Abstract
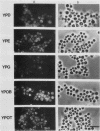
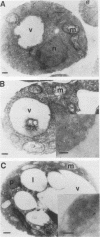
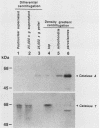
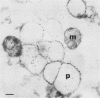
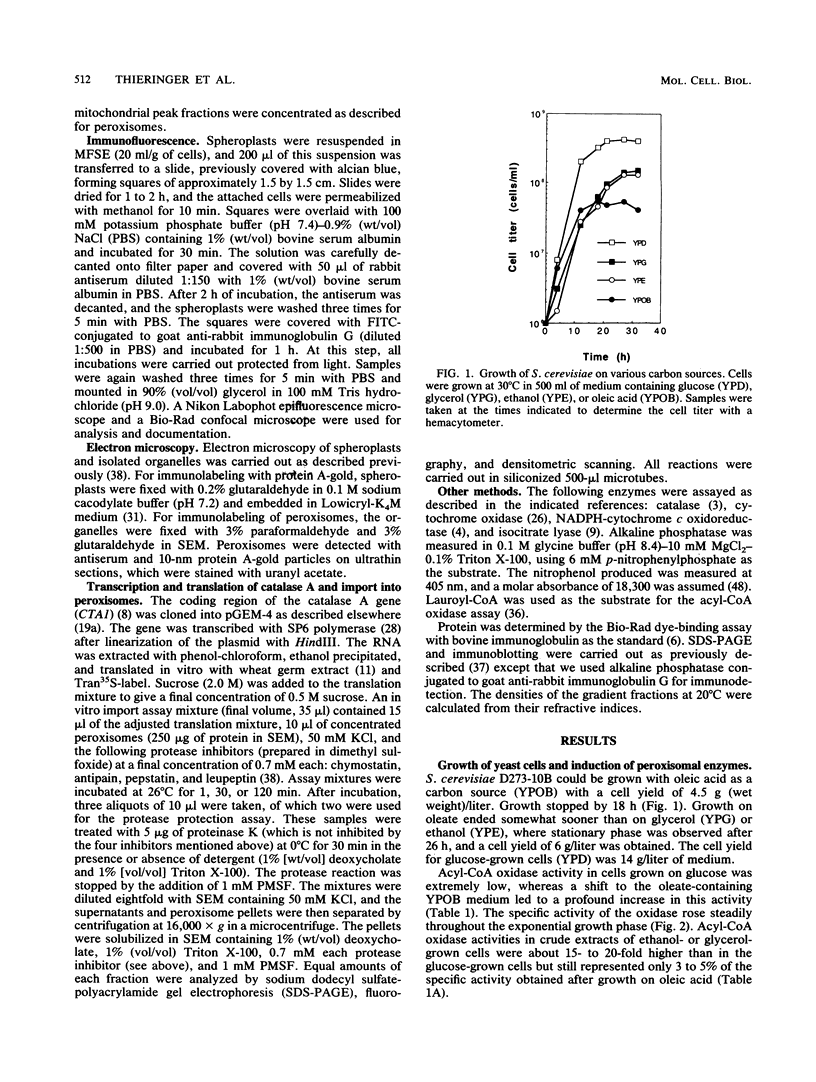
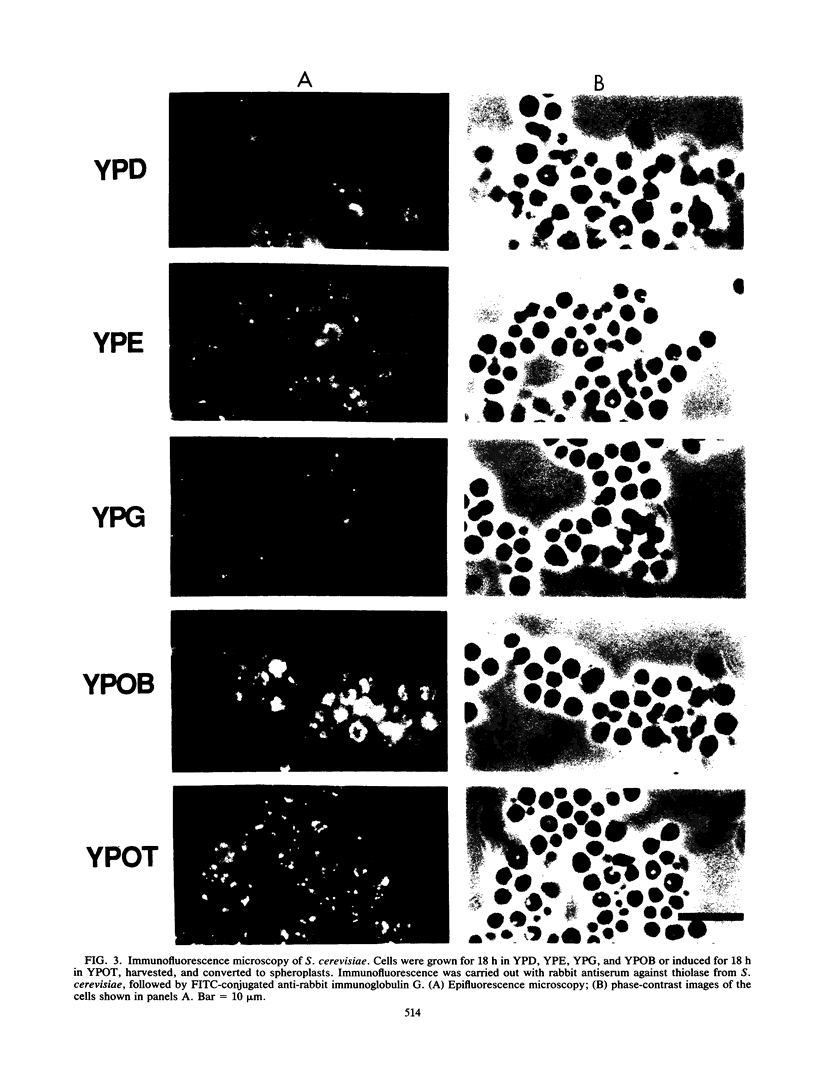
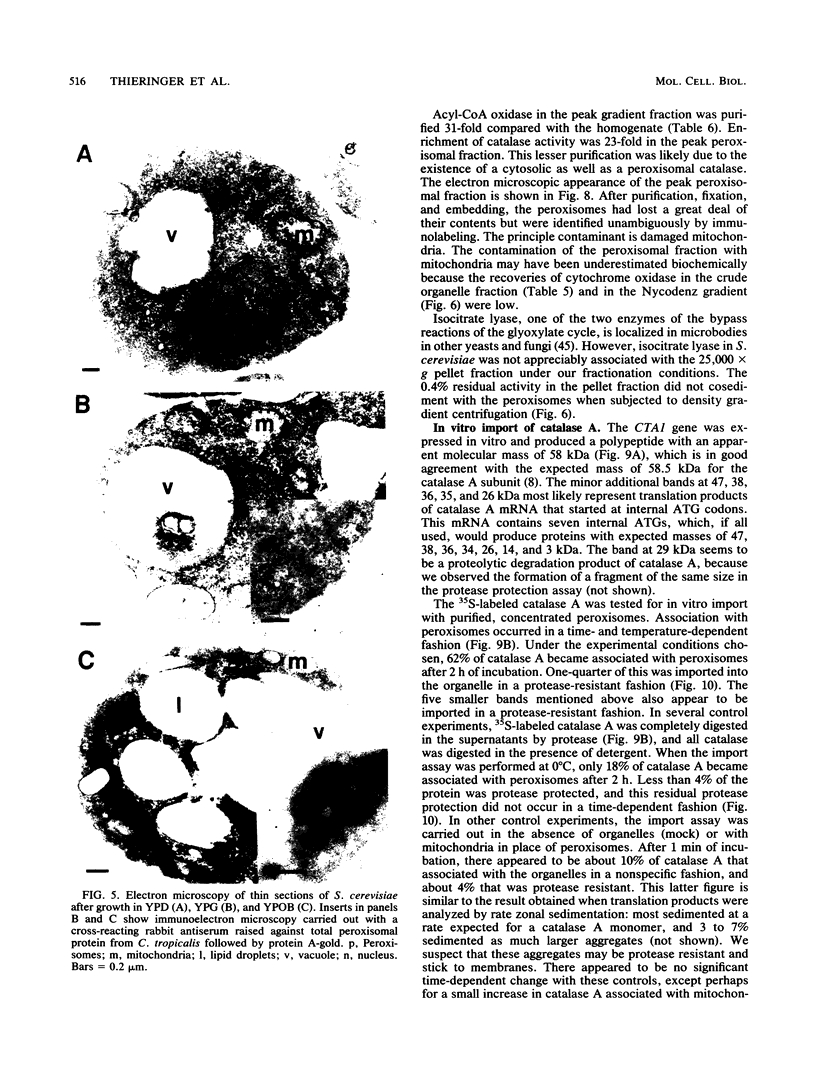
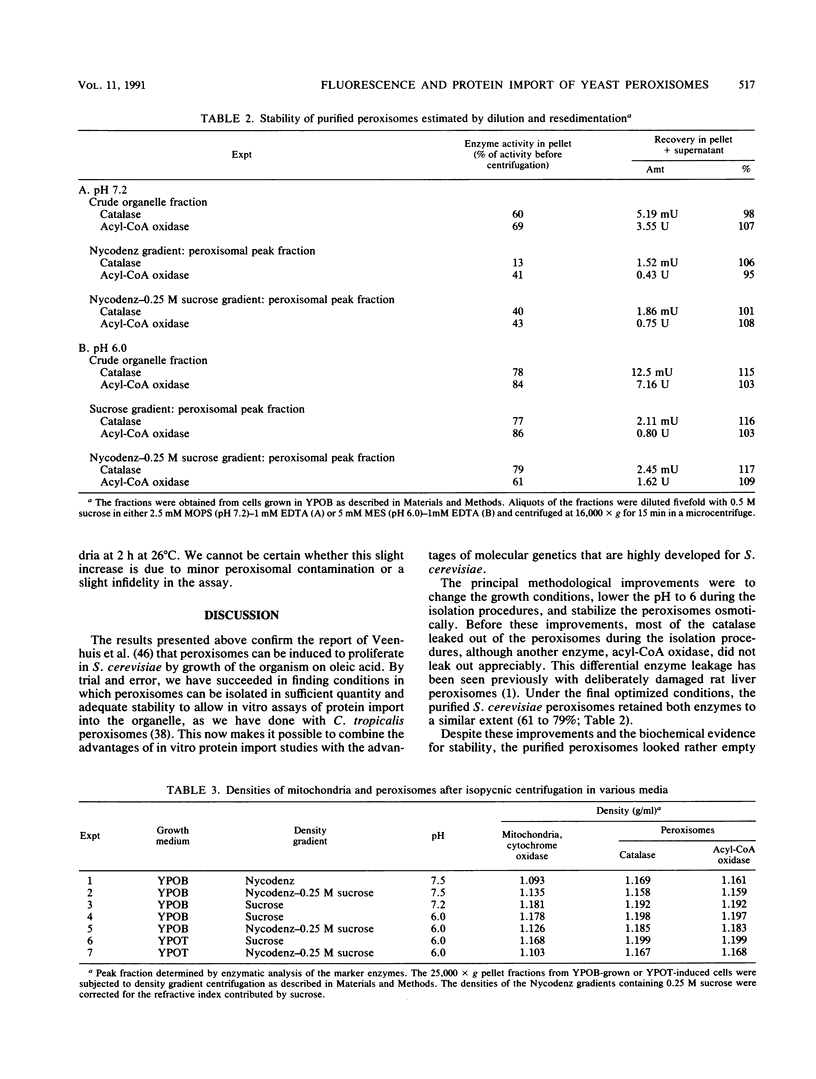
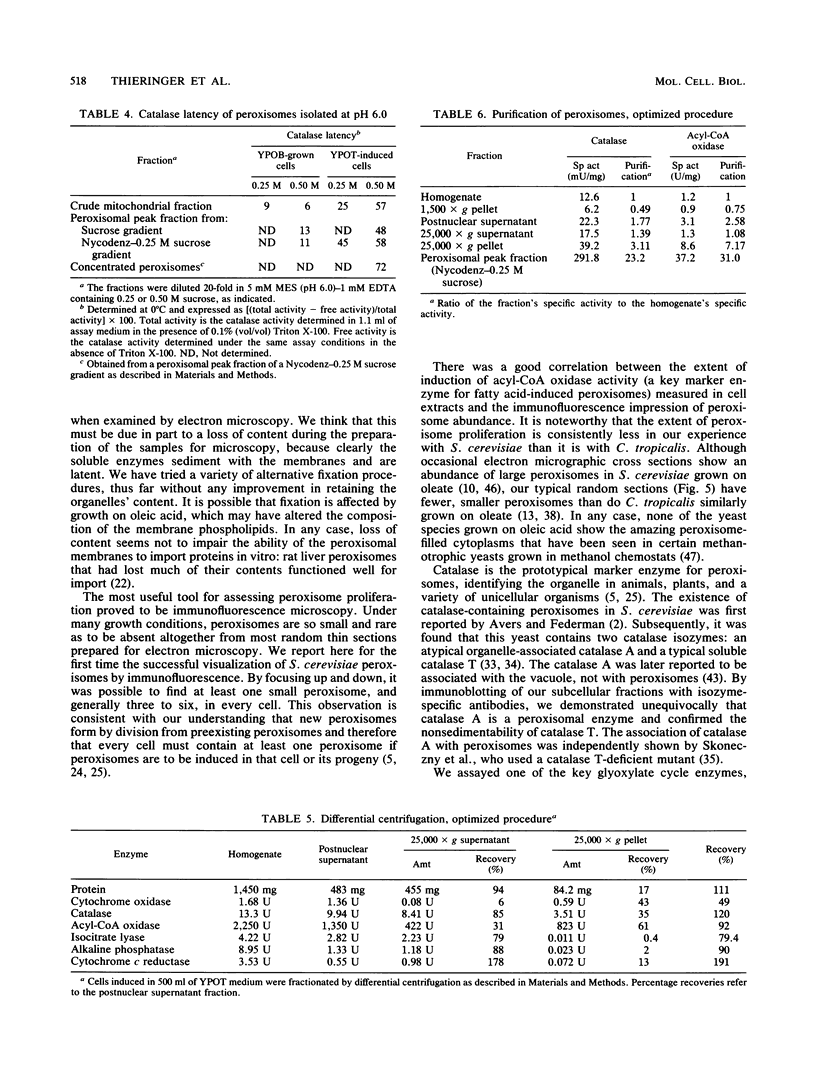
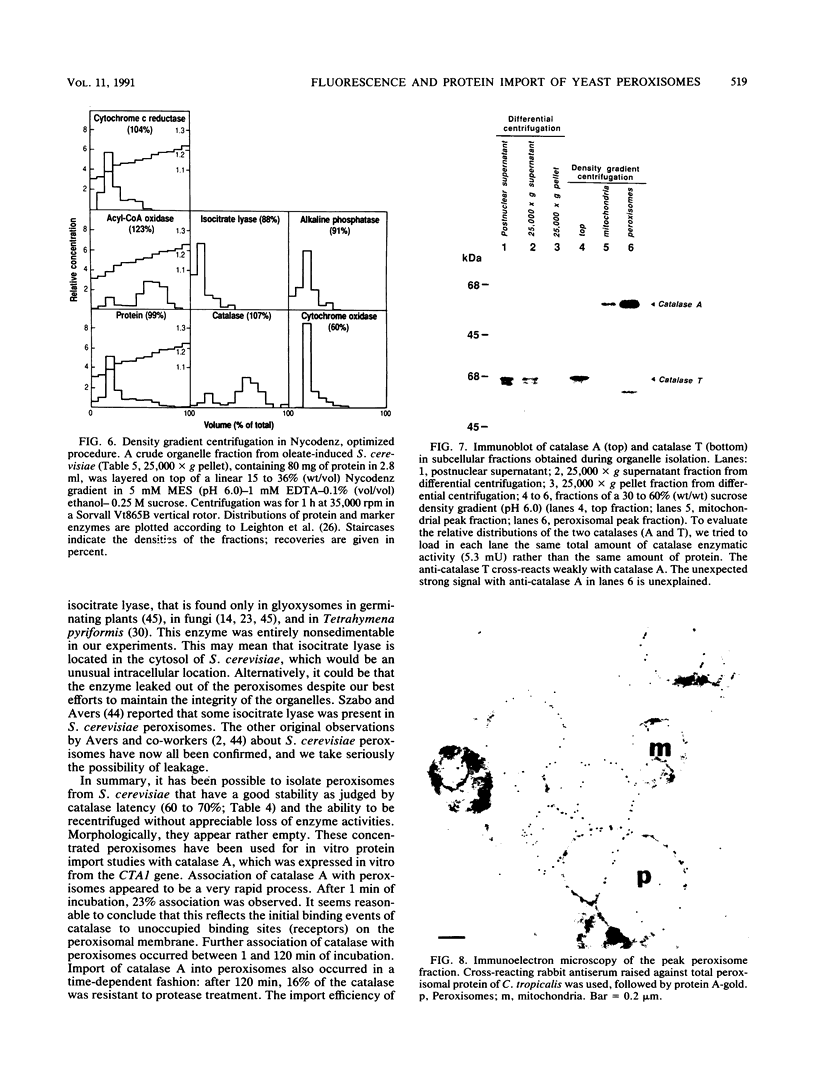
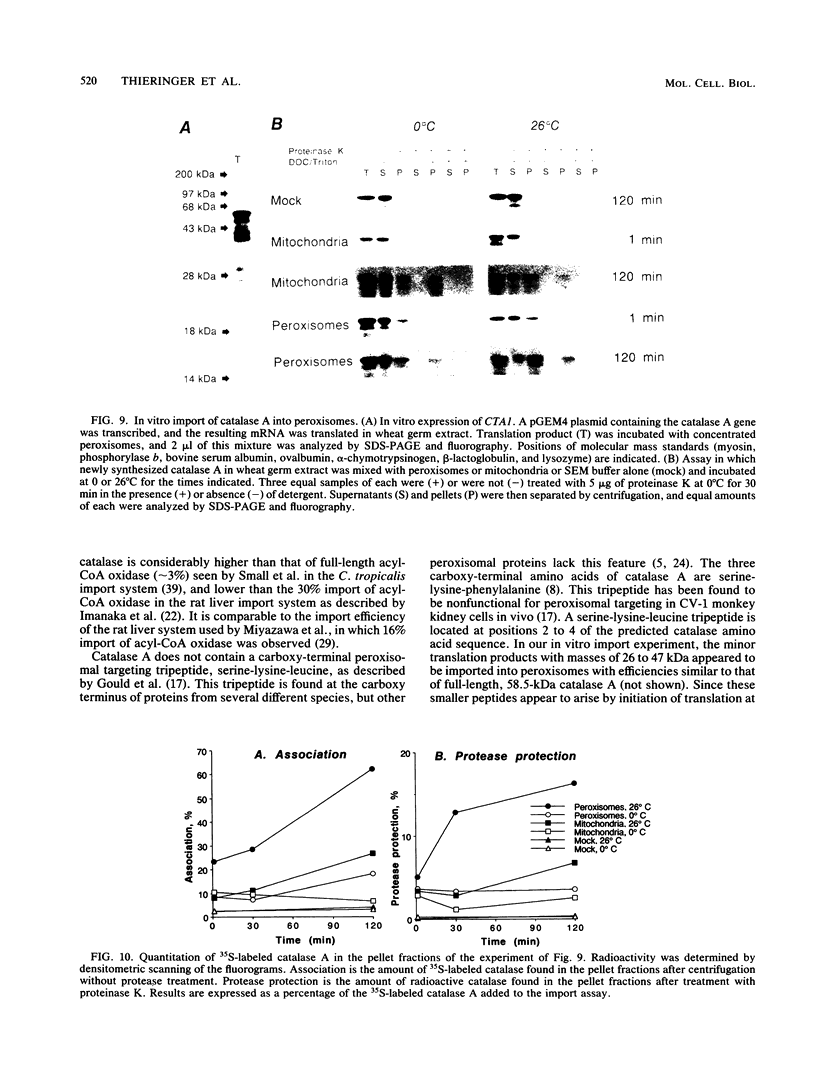
To isolate peroxisomes from Saccharomyces cerevisiae of a quality sufficient for in vitro import studies, we optimized the conditions for cell growth and for cell fractionation. Stability of the isolated peroxisomes was monitored by catalase latency and sedimentability of marker enzymes. It was improved by (i) using cells that were shifted to oleic acid medium after growth to stationary phase in glucose precultures, (ii) shifting the pH from 7.2 to 6.0 during cell fractionation, and (iii) carrying out equilibrium density centrifugation with Nycodenz containing 0.25 M sucrose throughout the gradient. A concentrated peroxisomal fraction was used for in vitro import of catalase A. After 2 h of incubation, 62% of the catalase was associated with, and 16% was imported into, the organelle in a protease-resistant fashion. We introduced immunofluorescence microscopy for S. cerevisiae peroxisomes, using antibodies against thiolase, which allowed us to identify even the extremely small organelles in glucose-grown cells. Peroxisomes from media containing oleic acid were larger in size, were greater in number, and had a more intense fluorescence signal. The peroxisomes were located, sometimes in clusters, in the cell periphery, often immediately adjacent to the plasma membrane. Systematic immunofluorescence observations of glucose-grown S. cerevisiae demonstrated that all such cells contained at least one and usually several very small peroxisomes despite the glucose repression. This finding fits a central prediction of our model of peroxisome biogenesis: peroxisomes form by division of preexisting peroxisomes; therefore, every cell must have at least one peroxisome if additional organelles are to be induced in that cell.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexson S. E., Fujiki Y., Shio H., Lazarow P. B. Partial disassembly of peroxisomes. J Cell Biol. 1985 Jul;101(1):294–304. doi: 10.1083/jcb.101.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avers C. J., Federman M. The occurrence in yeast of cytoplasmic granules which resemble microbodies. J Cell Biol. 1968 May;37(2):555–559. doi: 10.1083/jcb.37.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudhuin P., Beaufay H., Rahman-Li Y., Sellinger O. Z., Wattiaux R., Jacques P., De Duve C. Tissue fractionation studies. 17. Intracellular distribution of monoamine oxidase, aspartate aminotransferase, alanine aminotransferase, D-amino acid oxidase and catalase in rat-liver tissue. Biochem J. 1964 Jul;92(1):179–184. doi: 10.1042/bj0920179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P. Peroxisome biogenesis revisited. Biochim Biophys Acta. 1989 Jun 1;1008(1):1–13. doi: 10.1016/0167-4781(89)90163-2. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cohen G., Fessl F., Traczyk A., Rytka J., Ruis H. Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol Gen Genet. 1985;200(1):74–79. doi: 10.1007/BF00383315. [DOI] [PubMed] [Google Scholar]
- Cohen G., Rapatz W., Ruis H. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur J Biochem. 1988 Sep 1;176(1):159–163. doi: 10.1111/j.1432-1033.1988.tb14263.x. [DOI] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis M., Mertens D., Kunau W. H. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5419–5423. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson A. H., Blobel G. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Rachubinski R. A., Mortensen R. M., Lazarow P. B. Synthesis of 3-ketoacyl-CoA thiolase of rat liver peroxisomes on free polyribosomes as a larger precursor. Induction of thiolase mRNA activity by clofibrate. Biochem J. 1985 Mar 15;226(3):697–704. doi: 10.1042/bj2260697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Rachubinski R. A., Zentella-Dehesa A., Lazarow P. B. Induction, identification, and cell-free translation of mRNAs coding for peroxisomal proteins in Candida tropicalis. J Biol Chem. 1986 Nov 25;261(33):15787–15793. [PubMed] [Google Scholar]
- Garoff H. Using recombinant DNA techniques to study protein targeting in the eucaryotic cell. Annu Rev Cell Biol. 1985;1:403–445. doi: 10.1146/annurev.cb.01.110185.002155. [DOI] [PubMed] [Google Scholar]
- Goodman J. M., Scott C. W., Donahue P. N., Atherton J. P. Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. J Biol Chem. 1984 Jul 10;259(13):8485–8493. [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989 May;108(5):1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Schneider M., Howell S. H., Garrard L. J., Goodman J. M., Distel B., Tabak H., Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990 Jan;9(1):85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen H., Roggenkamp R. Functional complementation of catalase-defective peroxisomes in a methylotrophic yeast by import of the catalase A from Saccharomyces cerevisiae. Eur J Biochem. 1989 Sep 1;184(1):173–179. doi: 10.1111/j.1432-1033.1989.tb15004.x. [DOI] [PubMed] [Google Scholar]
- Hartig A., Ogris M., Cohen G., Binder M. Fate of highly expressed proteins destined to peroxisomes in Saccharomyces cerevisiae. Curr Genet. 1990 Jul;18(1):23–27. doi: 10.1007/BF00321111. [DOI] [PubMed] [Google Scholar]
- Hartig A., Ruis H. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and deduced amino-acid sequence of yeast catalase T. Eur J Biochem. 1986 Nov 3;160(3):487–490. doi: 10.1111/j.1432-1033.1986.tb10065.x. [DOI] [PubMed] [Google Scholar]
- Hörtner H., Ammerer G., Hartter E., Hamilton B., Rytka J., Bilinski T., Ruis H. Regulation of synthesis of catalases and iso-1-cytochrome c in Saccharomyces cerevisiae by glucose, oxygen and heme. Eur J Biochem. 1982 Nov;128(1):179–184. doi: 10.1111/j.1432-1033.1982.tb06949.x. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Small G. M., Lazarow P. B. Translocation of acyl-CoA oxidase into peroxisomes requires ATP hydrolysis but not a membrane potential. J Cell Biol. 1987 Dec;105(6 Pt 2):2915–2922. doi: 10.1083/jcb.105.6.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kionka C., Kunau W. H. Inducible beta-oxidation pathway in Neurospora crassa. J Bacteriol. 1985 Jan;161(1):153–157. doi: 10.1128/jb.161.1.153-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow P. B. Peroxisome biogenesis. Curr Opin Cell Biol. 1989 Aug;1(4):630–634. doi: 10.1016/0955-0674(89)90026-4. [DOI] [PubMed] [Google Scholar]
- Leighton F., Poole B., Beaufay H., Baudhuin P., Coffey J. W., Fowler S., De Duve C. The large-scale separation of peroxisomes, mitochondria, and lysosomes from the livers of rats injected with triton WR-1339. Improved isolation procedures, automated analysis, biochemical and morphological properties of fractions. J Cell Biol. 1968 May;37(2):482–513. doi: 10.1083/jcb.37.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin A. S., Hines V., Small G. M. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol Cell Biol. 1990 Apr;10(4):1399–1405. doi: 10.1128/mcb.10.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa S., Osumi T., Hashimoto T., Ohno K., Miura S., Fujiki Y. Peroxisome targeting signal of rat liver acyl-coenzyme A oxidase resides at the carboxy terminus. Mol Cell Biol. 1989 Jan;9(1):83–91. doi: 10.1128/mcb.9.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Hogg J. F., De Duve C. Distribution of tricarboxylic acid cycle enzymes and glyoxylate cycle enzymes between mitochondria and peroxisomes in Tetrahymena pyriformis. J Biol Chem. 1968 Oct 25;243(20):5385–5395. [PubMed] [Google Scholar]
- Santos M. J., Imanaka T., Shio H., Lazarow P. B. Peroxisomal integral membrane proteins in control and Zellweger fibroblasts. J Biol Chem. 1988 Jul 25;263(21):10502–10509. [PubMed] [Google Scholar]
- Schekman R. Protein localization and membrane traffic in yeast. Annu Rev Cell Biol. 1985;1:115–143. doi: 10.1146/annurev.cb.01.110185.000555. [DOI] [PubMed] [Google Scholar]
- Seah T. C., Bhatti A. R., Kaplan J. G. Novel catalatic proteins of bakers' yeast. I. An atypical catalase. Can J Biochem. 1973 Nov;51(11):1551–1555. doi: 10.1139/o73-208. [DOI] [PubMed] [Google Scholar]
- Seah T. C., Kaplan J. G. Purification and properties of the catalase of bakers' yeast. J Biol Chem. 1973 Apr 25;248(8):2889–2893. [PubMed] [Google Scholar]
- Skoneczny M., Chełstowska A., Rytka J. Study of the coinduction by fatty acids of catalase A and acyl-CoA oxidase in standard and mutant Saccharomyces cerevisiae strains. Eur J Biochem. 1988 Jun 1;174(2):297–302. doi: 10.1111/j.1432-1033.1988.tb14097.x. [DOI] [PubMed] [Google Scholar]
- Small G. M., Burdett K., Connock M. J. A sensitive spectrophotometric assay for peroxisomal acyl-CoA oxidase. Biochem J. 1985 Apr 1;227(1):205–210. doi: 10.1042/bj2270205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small G. M., Imanaka T., Lazarow P. B. Immunoblotting of hydrophobic integral membrane proteins. Anal Biochem. 1988 Mar;169(2):405–409. doi: 10.1016/0003-2697(88)90304-1. [DOI] [PubMed] [Google Scholar]
- Small G. M., Szabo L. J., Lazarow P. B. Acyl-CoA oxidase contains two targeting sequences each of which can mediate protein import into peroxisomes. EMBO J. 1988 Apr;7(4):1167–1173. doi: 10.1002/j.1460-2075.1988.tb02927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak W., Fessl F., Rytka J., Traczyk A., Skoneczny M., Ruis H. Isolation of the catalase T structural gene of Saccharomyces cerevisiae by functional complementation. Mol Cell Biol. 1983 Sep;3(9):1545–1551. doi: 10.1128/mcb.3.9.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susani M., Zimniak P., Fessl F., Ruis H. Localization of catalase A in vacuoles of Saccharomyces cerevisiae: evidence for the vacuolar nature of isolated "yeast peroxisomes". Hoppe Seylers Z Physiol Chem. 1976 Jul;357(7):961–970. doi: 10.1515/bchm2.1976.357.2.961. [DOI] [PubMed] [Google Scholar]
- Szabo A. S., Avers C. J. Some aspects of regulation of peroxisomes and mitochondria in yeast. Ann N Y Acad Sci. 1969 Dec 19;168(2):302–312. doi: 10.1111/j.1749-6632.1969.tb43117.x. [DOI] [PubMed] [Google Scholar]
- Tolbert N. E. Metabolic pathways in peroxisomes and glyoxysomes. Annu Rev Biochem. 1981;50:133–157. doi: 10.1146/annurev.bi.50.070181.001025. [DOI] [PubMed] [Google Scholar]
- Veenhuis M., Mateblowski M., Kunau W. H., Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987 Jun;3(2):77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- Veenhuis M., van Dijken J. P., Harder W. Cytochemical studies on the localization of methanol oxidase and other oxidases in peroxisomes of methanol-grown Hansenula polymorpha. Arch Microbiol. 1976 Dec 1;111(1-2):123–135. doi: 10.1007/BF00446559. [DOI] [PubMed] [Google Scholar]
- Zimniak P., Hartter E., Woloszczuk W., Ruis H. Catalase biosynthesis in yeast: formation of catalase A and catalase T during oxygen adaptation of Saccharomyces cerevisiae. Eur J Biochem. 1976 Dec 11;71(2):393–398. doi: 10.1111/j.1432-1033.1976.tb11126.x. [DOI] [PubMed] [Google Scholar]