Abstract
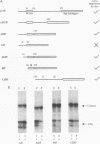
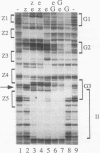
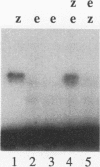
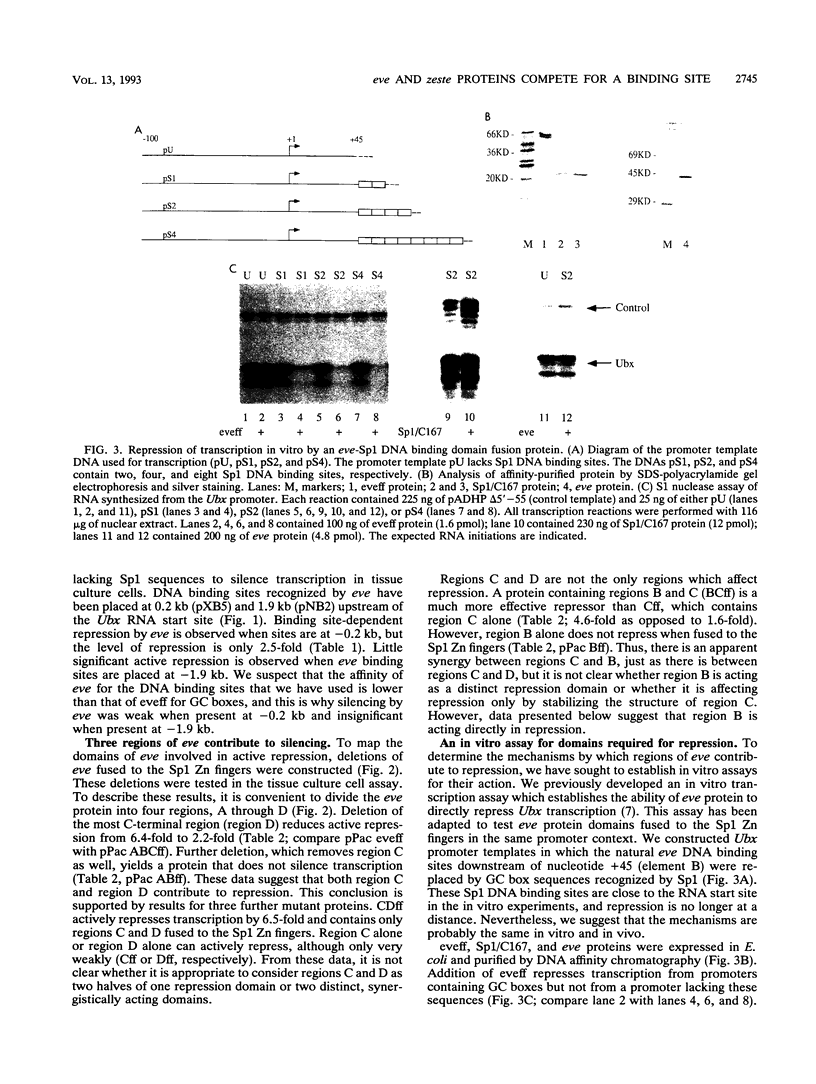
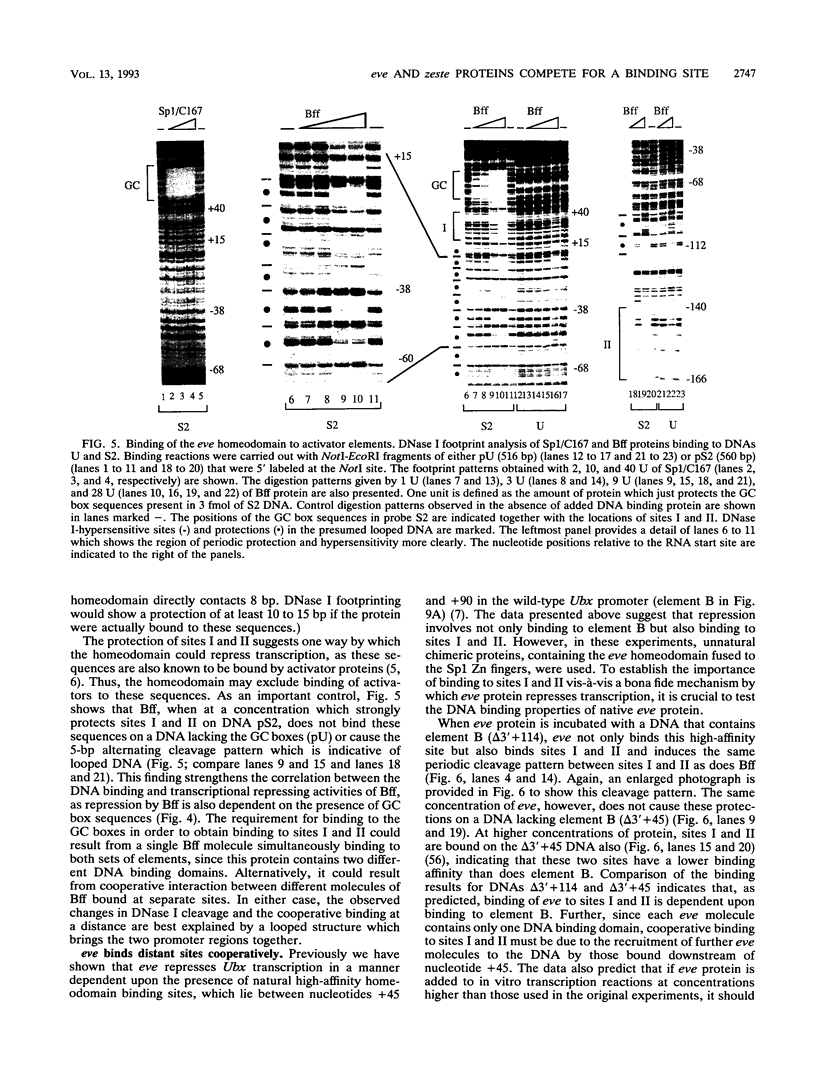
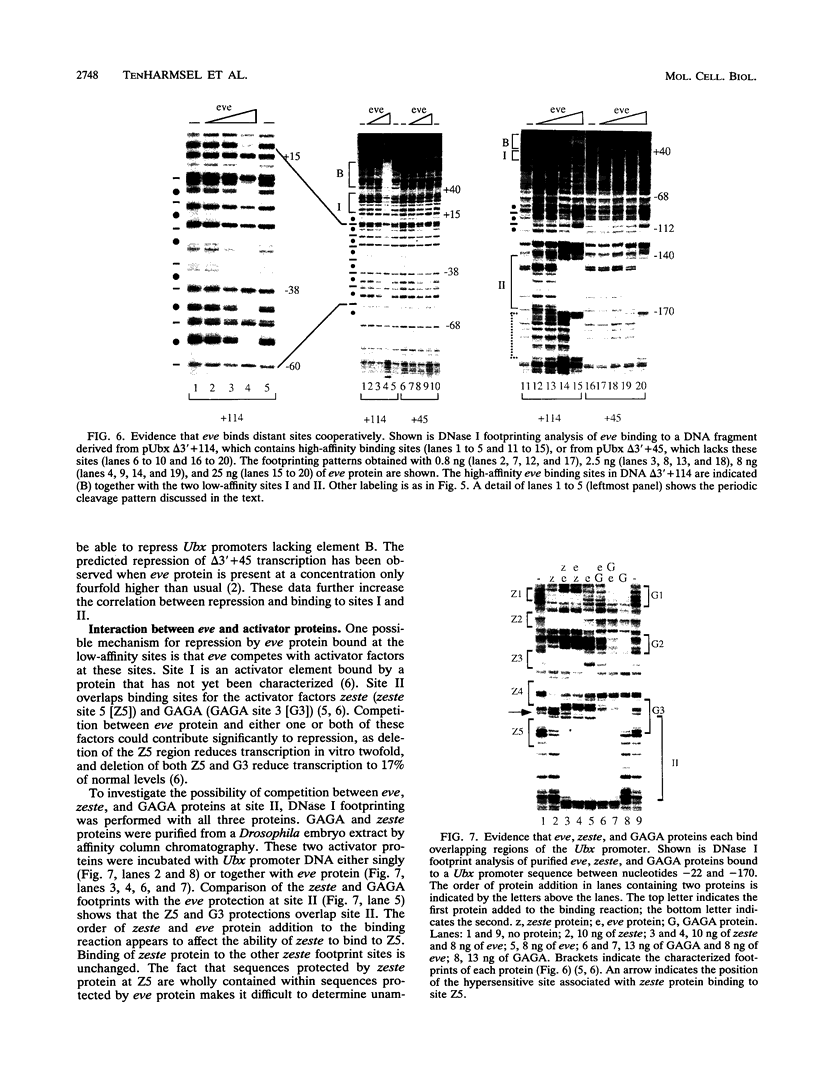
In this study, we examined how the Drosophila developmental control gene even-skipped (eve) represses transcription. Tissue culture cells were used to show that eve contains domains which inhibit transcriptional activators present at the Ultrabithorax (Ubx) proximal promoter when bound up to 1.5 kb away from these activators. Different portions of eve were fused to a heterologous DNA binding domain to show that three adjacent regions of eve contribute to silencing. There appear to be two mechanisms by which eve protein represses transcription. In this study, we used in vitro transcription and DNA binding experiments to provide evidence for one of these mechanisms. Repression in vitro correlates with binding of eve protein to two low-affinity sites in the Ubx proximal promoter. Occupancy of these low-affinity sites is dependent upon cooperative binding of other eve molecules to a separate high-affinity site. Some of these sites are separated by over 150 bp of DNA, and the data suggest that this intervening DNA is bent to form a looped structure similar to those caused by prokaryotic repressors. One of the low-affinity sites overlaps an activator element bound by the zeste transcription factor. Binding of eve protein is shown to exclude binding by zeste protein. These data suggest a mechanism for silencing whereby a repressor protein would be targeted to DNA by a high-affinity element, which itself does not overlap activator elements. Cooperative binding of further repressor molecules to distant low-affinity sites, and competition with activators bound at these sites lead to repression at a distance.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler S., Waterman M. L., He X., Rosenfeld M. G. Steroid receptor-mediated inhibition of rat prolactin gene expression does not require the receptor DNA-binding domain. Cell. 1988 Mar 11;52(5):685–695. doi: 10.1016/0092-8674(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Bickel S., Benson M., Pirrotta V., Tjian R. Zeste encodes a sequence-specific transcription factor that activates the Ultrabithorax promoter in vitro. Cell. 1988 Jun 3;53(5):713–722. doi: 10.1016/0092-8674(88)90089-x. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989 Aug 11;58(3):433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. Transcription factors and the control of Drosophila development. Trends Genet. 1989 Nov;5(11):377–383. doi: 10.1016/0168-9525(89)90173-x. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988 Jun 3;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Christen B., Bienz M. A cis-element mediating Ultrabithorax autoregulation in the central nervous system. Mech Dev. 1992 Nov;39(1-2):73–80. doi: 10.1016/0925-4773(92)90027-h. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Holtzman D. A., Jackson S. P., Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989 Dec 1;59(5):827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P., Dawid I. B. Transient expression of genes introduced into cultured cells of Drosophila. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M. I., Miner J. N., Yoshinaga S. K., Yamamoto K. R. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990 Sep 14;249(4974):1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989 Jan 12;337(6203):138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Baltimore D. Activation in vitro of NF-kappa B by phosphorylation of its inhibitor I kappa B. Nature. 1990 Apr 12;344(6267):678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Han K., Levine M. S., Manley J. L. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989 Feb 24;56(4):573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Scott M. P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990 Nov 30;63(5):883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Kraut R., Levine M., Rushlow C. A. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell. 1991 Jan 25;64(2):439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., O'Farrell P. H. Activation and repression of transcription by homoeodomain-containing proteins that bind a common site. Nature. 1988 Dec 22;336(6201):744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes J. B., O'Farrell P. H. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991 Jun;10(6):1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Johnson F. B., Krasnow M. A. Stimulation of transcription by an Ultrabithorax protein in vitro. Genes Dev. 1990 Jun;4(6):1044–1052. doi: 10.1101/gad.4.6.1044. [DOI] [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987 Jul 3;50(1):143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Courey A. J., Ladika J., Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988 Dec 16;242(4885):1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P. D., Rio D. C. Drosophila P-element transposase is a transcriptional repressor in vitro. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2613–2617. doi: 10.1073/pnas.88.7.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Knight J. D., Li R., Botchan M. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3204–3208. doi: 10.1073/pnas.88.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Saffman E. E., Kornfeld K., Hogness D. S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989 Jun 16;57(6):1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991 Apr;5(4):616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- Laney J. D., Biggin M. D. zeste, a nonessential gene, potently activates Ultrabithorax transcription in the Drosophila embryo. Genes Dev. 1992 Aug;6(8):1531–1541. doi: 10.1101/gad.6.8.1531. [DOI] [PubMed] [Google Scholar]
- Licht J. D., Grossel M. J., Figge J., Hansen U. M. Drosophila Krüppel protein is a transcriptional repressor. Nature. 1990 Jul 5;346(6279):76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Mandal N., Su W., Haber R., Adhya S., Echols H. DNA looping in cellular repression of transcription of the galactose operon. Genes Dev. 1990 Mar;4(3):410–418. doi: 10.1101/gad.4.3.410. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991 Nov;10(11):3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y., Horikoshi M., Roeder R. G., Desplan C. Binding site-dependent direct activation and repression of in vitro transcription by Drosophila homeodomain proteins. Cell. 1990 May 4;61(3):475–484. doi: 10.1016/0092-8674(90)90529-n. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Rio D. C., Tjian R. SV40 T antigen binding site mutations that affect autoregulation. Cell. 1983 Apr;32(4):1227–1240. doi: 10.1016/0092-8674(83)90305-7. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972 Apr;27(2):353–365. [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Struhl G., White R. A. Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell. 1985 Dec;43(2 Pt 1):507–519. doi: 10.1016/0092-8674(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Thali M., Müller M. M., DeLorenzi M., Matthias P., Bienz M. Drosophila homoeotic genes encode transcriptional activators similar to mammalian OTF-2. Nature. 1988 Dec 8;336(6199):598–601. doi: 10.1038/336598a0. [DOI] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. Alpha beta lineage-specific expression of the alpha T cell receptor gene by nearby silencers. Cell. 1989 Nov 17;59(4):649–655. doi: 10.1016/0092-8674(89)90010-x. [DOI] [PubMed] [Google Scholar]
- Winslow G. M., Hayashi S., Krasnow M., Hogness D. S., Scott M. P. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989 Jun 16;57(6):1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- Zuo P., Stanojević D., Colgan J., Han K., Levine M., Manley J. L. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991 Feb;5(2):254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]