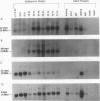
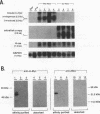
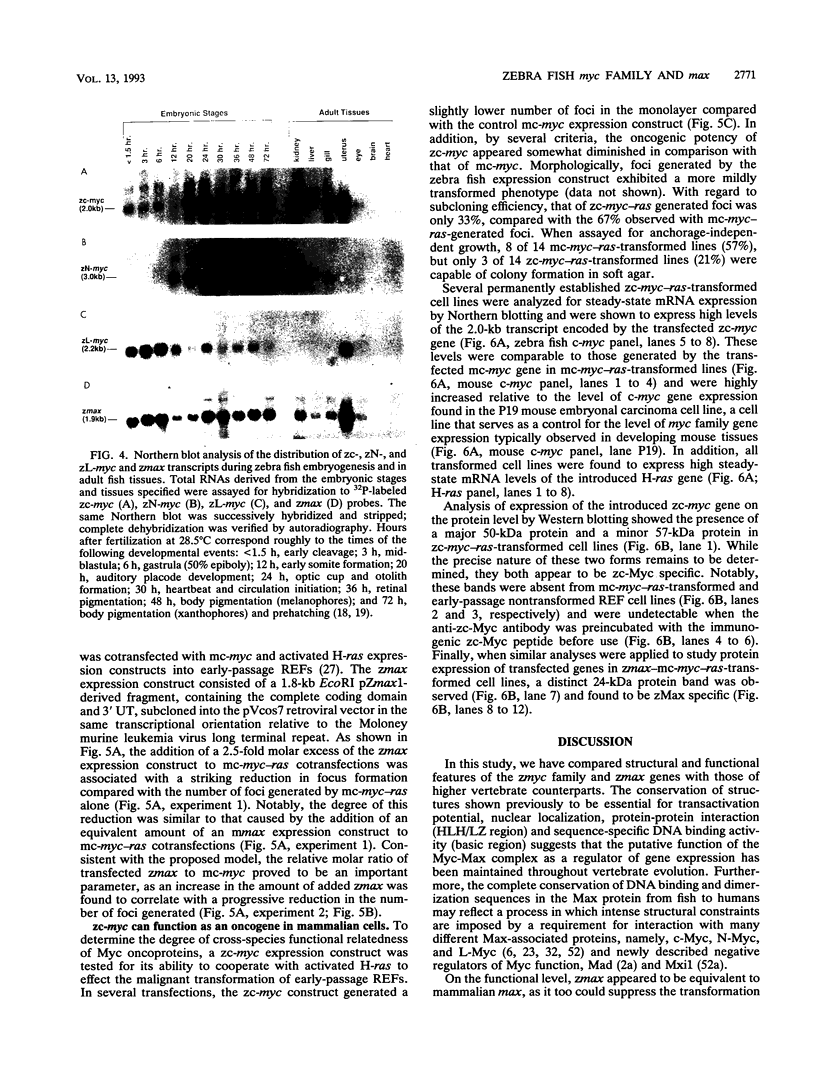
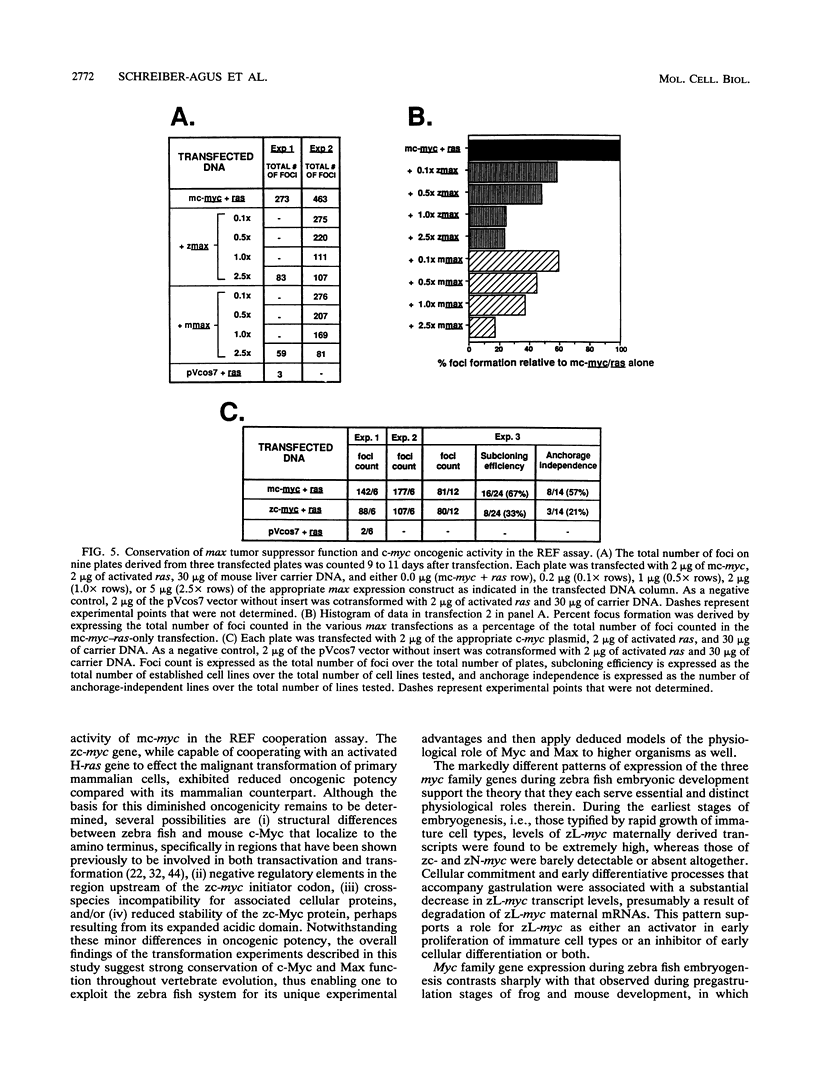
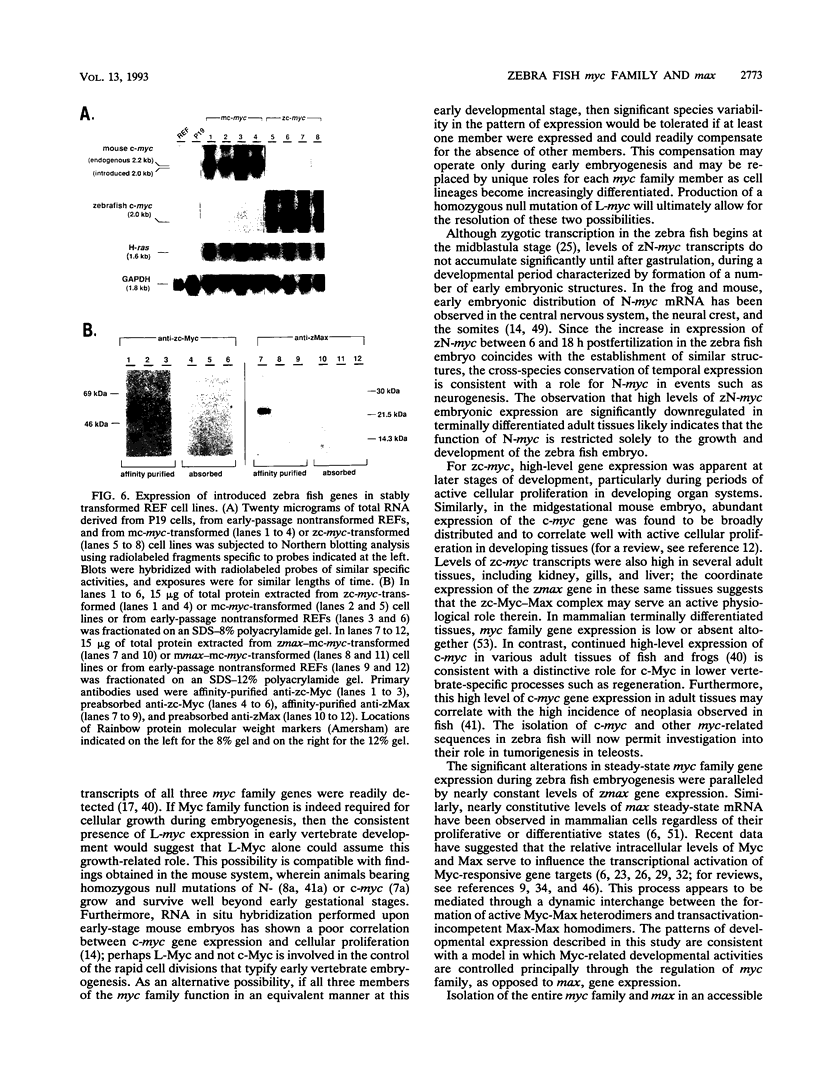
Abstract
To gain insight into the role of Myc family oncoproteins and their associated protein Max in vertebrate growth and development, we sought to identify homologs in the zebra fish (Brachydanio rerio). A combination of a polymerase chain reaction-based cloning strategy and low-stringency hybridization screening allowed for the isolation of zebra fish c-, N-, and L-myc and max genes; subsequent structural characterization showed a high degree of conservation in regions that encode motifs of known functional significance. On the functional level, zebra fish Max, like its mammalian counterpart, served to suppress the transformation activity of mouse c-Myc in rat embryo fibroblasts. In addition, the zebra fish c-myc gene proved capable of cooperating with an activated H-ras to effect the malignant transformation of mammalian cells, albeit with diminished potency compared with mouse c-myc. With respect to their roles in normal developing tissues, the differential temporal and spatial patterns of steady-state mRNA expression observed for each zebra fish myc family member suggest unique functions for L-myc in early embryogenesis, for N-myc in establishment and growth of early organ systems, and for c-myc in increasingly differentiated tissues. Furthermore, significant alterations in the steady-state expression of zebra fish myc family genes concomitant with relatively constant max expression support the emerging model of regulation of Myc function in cellular growth and differentiation.
Full text
PDF
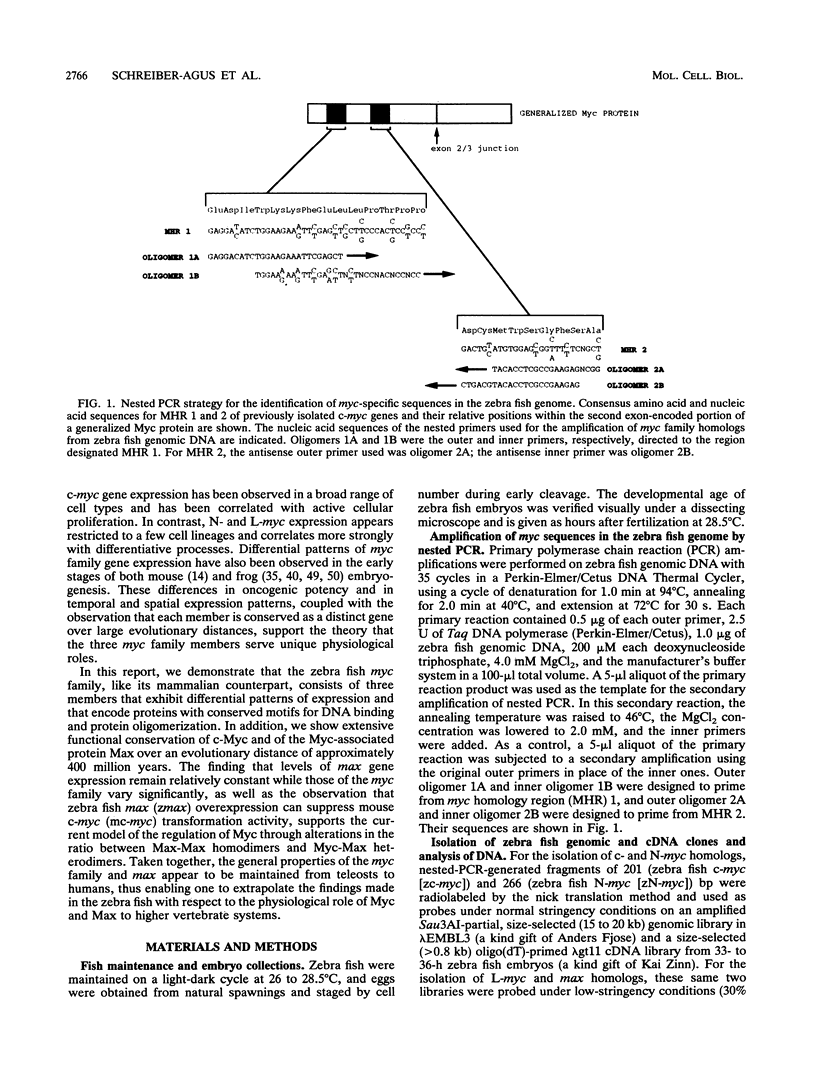

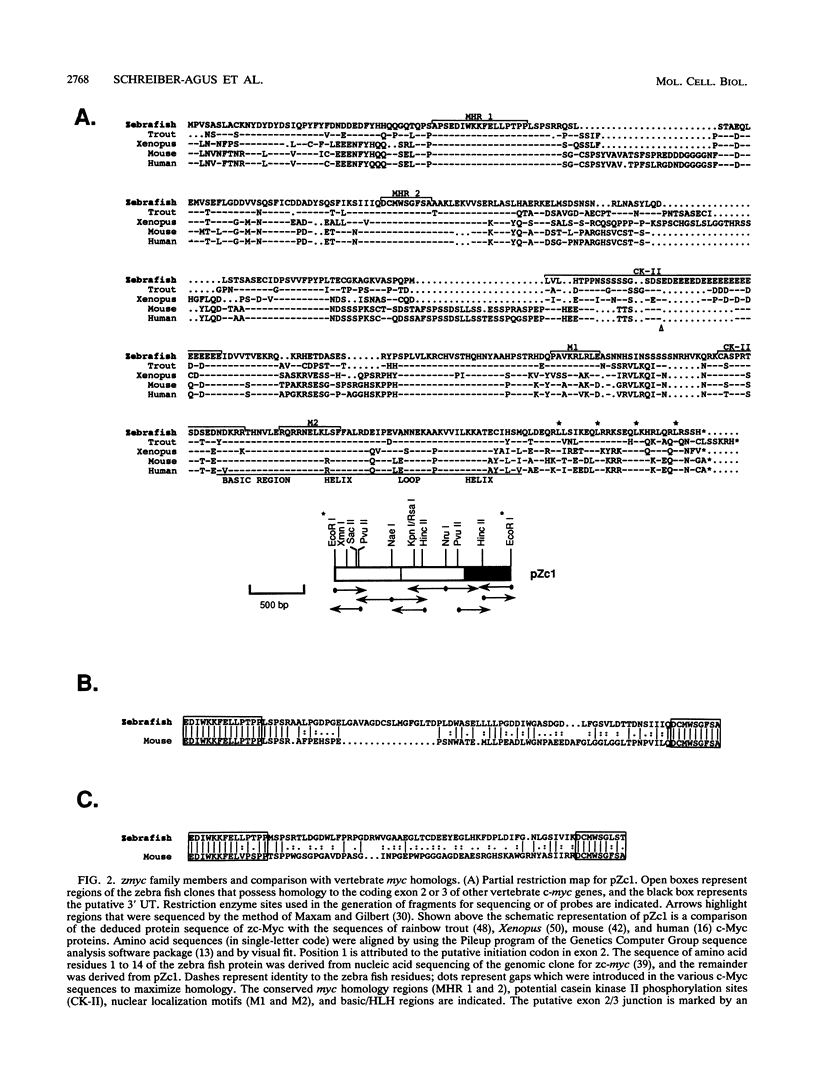




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson P., Goldfarb M. P., Weinberg R. A. A defined subgenomic fragment of in vitro synthesized Moloney sarcoma virus DNA can induce cell transformation upon transfection. Cell. 1979 Jan;16(1):63–75. doi: 10.1016/0092-8674(79)90188-0. [DOI] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Kretzner L., Eisenman R. N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993 Jan 29;72(2):211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., Dang C. V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992 Jul;12(7):3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberich S. J., Cole M. D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992 Feb;6(2):166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Lüscher B., Eisenman R. N. Myc and Max associate in vivo. Genes Dev. 1992 Jan;6(1):71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- Bonnieu A., Piechaczyk M., Marty L., Cuny M., Blanchard J. M., Fort P., Jeanteur P. Sequence determinants of c-myc mRNA turn-over: influence of 3' and 5' non-coding regions. Oncogene Res. 1988 Sep;3(2):155–166. [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J., Malynn B. A., Fisher P., Stewart V., Jeannotte L., Goff S. P., Robertson E. J., Alt F. W. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992 Dec;6(12A):2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Cole M. D. Myc meets its Max. Cell. 1991 May 31;65(5):715–716. doi: 10.1016/0092-8674(91)90377-b. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Hatton K. S., Tesfaye A., Yancopoulos G. D., Alt F. W. The human myc gene family: structure and activity of L-myc and an L-myc pseudogene. Genes Dev. 1987 Dec;1(10):1311–1326. doi: 10.1101/gad.1.10.1311. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho R. A., Schreiber-Agus N., Alt F. W. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs K. M., Martin G. R., Bishop J. M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989 Jun;3(6):860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Fasano O., Taparowsky E., Fiddes J., Wigler M., Goldfarb M. Sequence and structure of the coding region of the human H-ras-1 gene from T24 bladder carcinoma cells. J Mol Appl Genet. 1983;2(2):173–180. [PubMed] [Google Scholar]
- Gazin C., Dupont de Dinechin S., Hampe A., Masson J. M., Martin P., Stehelin D., Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984 Feb;3(2):383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The nematode Caenorhabditis elegans. Science. 1988 Jun 10;240(4858):1448–1453. doi: 10.1126/science.3287621. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B. Genetics and early development of zebrafish. Trends Genet. 1989 Aug;5(8):283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Legouy E., DePinho R., Zimmerman K., Collum R., Yancopoulos G., Mitsock L., Kriz R., Alt F. W. Structure and expression of the murine L-myc gene. EMBO J. 1987 Nov;6(11):3359–3366. doi: 10.1002/j.1460-2075.1987.tb02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McBurney M. W., Jones-Villeneuve E. M., Edwards M. K., Anderson P. J. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982 Sep 9;299(5879):165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Mukherjee B., Morgenbesser S. D., DePinho R. A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992 Aug;6(8):1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- Mäkelä T. P., Koskinen P. J., Västrik I., Alitalo K. Alternative forms of Max as enhancers or suppressors of Myc-ras cotransformation. Science. 1992 Apr 17;256(5055):373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. A new bind for Myc. Trends Genet. 1992 Mar;8(3):91–96. doi: 10.1016/0168-9525(92)90196-b. [DOI] [PubMed] [Google Scholar]
- Principaud E., Spohr G. Xenopus laevis c-myc I and II genes: molecular structure and developmental expression. Nucleic Acids Res. 1991 Jun 11;19(11):3081–3088. doi: 10.1093/nar/19.11.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Hopkins N. Of fin and fur: mutational analysis of vertebrate embryonic development. Genes Dev. 1992 Jan;6(1):1–13. doi: 10.1101/gad.6.1.1. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Drosophila melanogaster as an experimental organism. Science. 1988 Jun 10;240(4858):1453–1459. doi: 10.1126/science.3131880. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N., Torres R., Horner J., Lau A., Jamrich M., DePinho R. A. Comparative analysis of the expression and oncogenic activities of Xenopus c-, N-, and L-myc homologs. Mol Cell Biol. 1993 Apr;13(4):2456–2468. doi: 10.1128/mcb.13.4.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton B. R., Perkins A. S., Tessarollo L., Sassoon D. A., Parada L. F. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992 Dec;6(12A):2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Walker C., Dower N., Knauber D., Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature. 1981 May 28;291(5813):293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- Torres R., Schreiber-Agus N., Morgenbesser S. D., DePinho R. A. Myc and Max: a putative transcriptional complex in search of a cellular target. Curr Opin Cell Biol. 1992 Jun;4(3):468–474. doi: 10.1016/0955-0674(92)90013-3. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Kao T. H., Reece K. S., Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 1985 Apr 11;13(7):2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beneden R. J., Watson D. K., Chen T. T., Lautenberger J. A., Papas T. S. Cellular myc (c-myc) in fish (rainbow trout): its relationship to other vertebrate myc genes and to the transforming genes of the MC29 family of viruses. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3698–3702. doi: 10.1073/pnas.83.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize P. D., Vaughan A., Krieg P. Expression of the N-myc proto-oncogene during the early development of Xenopus laevis. Development. 1990 Nov;110(3):885–896. doi: 10.1242/dev.110.3.885. [DOI] [PubMed] [Google Scholar]
- Vriz S., Taylor M., Méchali M. Differential expression of two Xenopus c-myc proto-oncogenes during development. EMBO J. 1989 Dec 20;8(13):4091–4097. doi: 10.1002/j.1460-2075.1989.tb08593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. J., Le Beau M. M., Diaz M. O., Hay N. Expression, regulation, and chromosomal localization of the Max gene. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3111–3115. doi: 10.1073/pnas.89.7.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A., Cziepluch C., Hamann U., Schürmann J., Schwab M. The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells. EMBO J. 1991 Dec;10(12):3703–3712. doi: 10.1002/j.1460-2075.1991.tb04938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos A. S., Gyuris J., Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993 Jan 29;72(2):223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]