3-L1 and 10T1/2 cells were cultured at an initial plating density of 3×105 cells in complete growth medium containing 10% FBS per well of a 6-well plate. At 100% confluence, cells received fresh complete medium. After being maintained at confluence for 2 days, cells were treated with 1.74 µM insulin, 1 µM dexamethasone, 0.5 µM methylisobutylxanthine, or combinations of these agents. The initiation of treatment is designated as zero hour (0 h) and cells were harvested at designated time points. Down panel, experimental designs for luciferase assays. Transient transfection of Cyp1b1 reporters into 3T3-L1 and 10T1/2 cells were performed with TransIT-LT1 (Mirus Bio Corporation, Madison, WI). Approximately 24 h prior to transfection, cells were plated at a cell density of 1×105 cells in complete growth medium per well of a 12-well plate. Each transfection for a well requires 0.83 µg of DNA and 2.5 µl of TransIt-LT1. Cotransfection of mouse Pax6 expression vector pmPax6 with Cyp1b1 reporters was carried out at the ratio 0.38 µg: 0.44 µg (molar ratio at 1 : 1, Pax6 vector : reporter). Cells were incubated with TransIT-LT1-DNA mixture for 48 h, and then started DM treatments. The initiation of treatment is designated as zero hour (0 h), and cells were harvested at designated time points. Luciferase activity was normalized by Dual-luciferase Reporter Assay System (Promega, Madison, WI). (B) Time course for DM stimulation of Cyp1b1 and PPARγ protein expression in 3T3-L1 cells after DM treatment. Total cellular protein was separated by SDS-PAGE and immunoblotted with rabbit anti-mouse Cyp1b1 and PPARγ antibodies. This is representative of one of two experiments with similar results. Note: the PPARγ bands separate into multiple PPARγ1 forms probably derived from post translational modifications and the PPARγ2 form predominates in adipocytes but is much less mobile due to an extra 28 AA at the N-terminus. (C) Cyp1b1 and PPARγ expression in 3T3-L1 cells after treatments of Dexamethasone (D), Methyl isobutylxanthine (M) and insulin (I) singly and in combinations for 48 h. This is representative of one of two experiments with similar results. (D) Time courses for induction of three Cyp1b1 promoter reporters transfected into 3T3-L1 cells after DM stimulation, measured as luciferase activity. The cells transfected with luciferase reporters were treated with DM for the indicated time (0 to 48 h). Luciferase activities represent the averages of at least three separate transfections and are presented as the mean relative expression ± S.D. The average luciferase activities of pMo6.7 in untreated 3T3-L1 and 10T1/2 cells are assigned a value of 100.
Cyp1b1 and PPARγ expression in 3T3-L1 cells
The expression of Cyp1b1 and the key differentiation mediator PPARγ1 in pre-adipocyte 3T3-L1 cells were increased with time after addition of a DM combination (Fig. 1B). Inclusion of insulin in the differentiation cocktail had no effect while each component was separately ineffective (Fig. 1C). These increases in PPARγ expression are consistent with previous reports for these cells [45].
In order to characterize the regulation of this Cyp1b1 in 3T3-L1 cells, we examined the response of a 6.7 kb mouse Cyp1b1 reporter (pMo6.7) in comparison to the Cyp1b1 protein expression (Fig. 1B and 1D). Expression of the pMo6.7 reporter mRNA responded to the adipogenic cocktail (DM) with a significant twofold stimulation within 12 h at which time Cyp1b1 protein expression appeared to be unaffected (Fig. 1B and 1D). The shorter 1.1 kb mouse Cyp1b1 luciferase reporter (pMo1.1) was much less responsive. This suggests that the DM-responsive element is located at sites distal to the conserved AhR-response enhancer region (AhER) centered at −1.0 kb [29]. There was no stimulation of the pMo0.2 reporter (Fig. 1D), emphasizing the importance of the upstream enhancer elements.
Far Upstream Enhancer Region does not contribute to DM stimulation of Cyp1b1
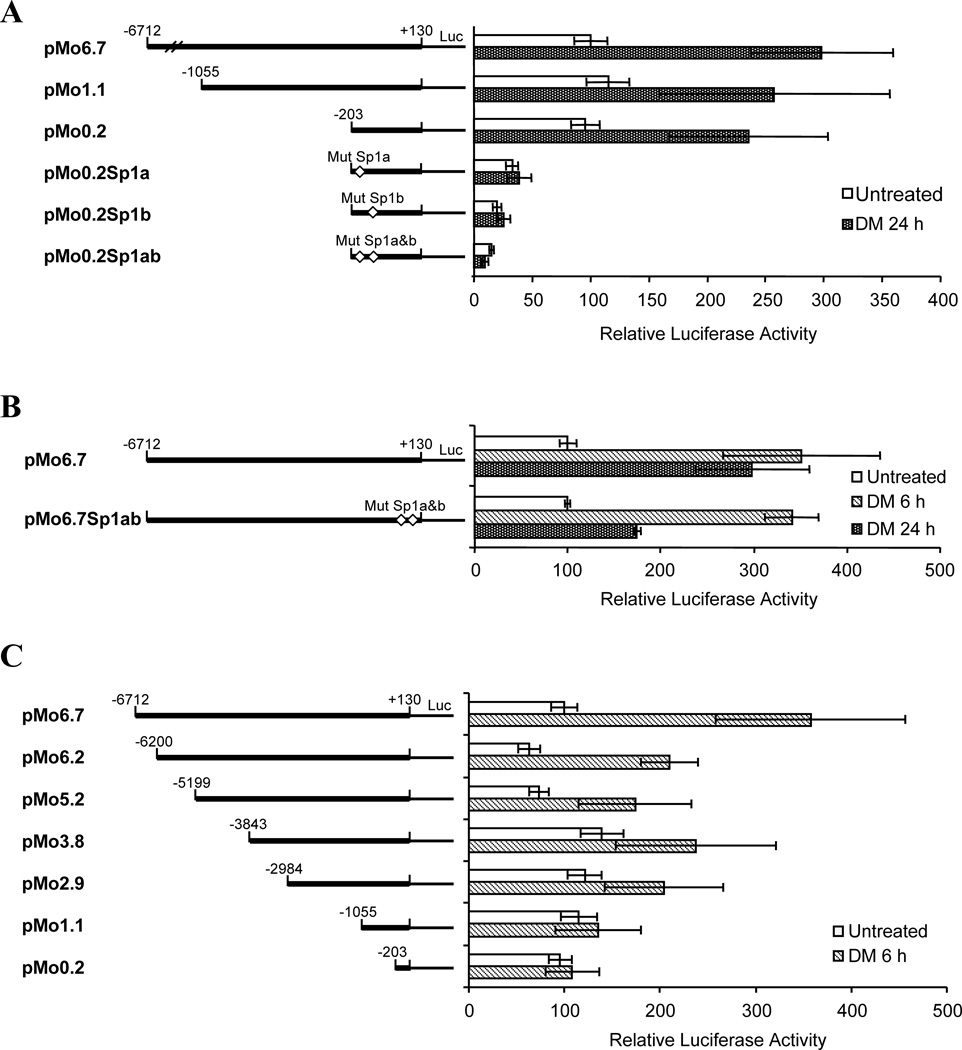
We have identified a far upstream enhancer (FUER) in the rat Cyp1b1 promoter that mediates responses to protein kinase A in adrenal cells via synergy between three SF-1 sites and a 3’-CRE element [17] (rat −5298 to −5110) (Fig. 2A). Utilizing a rat Cyp1b1 luciferase reporter (pRat7.0) equivalent to the mouse flanking region pMo6.7, the responses of these constructs to DM stimulation in 3T3 L1 cells were compared (Table 2). A similar pattern of species-specific responses was seen between the mouse and rat reporter constructs when they were transfected into 10T1/2 cells. The responses to DM were greater in rats than mice, and occurred more rapidly than in 3T3-L1 cells. A maximum level of luciferase activity was attained in 3–6 h and was sustained out to 24 h (Table 2; Fig. 5B). When the rat FUER was linked to only the truncated 0.2 kb proximal promoter (pRatFUER) DM stimulation was similar to that of the Mo6.7 reporter in both cell lines (Table 2).
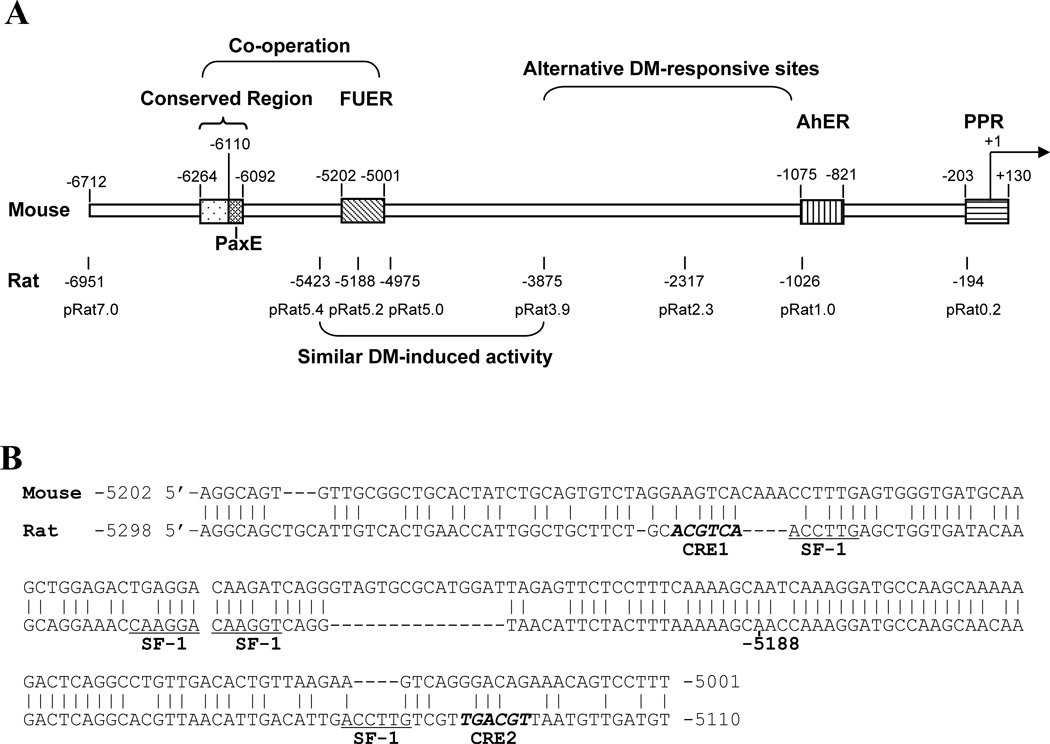
Figure 2. DM stimulation of Cyp1b1 in 3T3-L1 cells depends on a conserved 19 base sequence 6 kb upstream and on proximal Sp1 sites, but not on the FUER.
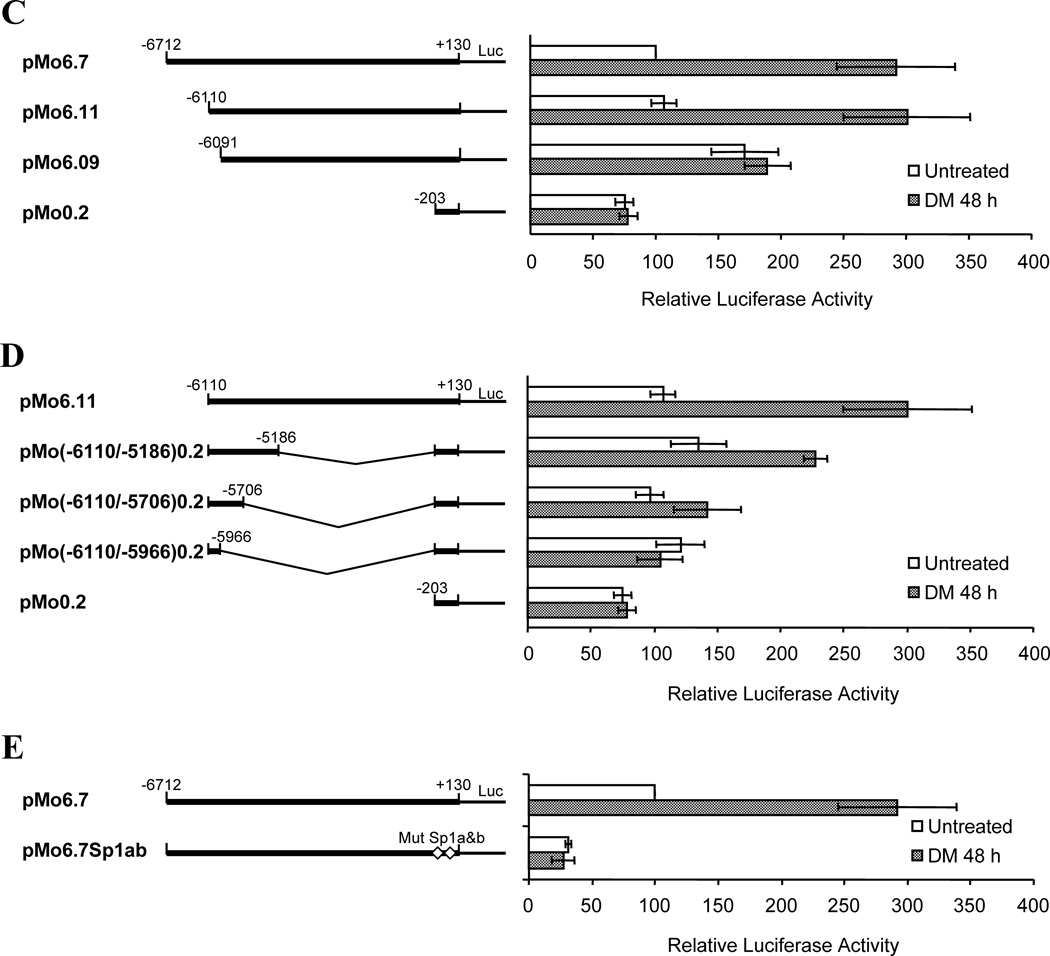
(A) Cyp1b1 regulatory regions in the 5’-flanking sequence and the sites of termination of mouse reporters in relation to enhancer regions. The region starting at −6264 is shown in Figure 3. (B) Alignment of mouse and rat homologous FUER sequences. PKA responsive elements in the rat are compared (Two CRE and Four SF-1 binding sites). (C) Comparison of sequential deletion Cyp1b1 reporter constructs yields identification of a 19 bp sequence critical for the response to stimulation by DM. (D) Comparison of sequential deletion Cyp1b1 reporter constructs of sequences downstream of a key 19 bp regulatory sequence on the response of Cyp1b1 to DM. (E) Mutation of Sp1 sites (Sp1a and Sp1b) in the Cyp1b1 promoter demonstrates key role of these regulatory elements in both basal activity and DM responsiveness. For all experiments shown, the 3T3-L1 cells were transfected with luciferase reporters and treated with DM for 48 h. Luciferase activities represent the averages of at least three separate transfections. Data are presented as the mean relative expression ± S.D. compared to unstimulated pMo6.7 or pMo6.11.
Table 2.
Rat Cyp1b1 promoter activity is higher than mouse Cyp1b1 promoter activity after DM treatment.
| Reporters | 3T3-L1 (DM 24 h) | C3H10T1/2 (DM 3 h) | C3H10T1/2 (DM 24 h) |
|---|---|---|---|
| Rat | |||
| pRat7.0 | 6.5±0.5 | 11.7* | 6.3±0.7 |
| pRat5.4 | ND | 3.3* | ND |
| pRat5.2 | ND | 4.7* | ND |
| pRat5.0 | ND | 7.3* | ND |
| pRat3.9 | ND | 5.5* | ND |
| pRat2.3 | ND | 2.7* | ND |
| pRat1.0 | 0.8±0.1 | 1.4* | 2.6±0.6 |
| pRat0.2 | 0.6±0.1 | 1.0±0.2 | 1.2±0.2 |
| pRatFUER | ND | 3.9±0.2 | 2.7±0.4 |
| Mouse | |||
| pMo6.7 | 2.2±0.2 | 2.9±0.4 | 3.0±0.4 |
The data shown are the means ± S.E.M from three separate experiments except indicated with asterisk.
ND, not done.
data from one experiment.
Figure 5. Time courses for Cyp1b1 and PPARγ expression following adipogenic stimulation of mouse embryo 10T1/2 cells.
(A) Cyp1b1 and PPARγ expression in 10T1/2 cells after DM treatment measured as protein and mRNA. 10T1/2 cells were treated with DM for indicated times. Total cellular protein was separated by SDS-PAGE and immunoblotted with rabbit anti-mouse Cyp1b1 and PPARγ antibodies. Data shown are representative of one of two experiments with similar results. Cyp1b1 mRNA quantification was carried out via real-time-PCR. Relative mRNA expressions are shown as the mean ± S.D from triplicate samples. (B) Induction of luciferase activity of pMo6.7, pMo1.1 and pMo0.2 by DM treatment in 10T1/2 cells. The cells transfected with luciferase reporters were treated with DM for indicated time (0 to 24 h). Luciferase activities represent the averages of at least three separate transfections times and data are presented as the mean relative expression ± S.D.
Interestingly, sequence analysis of the mouse FUER (mouse −5202 to −5001) showed that each of the functionally important SF-1 sequences exhibited base substitutions compared to the orthologous rat SF-1 and CREB sites (Fig. 2B). We have previously shown that such mutations remove PKA responsiveness [17]. We therefore shortened the rat Cyp1b1 reporter to either include (pRat5.4) or exclude the FUER (pRat 5.0, pRat3.9) (Fig. 2A). Sequential deletions of the rat Cyp1b1 reporters responded less to DM than the full length pRat 7.0 reporter but deletion of the functional FUER did not obliterate all responsiveness (Table 2). These results suggest that there exists some redundancy between the FUER and more proximal sequences with regard to responsiveness of the Cyp1b1 promoter to stimulation by DM.
The DM stimulation of mouse Cyp1b1 in 3T3-L1 cells depends on a 19 base distal sequence and proximal Sp1 sites
To further identify elements contributing to the DM response we carried out a more detailed deletion analysis of the 6.7 kb mouse Cyp1b1 promoter reporters using 3T3-L1 cells. Deletion from −6.7 to −6.110 kb did not affect activity whereas deletion to −6.091 kb eliminated the response to DM. This identifies a 19 bp region (−6110 to −6092) as essential for the DM response (Fig. 2C). The immediate upstream sequence (−6712 to −6111) did not appear to contribute significantly. The 19 bp sequence plus a further 144 bases of downstream sequence was linked directly to the minimal 0.2 kb proximal promoter (−6110 to −5966). This reporter was found to be unresponsive to DM and had only minimal activity above that of the proximal promoter (Fig. 2D). A longer downstream segment (−6110 to −5706) was also ineffective. However, when the reporter was extended from the 19 base sequence to include an additional downstream 0.9 kb (−6110 to −5186) enhancer activity regained about half of the maximum responsiveness to DM stimulation (Fig. 2D). This 19 bp sequence thus functions with additional complexes formed upstream of −5186.
The proximal promoter contains two Sp1 sites that are essential for basal, cAMP and TCDD stimulated Cyp1b1 reporter activity [16, 17, 34]. Mutation of these sequences also completely removed most basal activity and all DM stimulatory activity from the pMo6.7 reporter (Fig. 2E), strongly suggesting that the Sp1 sites cooperate with upstream enhancer sequences in this DM response.
Characterization of the 19 base element as a Pax6 binding site (PaxE)
The 19 base mouse sequence which is essential for DM responsiveness includes an 18 base sequence that is completely conserved in the rat (Fig. 3A). A search for known DNA recognition elements in the Cyp1b1 flanking region revealed a close match with the binding site for the Pax6 paired domain (PD) [46] (Fig. 3B). Sequence comparisons with the flanking regions of four mouse genes that are stimulated by Pax6 (Glucagon, δ-crystallin, optimedin A, and Reelin) [39, 47, 48] revealed similar sequence homologies (Fig. 3B). We have denoted this 18 base conserved sequence in the mouse Cyp1b1 promoter region as the Pax6 enhancer (PaxE). The variability in these PaxE-like sites suggests flexibility in the Pax6 recognition element. Although the upstream 602 bases do not increase the reporter activity, the immediate 154 base mouse upstream segment (−6264 to −6111) has only 12 mismatches with the corresponding rat sequence (92 percent conserved) (Fig. 3A). This sequence contains a TAATTA core homeobox sequence that can potentially bind to Pax6 through a C-terminal homeodomain (HD) or compete with a homeobox protein [42, 43, 49] (Fig. 3A).
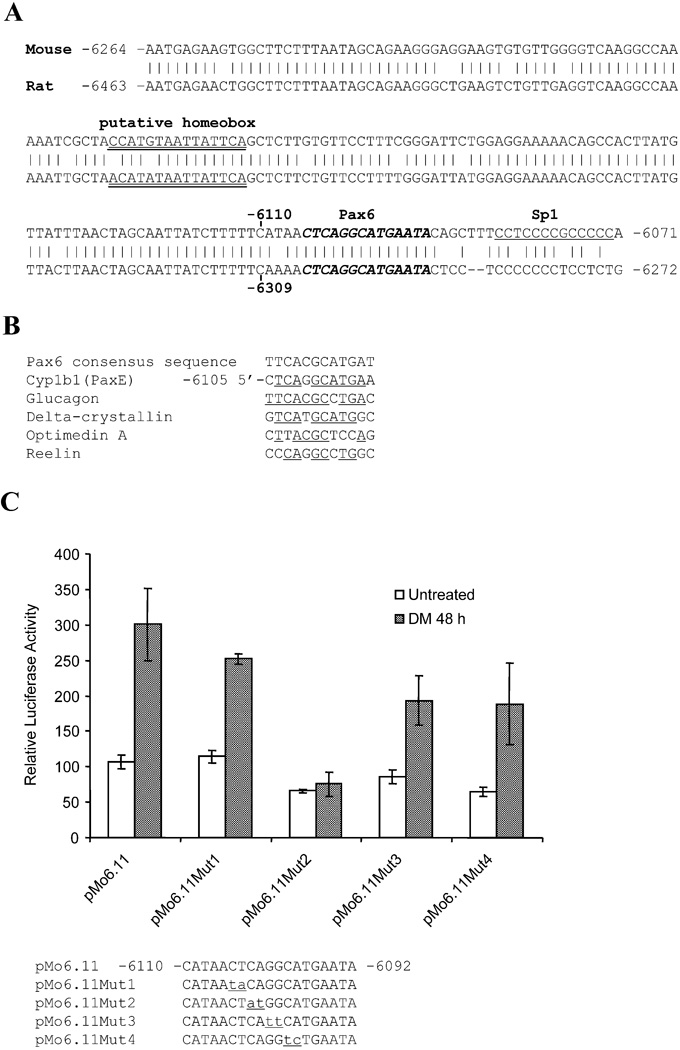
Figure 3. Pax6 binding element is located within a 19 base sequence designated as PaxE.
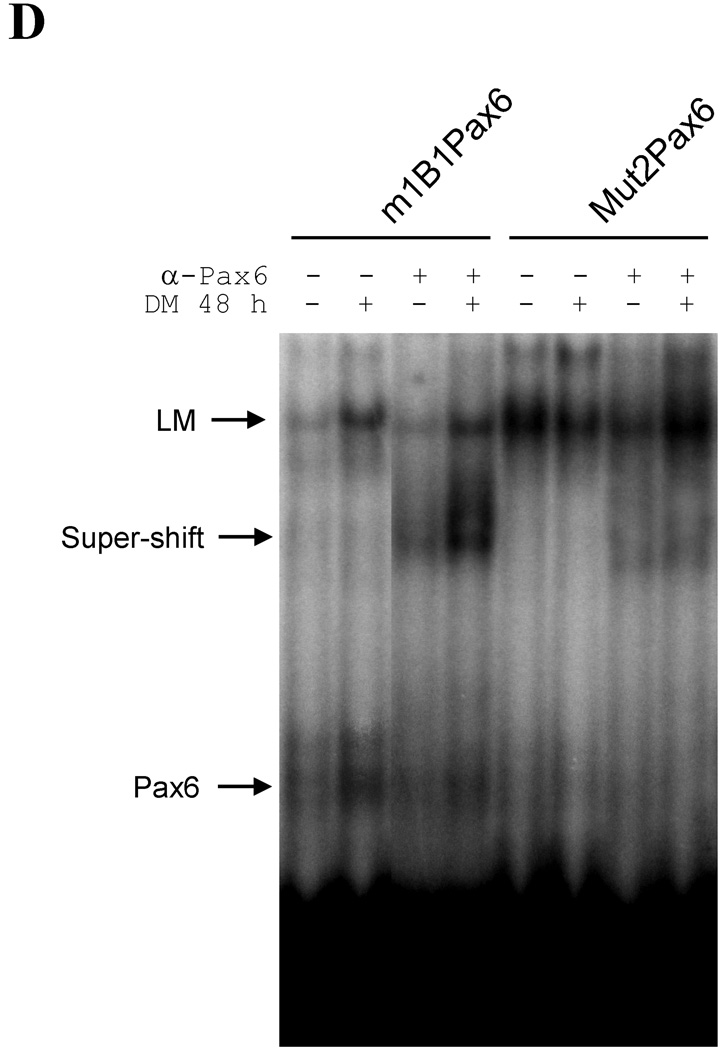
(A) Alignment of 193 base mouse sequence that includes PaxE with homologous rat Cyp1b1 sequence. (B) Alignment of the putative Pax6 sequence with well-defined Pax6 sequences from four mouse genes [40, 41, 47, 48]. (C) Comparison of the effects of two base substitutions in PaxE, using the same methods as described in Figure 2. Data are presented as the mean relative expression ± S.D. compared to unstimulated pMo6.11. (D) PaxE electrophoretic mobility assay using nuclear extracts from 3T3-L1 cells after 48 h of control or DM treatment. Wild type PaxE was also compared with the Mut2 mutation. The effect of pre-incubating the nuclear extract with anti-Pax6 antibody reveals the presence of Pax6 demonstrated binding of Pax6 to PaxE which is enhanced by DM treatment but obliterated by mutation of two key bases (Mut2). EMSA data are shown from one of two independent experiments that provided similar complexes. The Pax6 complexes (antibody suppressed), the supershifted complex (antibody stimulated) and low mobility (LM) complexes (antibody independent) are indicated.
To examine the functional role of these sequences, localized mutations of PaxE were generated by sequentially changing two bases in pMo6.11 (Fig. 3C). The impact of these mutations on the DM stimulation of 3T3-L1 cells showed that sites −6102/−6103 (Mut2) located at the center of the conserved PaxE element are critical for Cyp1b1 promoter activity. By contrast, six adjacent bases (Mut1, Mut3 and Mut4) are much less important (Fig. 3C).
DM activates complex formation of Pax6 with PaxE
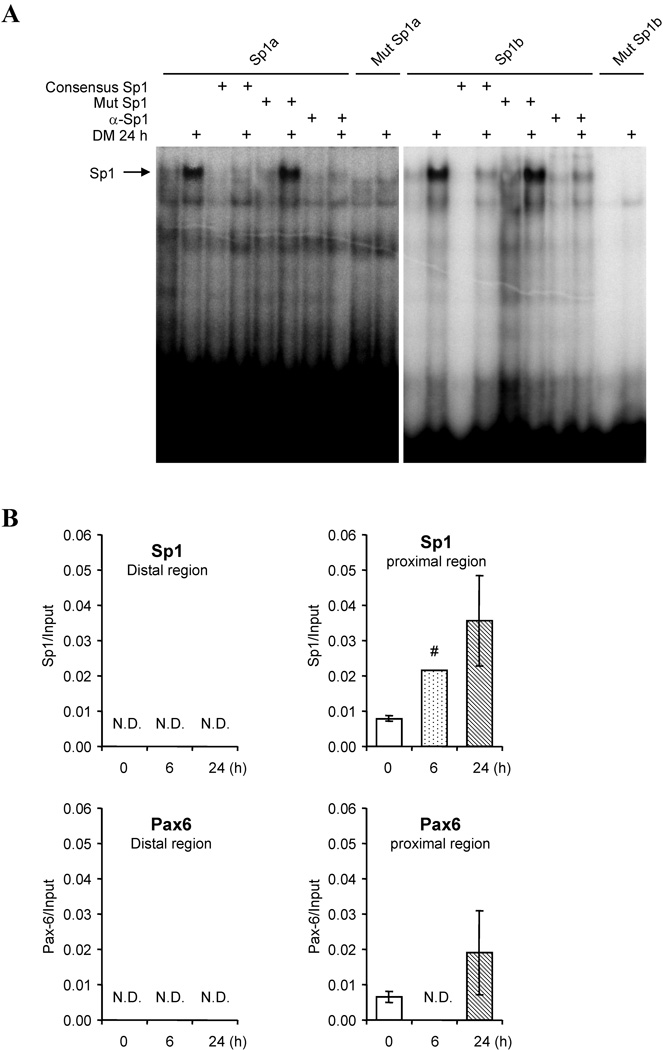
To examine whether DM stimulates Pax6 to binds directly with the PaxE sequences in the Cyp1b1 promoter, gel shift assays were conducted. Pax6 present in nuclear extracts from 3T3-L1 cells bound directly to the PaxE sequence as shown by the decrease in mobility after addition of an anti-Pax6 antibody to the extracts (α-Pax 6) (Fig. 3D). This doublet complex and the supershifted α-Pax6 complexes were each appreciably stimulated by DM treatment. The substantial increase in the amount of supershifted complex indicates stabilization by the antibody. Mutation at position 2 of PaxE (which diminishes promoter activity) substantially attenuated this complex formation, including the supershifted complex.
PaxE also formed a significant amount of a very low mobility complex (LM), which was increased by DM but did not contain Pax6, as evidenced by insensitivity to α-Pax 6. The basal level of the LM complex was also appreciably enhanced when the PaxE interaction with Pax6 was decreased by introduction of mutation 2. The decreased mobility of the LM complex suggests participation of larger proteins and/or additional proteins. This effect of mutation 2 suggests that there is competition for PaxE between a Pax6 complex and this LM complex, which is apparently favored by the mutation at position 2.
Transfection with Pax6
To assess the quantitative effect of Pax6 levels on Cyp1b1 expression we transfected heterologous Pax6 into 3T3-L1 cells which were then also treated with DM. We tested the interaction of Pax6 with distal and proximal sequences in the Cyp1b1 promoter by cotransfecting a CMV-Pax6 expression vector (pmPax6 or a control vector) with a Cyp1b1 luciferase reporter (pMo6.11 wild-type or various mutations/deletions) or a pMo0.2 reporter (wild-type or Sp1 mutations) [44]. Activities were compared with and without a 48 h DM treatment. Both DM addition and Pax6 transfection stimulated reporter activity by 1.8 and 1.55 times, respectively (Table 3, Table 4). Results deviated somewhat from those shown in Figure 3C which were conducted with earlier passages on 3T3-L1 cells and different serum in the media. Additional DNA was also transfected to control for the Pax6 vector.
Table 3.
The 48 hours DM induction of mouse Cyp1b1 luciferase reporters with or without cotransfection with a mouse Pax6 expression vector (pmPax6) in 3T3-L1 cells.
| pMo6.11 | pMo6.11Mut1 | pMo6.11Mut2 | pMo0.2 | |
|---|---|---|---|---|
| Control | 100±9 | 67±5 | 66±3 | 70±6 |
| DM | 180±13** | 134±12* | 120±5* | 102±3 |
| PAX6 | 155±10** | 147±8** | 143±15* | 124±9 |
| PAX6+DM | 322±9** | 309±17** | 328±10** | 282±16** |
The data shown are the means ± S.E.M from three separate experiments.
P value is determined by comparison to pMo6.11 control.
P < 0.05;
P <0.01.
Table 4.
The 48 hours DM induction of mouse Cyp1b1 luciferase reporters with or without cotransfection with a mouse Pax6 expression vector (pmPax6) in 3T3-L1 cells.
| pMo6.11 | pMo0.2 | pMo0.2Sp1a | pMo0.2Sp1b | pMo0.2Sp1ab | |
|---|---|---|---|---|---|
| Control | 100±9 | 70±6** | 25±0.8** | 9±0.4** | 5±0.3** |
| DM | 180±13## | 102±3## | 31±1.2## | 11±0.4## | 6±0.3# |
| PAX | 155±10## | 124±9## | 48±3.5## | 16±1.3## | 8±0.6## |
| PAX+DM | 322±9## | 282±16## | 96±3.4## | 31±2.6## | 16±1.9## |
The data shown are the means ± S.E.M from three separate experiments.
P value is determined by comparison to pMo6.11 control (**, P < 0.01) or their corresponding wildtype reporters (#, P < 0.05; ##, P < 0.01).
Basal activities as well as responses of the Cyp1b1 reporters to DM stimulation (Table 3) were suppressed by each of the PaxE mutations to a level equivalent to the activity of the proximal promoter pMo0.2. The DM stimulated levels also similarly declined (Table 3). The mutations in PaxE therefore remove most of the additional activity provided to the proximal promoter by the 6 kb of upstream sequence under either basal conditions or following DM stimulation. Therefore, the effects of these mutations suggest that PaxE is functioning similarly under both basal and stimulatory conditions consistent with the EMSA experiments which show Pax6 complex formation under both conditions (Fig. 3D).
By contrast, after transfection with pmPax6, Cyp1b1 reporter activities were insensitive to the mutations, especially in combination with DM (Table 3). The combination of DM addition and Pax6 transfection produced a synergistic 3.2 fold stimulation. However, there was almost as much activity after equivalent stimulations of the proximal promoter. This suggests that this ineffectiveness of the distal mutations arises because the activity is almost entirely generated from interaction of Pax6 with the proximal promoter. This proximal promoter activity of Pax6 is evidently further stimulated by activation by DM.
Stimulation by either DM addition or Pax6 transfection was almost completely blocked by mutation of the two Sp1 sites (Table 4). These data suggest that there is a low affinity interaction of transfected Pax6 with Sp1 in the proximal promoter, either through direct interaction with DNA or with the Sp1 protein.
Chromatin Immunoprecipitation (ChIP) of Pax6 and Sp1 complexes
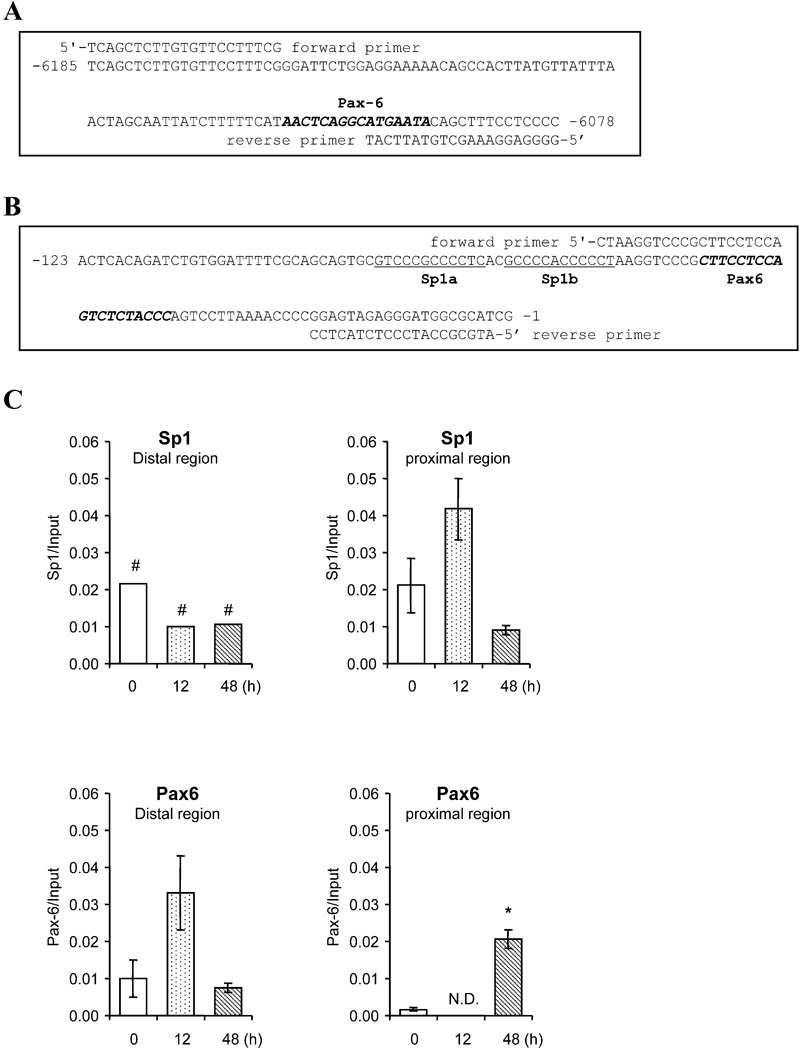
The role of Pax6 and Sp1 elements in the regulation of Cyp1b1 was also assessed by ChIP experiments. The binding of SP1 and Pax6 to the proximal promoter and to the distal PaxE region of Cyp1b1 were each confirmed via ChIP analyses utilizing the primers shown in Fig. 4A and 4B. Size fractionation showed that the ChIP fragments of DNA were typically about 200–500 bp in length. The distal primer pair targets immunoprecipitated DNA fragments that include the sequence −6078 to −6185 corresponding to the PaxE site, while the proximal primer pair targets the sequence immediately upstream of the transcription start site.
Figure 4. Chromatin immunoprecipitation (ChIP) assays of Pax6 and Sp1 binding to distal and proximal regions of the mouse Cyp1b1 5’-flanking sequence.
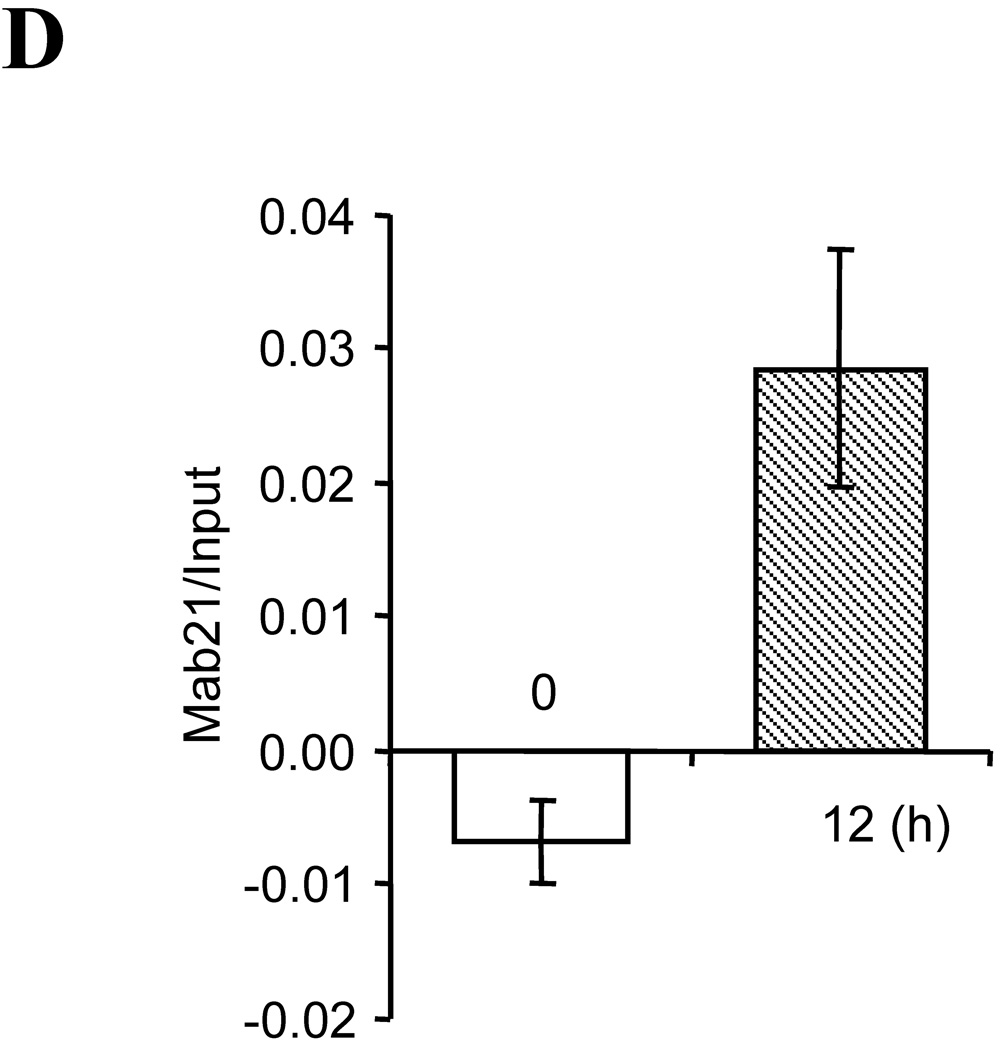
(A, B) Primer sets used to amplify distal and proximal regions. Pax6 and Sp1 elements (Sp1a and Sp1b) are shown. (C) The chromatin immunoprecipitation assays for Pax6 and Sp1 binding with 3T3-L1 cells untreated or treated with DM for 12 h or 48 h. Cell preparation of DM treatments is shown in Figure 1A. (D) Pax6 binding to Mab21/2 gene (positive control). Chromatin fragments were immunoprecipitated with anti-Pax6 antibody, and the Pax6 sequence were amplified and quantitated by real-time PCR using primers sets shown in Figure A and B. Data are the mean ± S.D. of three independent experiments except where indicated. *, p<0.05, compared to control. #, data from a single experiment.
DM stimulated an early binding of Sp1 (after 12 h) to the proximal promoter region (Fig. 4C), followed by subsequent binding of Pax6 in this region (after 48 h). In the distal region, an initial Sp1 binding (there is an Sp1 sequence adjacent to PaxE), occurred but decreased with time (Fig. 4C). On the other hand, Pax6 binding to the distal region increased after 12 h, prior to the increase in the proximal region at 48 h (Fig. 4C). Pax6 may therefore first bind to the upstream enhancer before shifting to a binding site on the proximal promoter, subsequent to a transfer of Sp1 from a distal site.
We also tested the DNA that was immunoprecipitated by anti-Pax6 antibody for other Pax6 responsive genes. PCR analyses showed binding to a segment of the Mab21 proximal promoter, a gene known to participate in eye development that contains a Pax6 element [50]. DM produced appreciable stimulated of Mab21 levels (Fig. 4D).
Stimulation of Cyp1b1 in 10T1/2 cells
Although 10T1/2 cells share many similar overall responses to the DM stimulus with 3T3-L1 cells, there are appreciable differences in the respective mechanisms. Treatment of 10T1/2 cells with DM concomitantly increased both Cyp1b1 and PPARγ1 expression during a 48 h post treatment (Fig. 5A), while a further 48 h of insulin treatment then stimulates formation of lipid droplets [32, 33]. However the full length pMo6.7 Cyp1b1 reporter (Fig. 5B) responded to DM treatment and also reached a maximum in approximately 6 h, which is much faster than in 3T3-L1 cells (Figure 1). Deletion mutants (pMo1.1and pMo0.2) failed to respond to stimulation by DM. A shift in mechanism between 6 and 24 h is suggested by the increased response of the proximal promoter reporter (pMo0.2) at later times relative to that of pMo6.7, unlike the shifts observed in Figure 1 for 3T3-L1 cells. Figure 6A confirms that 24 h of DM treatment stimulates promoter activity in 10T1/2 cells without any contributions from regulatory elements upstream from the proximal promoter, including apparently from PaxE. However, like the 3T3-L1 cells, this response depends on Sp1 sites in the proximal promoter (Fig. 6B & Table 4). By contrast, promoter activity 6 h after DM treatment (Fig. 6B) is insensitive to Sp1 mutations but is highly dependent on far upstream sequences (Fig. 6C). This DM stimulation of Cyp1b1 promoter activity 6 h after DM treatment in 10T1/2 cells decreases by about 50 percent compared to the full length reporter, when the PaxE sequence is deleted in pMo5.2 (Fig. 6C).
Figure 6. Cyp1b1 promoter activity after DM stimulation of 10T1/2 cells is regulated differently at 6 and 24 h.
(A) Promoter deletions used to test the role of the upstream and proximal regions in the DM stimulation of 10T1/2 cells. Two Sp1 mutations test the roles of Sp1a and Sp1b in the proximal promoter region. (B) The Sp1 sites contribute to the DM late response of pMo6.7 (p<0.01) after 24 h but not after 6 h, (C) Deletion analyses conducted to test the role of upstream sequences in DM stimulation of the Cyp1b1 promoter in 10T1/2 cells. Luciferase activities represent the averages of at least three separate transfections and data are presented as the mean relative expression ± S.D.
Sp1 is a particularly complex transcription factor of over 750 amino acids which binds GC rich DNA sites through a C-terminal zinc finger domain and is modified in many ways including by multiple kinases at 14 S/T sites [51]. Cyp1b1 has two Sp1 sites in the proximal promoter region that are commonly essential for transcription [5]. EMSA assays confirmed that Sp1 binding to both Sp1a and Sp1b sites was appreciably stimulated by 24 h of DM treatment of 10T1/2 cells (Fig. 7A). Excess Sp1 oligonucleotide competed the small amounts of 32P-Sp1a or Sp1b probes, and mutations in the competing Sp1 oligonucleotide obliterated the binding to the probe, confirming specificity of the binding interactions. Additional ChIP assays showed that DM stimulated Sp1 binding to the proximal promoter over time, but not to the distal region (Fig. 7B). In contrast, Pax6 binding was minimal in either region over the same treatment time span (Fig. 7B). Together these data indicate a rapid stimulation of Cyp1b1 in the multi-potential 10T1/2 cells, unlike DM responses in 3T3-L1 cells, is independent of Sp1 but dependent on upstream interactions. This upstream participation however seems to be independent of PaxE and Pax6. Importantly, this fast response mechanism appears to be absent in 3T3-L1 cells.
Figure 7. DM activates the binding of Sp1 but not Pax6 to the Cyp1b1 promoter in 10T1/2 cells.
(A) EMSA assessment of SP1 binding to proximal promoter sites using nuclear extracts from either untreated or DM treated (24 h) 10T1/2 cells. Sp1a, Sp1b and corresponding mutant oligonucleotides were tested directly and with gel mobility super shifts by anti-Sp1 antibody. Data shown are representative of one of two duplicate experiments with similar results. (B) Chromatin immunoprecipitation (ChIP) assays with distal and proximate regions of mouse gene 5-flanking sequence with 10T1/2 nuclear extracts. Primer sets used to amplify distal and proximate regions are shown in Figure 4A and 4B. The chromatin immunoprecipitation assay was performed with 10T1/2 cells untreated or treated with DM for 6 h or 24 h. Data are the mean ± S.D. of three independent experiments except where indicated. #, data from a single experiment.
This fast activation in 10T1/2 cells slowly transforms to a predominant dependence on the proximal binding of Sp1 that is shown by the EMSA complexes. The late binding of Pax6 in the proximal region suggested by the ChIP analysis may reflect a proximal Sp1/Pax6 interaction of the type identified through heterologous transfection of Pax6 (Table 3 & 4). Many interactions of Sp1 with other transcriptions factors have been reported [52]. The ChIP assays confirm that DM stimulated Sp1 binding to the proximal promoter (Fig. 7B).
Discussion
Cyp1b1 exhibits a specific spatial-temporal pattern of expression during mouse embryogenesis, notably in the neural crest and hindbrain [13, 35, 53]. Neural crest cells migrate during embryogenesis to provide a major source not only of neurons but of mesenchymal progenitor cells for generation of bone, cartilage and fat [54]. The mouse multi-potential 10T1/2 cell line and the more adipogenic 3T3-L1 line which each expresses Cyp1b1 provide models for cells derived from neural crest through this mesenchymal development [55]. We have previously shown that 10T1/2 cells exhibit a substantial increase in Cyp1b1 expression following adipogenic stimulation by the DM combination [22, 33]. This increase precedes the increase in PPARγ, the central regulator of adipogenesis [21]. Here we show that in 3T3-L1 cells there is a comparable increase in Cyp1b1 expression following the same adipogenic stimulation, but which is slower than in 10T1/2 cells and delayed temporally relative to the increase in PPARγ
Interestingly, different mechanisms for the stimulation of Cyp1b1 expression appear to be involved in each cell type. The slower stimulation of Cyp1b1 expression in 3T3-L1 by the adipogenic DM mixture depends on an 18 base sequence which is located 6.1 kb upstream of the transcription start site. This sequence, although highly conserved between mice and rats, is only minimally involved in the equivalent 10T1/2 response (Fig. 3 and 6). We have now shown that this 18 base sequence binds the major developmental regulator Pax6. This transcription factor, like Cyp1b1, is expressed in the neural crest, hind brain and eye and is critical to developmental processes at these sites [49]. This Pax6 element (PaxE) binds to the paired domain (PD) which is located near the N-terminus of Pax6 [46] (Fig. 3B). The PD mediates the stimulation of transcription for several Pax6 responsive genes (Glucagon, δ-cryystallin, optimedin A, and Reelin) [40, 47, 48]. Pax proteins also have a central homeobox domain (HD) [56, 57] which may bind to the TAATTA core homeobox element located in the highly conserved 155 base sequence (>90 percent) upstream of PaxE (Fig. 3A). Homeobox elements mediate transcription produced by several developmental regulators including Hox proteins. Importantly, Hoxb1 is strongly expressed in the mouse neural crest rhombomere 4 immediately prior to Cyp1b1 [58]. These two proteins are spatially co-localized while Pax6 appears at the rhombomere periphery [35] (M. McGuire, unpublished).
These same promoter analyses show that 3T3-L1 stimulation also depends on the Sp1 elements in the proximal promoter (Fig. 2E). In 10T1/2 cells DM rapidly activates the Cyp1b1 promoter activity within 3h through a process that depends on far upstream sequences (Fig. 6). Promoter deletion experiments and ChIP analyses each suggest that Pax6 and PaxE contribute much less to Cyp1b1 stimulation in 10T1/2 cells.
We initially considered that the FUER which mediates the strong adrenal response of rat Cyp1b1 to cAMP [17] might also contribute to this rapid response to DM in 10T1/2 cells (Table 2). This possibility was supported by the finding that FUER is indeed activated early in this stimulation of 10T1/2 cells, probably because DM activates either SF-1 (Nr5a1) or LRH-1 (Nr5a2) provided by transcription factors which each mediate the FUER response [17, 59]. However, deletion of either FUER from the rat Cyp1b1 promoter or the equivalent mouse FUER region (−5202 to −5001) did not affect the DM stimulation (Fig. 6C). In addition , each of the active SF-1 and CREB elements that mediate the PKA stimulus in the rat FUER differs at critical bases in the mouse sequence [17] (Fig. 2B).
DM stimulates the binding of very low levels of Pax6 protein to PaxE in 3T3-L1 cells which were only detectable through the high sensitivity provided by EMSA and 32P-labeling (Fig. 3D). ChIP comparisons (Fig. 4C and 7B) confirm that Pax6 binding to the Cyp1b1 promoter increases following the DM stimulation of 3T3-L1 cells, but is greater in the proximal promoter than in the distal region around PaxE. The EMSA complex is sensitive to the Mut2 mutation which blocks reporter activity and is not only bound but also stabilized by the Pax6 antibody (α-Pax6). The DM activation of Pax6 in this complex may involve kinase modification. The C-terminus of Pax6 comprises a proline-serine-threonine rich domain [60] that can be phosphorylated by several kinases including Mek/Erk Map kinase which is activated during early adipogenesis [8, 61]. Darpp-32, an inhibitor of PP1, a serine protein phosphatase which reverses Pax6 phosphorylation, increases during 3T3-L1 adipogenesis [60, 62].
The Pax6 complex also competes for the PaxE site with a low mobility (LM) complex which does not include Pax6 but which is similarly stimulated by DM (Fig. 3D). The LM complex formation is increased by the Mut2 mutation, suggesting that this mutation decreases the affinity of the 18 base sequence for Pax6 while increasing the affinity for alternative binding proteins. The individual components of the PaxE and LM complexes will be important to identify to further understand the mechanisms by which PAX6 contributes to the regulation of Cyp1b1 and the physiologic role of Pax6 and the PaxE binding site. We have previously described a similar competition between DNA binding proteins for a pair of AhR recognition sites in the AhER of the Cyp1b1 promoter [16]. Streptavidin beads combined with specific biotin-labeled oligonucleotides selectively precipitated the respective unique complexes. This approach could be used in combination with mass spectrometry to identify the individual protein components of PaxE complexes in future studies. They are however beyond the scope of the present study.
Pax6 functions in combination with Sp1 complexes in the proximal promoter. The DM stimulated Cyp1b1 promoter activity in 3T3-L1 cells is completely dependent on these Sp1 sites (Fig. 2E). Heterologous expression experiments established that Pax6 directly stimulates the Cyp1b1 proximal promoter in 3T3-L1 cells (Tables 3 and 4) consistent with the ChIP detection of Pax6 binding in this region. This stimulation by heterologous Pax6 also depends on Sp1 but is insensitive to PaxE mutations and may therefore replace the enhancer effects of PaxE. The Pax6 stimulation of the neuro-regulator reelin also requires Sp1 binding in the proximal promoter via a Pax6 element adjacent to the Sp1 element [40]. No equivalent sequence is however evident in the proximal promoter of Cyp1b1 (Fig. 4B).
The 24 h DM stimulation in 10T1/2 cells also depends on the activation of the adjacent Sp1 sites in the proximal promoter. The Sp1 complexes which have been directly shown by EMSA for 10T1/2 cells (Fig. 7A) probably also mediate the Sp1 contributions to DM stimulation of Cyp1b1 in 3T3-L1 cells. By contrast, the distinctive acute stimulation in 10T1/2 cells is independent of Sp1, consistent with the time dependent increase in Sp1 binding to the proximal promoter shown by the ChIP analyses (Fig. 7B).
These Sp1 elements are also essential for basal and AhR-induced activity of human CYP1B1 [63] and for cAMP stimulation of rat Cyp1b1 [17]. Sp1 contains three Cys2–His2 zinc fingers which bind to GC-rich DNA sequences [51, 64] but which also interact with other transcription factors [52]. Sp1 undergoes O-glycosylation and phosphorylation [64] which may therefore contribute to the DM activation (Fig. 7). Further analysis of proximal promoter Pax6 /Sp1 interactions is clearly of interest, especially for the role of putative phosphorylation sites on Sp1 and identification of additional proteins contributing to the regulatory complexes. Sequence modification in the proximal promoter adjacent to the Sp1 sites could also be employed to identify further cis interactions between binding proteins, while gel shift and pull down approaches coupled with mass spectrometry could be used to further characterize direct interactions of Pax6 with Sp1[16, 40].
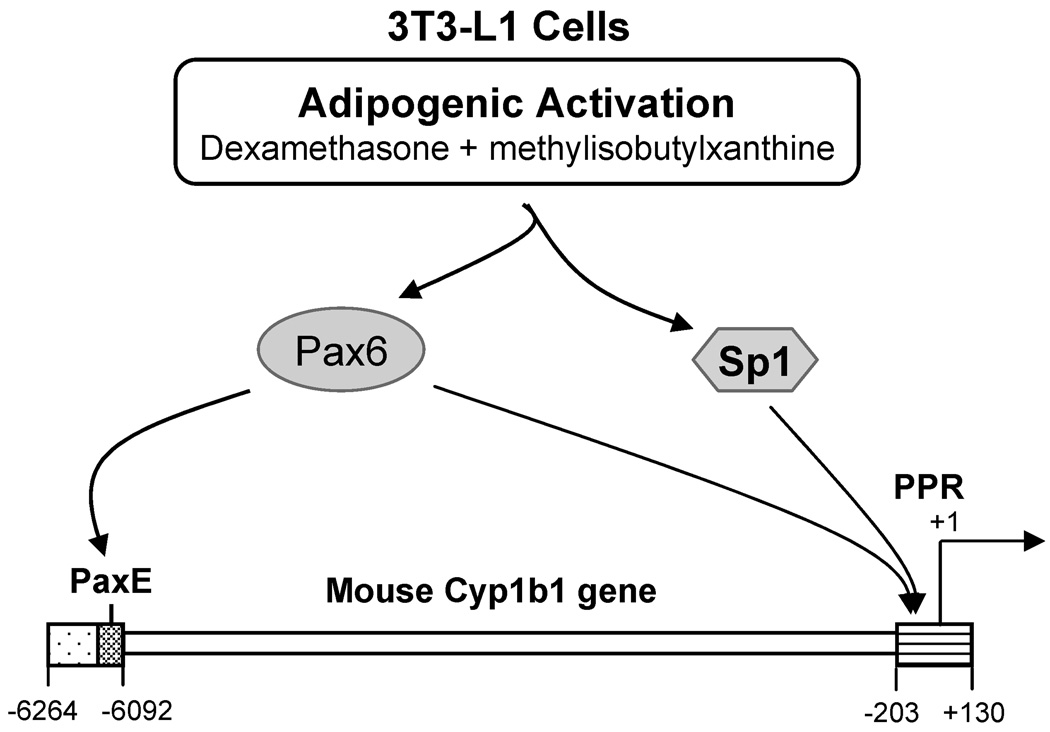
In summary, we have identified a Pax6 binding site in the far upstream Cyp1b1 5’-flanking sequence and also a partnership between Pax6 and Sp1 in the proximal promoter, each of which is stimulated by the adipogenic DM mixture in 3T3-L1 cells. This dual mechanism for Pax6 which involves distal and proximal sites is represented in Figure 8. The proximal process is aided by the elevated levels of Pax6 provided by transfection. The distal PaxE complex may however direct the Pax6 to the proximal Sp1. Differences in the Pax6 involvement in the related 10T1/2 and 3T3-L1 cells suggest appreciable plasticity in the regulation of Cyp1b1 during the fate processes emerging from the neural crest. The several unusual features of the Cyp1b1 gene suggest very special functions for Cyp1b1-mediated monooxygenase activity toward endogenous cell substrates during development. These gene features include the presence of only 2 introns and a very long conserved 3’-UTR [4, 5] and the presence of novel upstream regulatory regions including the AhER [16, 29], the FUER [17] and the Pax6 region described here. Although the upstream promoter regions of both mice and rats share the PaxE and adjacent sequences (Fig. 2A, Fig. 3A), they also differ significantly, especially with respect to the FUER at SF-1 and CREB sequences (Fig 2B). In comparison to the rodent promoter, the human Cyp1b1 promoter region does not retain either feature. The endogenous substrates for Cyp1b1 are critical to these activities. Conversion of retinaldehyde to retinoic acid by Cyp1b1 has been previously demonstrated and implicated in the neural crest changes [2, 36]. However, other substrates such as melatonin and estradiol may also involve Cyp1b1 during developmental processes [2, 65, 66]. Cyp1b1 in endothelial cells also suppresses oxidative stress and NF-κ B signaling while promoting vasculogenesis [3, 28]. The involvement of Cyp1b1 substrates in oxidative stress processes may contribute to fatty acid regulation and PPARγ. Notably, Pax6 directly binds to PPARγ and RXR in the regulation of glucagon expression in the pancreas [67]. PPARγ is expressed in endothelial cells [68] and in macrophage [69], each alongside Cyp1b1. The recurrent co-expression of Pax6, PPARγ and Cyp1b1 requires further investigation with respect to how their functions may interact.
Figure 8. Mechanism for activation of Cyp1b1 by Pax6 in mouse 3T3-L1 cells.
The developmental regulator Pax6 acts at two sites in the Cyp1b1 promoter when expression is increased in mouse 3T3-L1 cells by adipogenic stimuli. Co-operation of Pax6 with Sp1 in the proximal promoter region (PPR) is indicated. This co-operation is enhanced by a distal binding of Pax6 at a conserved 18 base element (PaxE).
Highlights.
Cyp1b1 is stimulated in multipotential mouse cell lines by adipogenic stimuli.
A far upstream element in the Cyp1b1 promoter binds the homeobox factor Pax6.
Pax6 binding to this conserved 18 base element is activated by adipogenic stimuli.
Higher levels of transfected Pax6 also stimulate the proximal promoter of Cyp1b1.
Pax6 partners with Sp1 in these effects of adipogenic stimuli on the Cyp1b1 promoter.
Acknowledgments
This work was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK072749]. We thank Meghan Maguire for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used:
AhER, AhR-response enhancer region; AhR, aryl hydrocarbon receptor; ChIP, chromatin immunoprecipitation; CRE, cAMP response element; CREB, CRE binding protein; CYP, cytochrome P450; D, dexamethasone; DM, dexamethasone and methylisobutylxanthine; ECL, enhanced chemiluminescence; EMSA, electrophoretic mobility shift assay; FUER, far upstream enhance region; h, hour(s); HD, homeobox domain; I, insulin; IDM, insulin, dexamethasone, and methylisobutylxanthine; M, methylisobutylxanthine; LRH-1, liver receptor homologue 1; MEF, mouse embryo fibroblast; PAH, polycyclic aromatic hydrocarbon; PaxE, mouse Cyp1b1 Pax6 element; PCR, polymerase chain reaction; PD, paired domain; PKA, protein kinase A; PPARγ, peroxisome proliferator-activated receptorγ; PPR, proximal promoter region; qPCR, quantitative real-time PCR; SF-1, steroidogenic factor 1; Sp1, stimulatory protein 1; TCDD, 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin; SREBP, Sterol regulatory element-binding protein.
References
- 1.Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Proc Natl Acad Sci U S A. 1999;96:1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers D, Wilson L, Maden M, Lumsden A. Development. 2007;134:1369–1383. doi: 10.1242/dev.02815. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y, Scheef EA, Wang S, Sorenson CM, Marcus CB, Jefcoate CR, Sheibani N. Blood. 2009;113:744–754. doi: 10.1182/blood-2008-03-145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savas U, Bhattacharyya KK, Christou M, Alexander DL, Jefcoate CR. J Biol Chem. 1994;269:14905–14911. [PubMed] [Google Scholar]
- 5.Sutter TR, Tang YM, Hayes CL, Wo YY, Jabs EW, Li X, Yin H, Cody CW, Greenlee WF. J Biol Chem. 1994;269:13092–13099. [PubMed] [Google Scholar]
- 6.Bhattacharyya KK, Brake PB, Eltom SE, Otto SA, Jefcoate CR. The Journal of biological chemistry. 1995;270:11595–11602. doi: 10.1074/jbc.270.19.11595. [DOI] [PubMed] [Google Scholar]
- 7.Murray GI, Melvin WT, Greenlee WF, Burke MD. Annu Rev Pharmacol Toxicol. 2001;41:297–316. doi: 10.1146/annurev.pharmtox.41.1.297. [DOI] [PubMed] [Google Scholar]
- 8.Tang QQ, Otto TC, Lane MD. Proc Natl Acad Sci U S A. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beedanagari SR, Taylor RT, Bui P, Wang F, Nickerson DW, Hankinson O. Mol Pharmacol. 2010;78:608–616. doi: 10.1124/mol.110.064899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarfarazi M. Hum Mol Genet. 1997;6:1667–1677. doi: 10.1093/hmg/6.10.1667. [DOI] [PubMed] [Google Scholar]
- 11.Stoilov I, Akarsu AN, Sarfarazi M. Hum Mol Genet. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- 12.Libby RT, Smith RS, Savinova OV, Zabaleta A, Martin JE, Gonzalez FJ, John SW. Science. 2003;299:1578–1581. doi: 10.1126/science.1080095. [DOI] [PubMed] [Google Scholar]
- 13.Stoilov I, Rezaie T, Jansson I, Schenkman JB, Sarfarazi M. Mol Vis. 2004;10:629–636. [PubMed] [Google Scholar]
- 14.Jonsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Toxicol Sci. 2007;100:180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- 15.Brake PB, Jefcoate CR. Endocrinology. 1995;136:5034–5041. doi: 10.1210/endo.136.11.7588239. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Zheng W, Jefcoate CR. Toxicol Appl Pharmacol. 2003;192:174–190. doi: 10.1016/s0041-008x(03)00276-x. [DOI] [PubMed] [Google Scholar]
- 17.Zheng W, Jefcoate CR. Mol Pharmacol. 2005;67:499–512. doi: 10.1124/mol.104.005504. [DOI] [PubMed] [Google Scholar]
- 18.Alexander DL, Ganem LG, Fernandez-Salguero P, Gonzalez F, Jefcoate CR. J Cell Sci. 1998;111(Pt 22):3311–3322. doi: 10.1242/jcs.111.22.3311. [DOI] [PubMed] [Google Scholar]
- 19.Green H, Meuth M. Cell. 1974;3:127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 20.Green H, Kehinde O. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 21.Tontonoz P, Hu E, Spiegelman BM. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 22.Cho YC, Zheng W, Yamamoto M, Liu X, Hanlon PR, Jefcoate CR. Arch Biochem Biophys. 2005;439:139–153. doi: 10.1016/j.abb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Kim JB, Wright HM, Wright M, Spiegelman BM. Proc Natl Acad Sci U S A. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita H, Azma T, Iranami H, Nakahata K, Kimoto Y, Dojo M, Yuge O, Hatano Y. J Pharmacol Exp Ther. 2006;318:312–318. doi: 10.1124/jpet.106.100958. [DOI] [PubMed] [Google Scholar]
- 26.Spink BC, Hussain MM, Katz BH, Eisele L, Spink DC. Biochem Pharmacol. 2003;66:2313–2321. doi: 10.1016/j.bcp.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Chiaro CR, Patel RD, Perdew GH. Mol Pharmacol. 2008;74:1649–1656. doi: 10.1124/mol.108.049379. [DOI] [PubMed] [Google Scholar]
- 28.Tang Y, Scheef EA, Gurel Z, Sorenson CM, Jefcoate CR, Sheibani N. Am J Physiol Cell Physiol. 2010;298:C665–C678. doi: 10.1152/ajpcell.00153.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Savas U, Alexander DL, Jefcoate CR. J Biol Chem. 1998;273:5174–5183. doi: 10.1074/jbc.273.9.5174. [DOI] [PubMed] [Google Scholar]
- 30.Shimba S, Todoroki K, Aoyagi T, Tezuka M. Biochem Biophys Res Commun. 1998;249:131–137. doi: 10.1006/bbrc.1998.9100. [DOI] [PubMed] [Google Scholar]
- 31.Shimba S, Wada T, Tezuka M. Journal of cell science. 2001;114:2809–2817. doi: 10.1242/jcs.114.15.2809. [DOI] [PubMed] [Google Scholar]
- 32.Hanlon PR, Cimafranca MA, Liu X, Cho YC, Jefcoate CR. Toxicol Appl Pharmacol. 2005;207:39–58. doi: 10.1016/j.taap.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Cho YC, Jefcoate CR. J Cell Biochem. 2004;91:336–353. doi: 10.1002/jcb.10743. [DOI] [PubMed] [Google Scholar]
- 34.Zheng W, Brake PB, Bhattacharyya KK, Zhang L, Zhao D, Jefcoate CR. Arch Biochem Biophys. 2003;416:53–67. doi: 10.1016/s0003-9861(03)00282-0. [DOI] [PubMed] [Google Scholar]
- 35.Chambers D, Wilson LJ, Alfonsi F, Hunter E, Saxena U, Blanc E, Lumsden A. Neural development. 2009;4:6. doi: 10.1186/1749-8104-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Howald WN, Juchau MR. Drug Metab Dispos. 2000;28:315–322. [PubMed] [Google Scholar]
- 37.Huang JI, Kazmi N, Durbhakula MM, Hering TM, Yoo JU, Johnstone B. J Orthop Res. 2005;23:1383–1389. doi: 10.1016/j.orthres.2005.03.008.1100230621. [DOI] [PubMed] [Google Scholar]
- 38.Rezaie T, Stoilov I, Sarfarazi M. Mol Vis. 2007;13:1446–1450. [PubMed] [Google Scholar]
- 39.Huang Y, Xie L. Mol Vis. 2010;16:341–352. [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Kundakovic M, Agis-Balboa RC, Pinna G, Grayson DR. J Neurochem. 2007;103:650–665. doi: 10.1111/j.1471-4159.2007.04797.x. [DOI] [PubMed] [Google Scholar]
- 41.Grapp M, Teichler S, Kitz J, Dibaj P, Dickel C, Knepel W, Kratzner R. Biochim Biophys Acta. 2009;1789:403–412. doi: 10.1016/j.bbagrm.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 43.Czerny T, Busslinger M. Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakai M, Serria MS, Ikeda H, Yoshida K, Imaki J, Nishi S. Nucleic Acids Res. 2001;29:1228–1237. doi: 10.1093/nar/29.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fajas L, Schoonjans K, Gelman L, Kim JB, Najib J, Martin G, Fruchart JC, Briggs M, Spiegelman BM, Auwerx J. Mol Cell Biol. 1999;19:5495–5503. doi: 10.1128/mcb.19.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 47.Grinchuk O, Kozmik Z, Wu X, Tomarev S. J Biol Chem. 2005;280:35228–35237. doi: 10.1074/jbc.M506195200. [DOI] [PubMed] [Google Scholar]
- 48.Muta M, Kamachi Y, Yoshimoto A, Higashi Y, Kondoh H. Genes Cells. 2002;7:791–805. doi: 10.1046/j.1365-2443.2002.00560.x. [DOI] [PubMed] [Google Scholar]
- 49.Smith SB, Ee HC, Conners JR, German MS. Mol Cell Biol. 1999;19:8272–8280. doi: 10.1128/mcb.19.12.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, Zavadil J, Cvekl A. PLoS One. 2009;4:e4159. doi: 10.1371/journal.pone.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu S. Gene. 508:1–8. doi: 10.1016/j.gene.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 52.Tan NY, Khachigian LM. Mol Cell Biol. 2009;29:2483–2488. doi: 10.1128/MCB.01828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamm ER. Ophthalmologe. 2011;108:610–614. 616–617. doi: 10.1007/s00347-010-2294-5. [DOI] [PubMed] [Google Scholar]
- 54.Lemos DR, Paylor B, Chang C, Sampaio A, Underhill TM, Rossi FM. Stem Cells. 30:1152–1162. doi: 10.1002/stem.1082. [DOI] [PubMed] [Google Scholar]
- 55.Kemaladewi DU, de Gorter DJ, Aartsma-Rus A, van Ommen GJ, ten Dijke P, t Hoen PA, Hoogaars WM. Faseb J. 26:1462–1472. doi: 10.1096/fj.11-191189. [DOI] [PubMed] [Google Scholar]
- 56.Breitling R, Gerber JK. Dev Genes Evol. 2000;210:644–650. doi: 10.1007/s004270000106. [DOI] [PubMed] [Google Scholar]
- 57.Jun S, Desplan C. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 58.Alexander T, Nolte C, Krumlauf R. Annu Rev Cell Dev Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 59.Kelly VR, Hammer GD. Mol Cell Endocrinol. 332:116–124. doi: 10.1016/j.mce.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan Q, Liu WB, Qin J, Liu J, Chen HG, Huang X, Chen L, Sun S, Deng M, Gong L, Li Y, Zhang L, Liu Y, Feng H, Xiao Y, Liu Y, Li DW. J Biol Chem. 2007;282:13954–13965. doi: 10.1074/jbc.M611476200. [DOI] [PubMed] [Google Scholar]
- 61.Hanlon PR, Zheng W, Ko AY, Jefcoate CR. Toxicol Appl Pharmacol. 2005;202:215–228. doi: 10.1016/j.taap.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 62.Mikkola I, Bruun JA, Bjorkoy G, Holm T, Johansen T. J Biol Chem. 1999;274:15115–15126. doi: 10.1074/jbc.274.21.15115. [DOI] [PubMed] [Google Scholar]
- 63.Tang YM, Wo YY, Stewart J, Hawkins AL, Griffin CA, Sutter TR, Greenlee WF. J Biol Chem. 1996;271:28324–28330. doi: 10.1074/jbc.271.45.28324. [DOI] [PubMed] [Google Scholar]
- 64.Bouwman P, Philipsen S. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 65.Chang TK, Chen J, Yang G, Yeung EY. J Pineal Res. 2010;48:55–64. doi: 10.1111/j.1600-079X.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 66.Ma X, Idle JR, Krausz KW, Gonzalez FJ. Drug Metab Dispos. 2005;33:489–494. doi: 10.1124/dmd.104.002410. [DOI] [PubMed] [Google Scholar]
- 67.Kratzner R, Frohlich F, Lepler K, Schroder M, Roher K, Dickel C, Tzvetkov MV, Quentin T, Oetjen E, Knepel W. Mol Pharmacol. 2008;73:509–517. doi: 10.1124/mol.107.035568. [DOI] [PubMed] [Google Scholar]
- 68.Kanda T, Brown JD, Orasanu G, Vogel S, Gonzalez FJ, Sartoretto J, Michel T, Plutzky J. J Clin Invest. 2009;119:110–124. doi: 10.1172/JCI36233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gautier EL, Chow A, Spanbroek R, Marcelin G, Greter M, Jakubzick C, Bogunovic M, Leboeuf M, van Rooijen N, Habenicht AJ, Merad M, Randolph GJ. J Immunol. 2012;189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]