Abstract
Background and Purpose
Induction of cellular migration is the primary effect of chemokine receptor activation. However, several chemokine receptor-like proteins bind chemokines without subsequent induction of intracellular signalling and chemotaxis. It has been suggested that they act as chemokine scavengers, which may control local chemokine levels and contribute to the function of chemokines during inflammation. This has been verified for the chemokine-like receptor proteins D6 and DARC as well as CCX-CKR. Here, we provide evidence for an additional biological function of human (h)CCX-CKR.
Experimental Approach
We used transfection strategies in HEK293 and human T cells.
Key Results
Co-expression of hCCX-CKR completely inhibits hCXCR3-induced chemotaxis. We found that hCCX-CKR forms complexes with hCXCR3, suggesting a relationship between CCX-CKR heteromerization and inhibition of chemotaxis. Moreover, negative binding cooperativity induced by ligands both for hCXCR3 and hCCX-CKR was observed in cells expressing both receptors. This negative cooperativity may also explain the hCCX-CKR-induced inhibition of chemotaxis.
Conclusions and Implications
These findings suggest that hCCX-CKR prevents hCXCR3-induced chemotaxis by heteromerization thus representing a novel mechanism of regulation of immune cell migration.
Keywords: chemokine receptors, T cells, heteromerization
Introduction
Chemokines, small proteins that regulate cell migration, play important roles in the immune system, angiogenesis, hematopoiesis, cell differentiation and development (Rossi and Zlotnik, 2000; Horuk, 2001). Chemokines and their receptors have been classified into four different subgroups based on the presence and position of conserved cysteine residues i.e. CXC, CC, C and CX3C (Murphy et al., 2000). Chemokine receptors are GPCRs that signal via G-proteins to various signalling pathways (Sanchez-Madrid and del Pozo, 1999; Thelen, 2001). In addition, atypical chemokine receptors represent a subfamily of chemokine-binding proteins that do not signal along classic GPCR-mediated pathways, but efficiently internalize their cognate ligands and act as chemokine scavengers (Hansell et al., 2006). In addition to DARC and D6, CCX-CKR belongs to this family of proteins and scavenges its ligands CCL21, CCL19, CCL25 and CXCL13 (Gosling et al., 2000; Townson and Nibbs, 2002; Comerford et al., 2006). CCX-CKR is widely expressed in several tissues, including the brain and is expressed in many immune cells such as microglia, dendritic cells and T cells together with other chemokine receptors (Gosling et al., 2000; Murphy, 2002; Townson and Nibbs, 2002; Zuurman et al., 2003; Brouwer et al., 2004). It is well known that chemokine receptors like other GPCRs can, when co-expressed, form heteromers that exhibit altered intracellular signalling properties (Rodriguez-Frade et al., 1999; Rios et al., 2001; Springael et al., 2005; 2006). Because CCX-CKR is found co-expressed with other chemokine receptors, we investigated whether co-expression of CCX-CKR might change the signalling of other chemokine receptors in addition to its function as scavenger.
Methods
Chemicals
Media, sera, and reagents used for cell culture were purchased from PAA Laboratories (Colbe, Germany). All other chemicals were from Sigma-Aldrich (Zwijndrecht, The Netherlands), unless mentioned otherwise.
Cell cultures
HEK293 cells and HEK293T (gift from Dr S. Carra, Department of Cell Biology, UMCG, Groningen, Netherlands) were cultured in growth medium [DMEM, supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% Pen/Strep (Invitrogen, Breda, The Netherlands) and 1% sodium pyruvate] and kept in a humidified atmosphere (5% CO2) at 37°C.
T cell isolation and activation
Peripheral blood mononuclear cells were isolated from heparinized blood taken from healthy donors using a Ficoll (GE Healthcare, Diegem, Belgium) gradient centrifugation for 15 min at 1000 g without brake. T cells were isolated by co-incubating the mononuclear cells with 2-(2-aminoethyl) isothiourea dihydrobromide (AET)-treated sheep red blood cells (Oxiod, Badhoevedorp, The Netherlands) for 15 min on ice. Erythrocyte rosettes were separated from the mononuclear cells by Ficoll gradient centrifugation for 15 min at 1000 g without brake. Subsequently, the sheep red blood cells were lysed using Gey's lysis buffer (7.0 g·L−1 NH4Cl, 0.37 g·L−1 KCl, 0.3 g·L−1 Na2HPO4.12H2O, 0.024 g·L−1 KH2PO4, 1.0 g·L−1 glucose, 10.0 mg·L−1 phenol red, 8.4 mg·L−1 MgCl2.6H2O, 7.0 mg·L−1 MgSO4.7H2O, 6.8 mg·L−1 CaCl2 and 45 mg·L−1 NaHCO3), yielding a population of 92.5 ± 4.6% CD3+ T cells. Freshly isolated T cells were used directly for further experiments or resuspended in growth medium [RPMI 1640 (Invitrogen), supplemented with 5% FCS (Invitrogen), 100 U·mL−1 penicillin, 100 μg·mL−1 streptomycin] and kept in a humidified atmosphere (5% CO2) at 37°C. T cells were activated by stimulating them with 2 μg·mL−1 phytohemagglutinin (PHA; Biotrading, Mijdrecht, The Netherlands) for 24 h and with 25 mL−1 IL-2 (Peprotech, London, UK) for another 3–5 days.
Plasmids
The following constructs were used for generating stable cell lines: pcDNA3.1 plasmid containing the full-length sequence of human CXCR3 [hCXCR3; kind gift of C. Tensen and B. Moser (Hensbergen et al., 2001)]; pcDNA3.1 containing full-length human CCX-CKR [hCCXCKR; kindly provided by R. Nibbs (Townson and Nibbs, 2002)]; pREP9 containing full-length CX3CR1 [kindly provided by P.M. Murphy (Combadiere et al., 1995)].
The following constructs were used for FRET experiments: hCXCR3, hCCX-CKR, GABAB-R1 and hCCR5 coupled both to Venus or Cyan Fluorescent Protein (CFP). pcDNA3.1 (+)-GABAB-R1 [kindly provided by B. van Lith (Molecular Pharmacology & DMPK, Schering-Plough Research Institute, Oss, The Netherlands)] was used as negative control. hCCR5 [Missouri S&T cDNA Resource Center (http://www.cdna.org)] served also as negative control. All four constructs were subcloned using primers described in Table 1. hCXCR3 and hCCX-CKR were inserted into pECFP-N1 and pVenus-N1 vector using EcoRI and SmaI as restriction enzymes. hCCR5 was inserted using HindIII and SmaI while hGABAB-R1 was inserted using EcoRI and SalI. hGABAB-R1 subcloning was done in presence of 10% glycerol. The pVenus-N1 vector was generated using the pECFP-N1 (Clonetech) as backbone. ECFP was cut out of the pECFP-N1 plasmid using AgeI and NotI. Venus was cut out of the pcDNA3.1 CFP-EPAC-Venus construct using XhoI. Venus was subcloned (see primers in Table 1) and inserted into the pECFP-N1 (after deletion of ECFP) using AgeI and NotI.
Table 1.
Primers used for cloning for FRET constructs
| Gene of interest | Forward primer | Reverse primer |
|---|---|---|
| CXCR3 | AAAGAATTCTAATGGTCCTTGAGGTGAGTGACC | AAACCCGGGCCAAGCCCGAGTAGGAGGCCTC |
| CCXCKR | AAAGAATTCTAATGGCTTTGGAACAGAACCAGTC | AAACCCGGGCAATGCTAAAAGTACTGGTTGGCTCT |
| GABAB-R1 | AAAGAATTCTGATGTTGCTGCTGCTGTTACTGGCG | AAAAGTCGACTGCTTATAAAGCAAATGCACTCGACTCC |
| CCR5 | AAAAAAGCTTATGGATTATCAAGTGTCAAGTCCAAT | AAACCCGGGCCAAGCCCACAGATATTTCCTGCTC |
| Venus | AAAACCGGTCGCCACCATGGTGAGCAAGGGCGAGGAGCT | AAAGCGGCCGCTTACTTGTACAGCTCGTCCATGCCGA |
The following constructs were used for transient transfection followed by chemotaxis assays to verify whether hCCX-CKR is affecting other chemokine receptors: hCCR2, hCCR4, hCCR5, hCCR6, hCCR7, hCCR9, hCCR10 and hCXCR4. These constructs have been purchased from: Missouri S&T cDNA Resource Center (http://www.cdna.org).
Generation of stable HEK293 cell lines
Cells were transfected with either hCXCR3, hCCX-CKR or both constructs using FugeneR (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. Stable polyclonal transfected cells were selected using 500 μg·mL−1 G-418 (Omnilabo, Breda, The Netherlands) for approximately 2 weeks. Hereafter, monoclonal cell lines were generated and maintained for maximum 15 passages.
Cell transfection
HEK293 and HEK293T cells were transfected using calcium/phosphate method. Briefly, cells were kept in DMEM without additives and where left with DNA precipitates for 5 h. After transfection, cells were rinsed with DMEM and put back in growth medium. Mock transfections were done using empty plasmids.
For radioligand-binding experiments, HEK293T cells were transiently transfected with 1 μg receptor-encoding plasmid DNA using 25-kDa linear polyethyleneimine (Polysciences, Eppelheim, Germany) as described previously (Verzijl et al., 2008). The total amount of transfected DNA was adjusted to 5 μg with the empty vector pcDEF3.
hCCX-CKR was silenced in freshly isolated T cells using pLKO.1-puro vectors containing DNA encoding for CCX-CKR short-hairpin silencing RNA (shRNA) (MISSION® shRNA NM_16557; Sigma-Aldrich). Five vectors for hCCX-CKR silencing were used for each different T cell donor. Plasmid containing GFP-encoding DNA (pmaxGFP; Lonza, Basel, Switzerland) was used as positive control for transfection and the pLKO.1-puro vector containing DNA encoding for non-targeting shRNA (MISSION® shRNA SHC002; Sigma-Aldrich) was used as negative control. T cells were transfected using the Amaxa nucleofector kit for human T cells (Lonza) according to manufacturer's instructions. Briefly, 2 × 106 to 5 × 106 T cells were resuspended in specific nucleofector solution and mixed with 2 μg vector DNA. Subsequently, the cells were nucleofected using the T cell-specific programme V-24 and transferred into growth medium. hCCX-CKR was overexpressed in PHA/IL-2-treated T cells using the same method. T cells were resuspended in specific nucleofector solution, mixed with 5 μg of hCCX-CKR plasmid DNA or pmaxGFP, and nucleofected by using the T cell specific programme T-20. Transfection efficiency and hCCX-CKR expression were assessed by flow cytometry 24 h (overexpression) or 48 h (silencing) post-nucleofection.
Flow cytometry
Expression of surface molecules by human T cells was determined using flow cytometry. Cells were incubated with predetermined optimal dilutions of primary antibody diluted in PBS containing 0.5% BSA and 0.01% sodium azide for 10 min at RT. Primary antibodies were phycoerythrine (PE)-conjugated anti-CXCR3 (clone 49801; R&D Systems, Abingdon, UK), FITC-conjugated anti-CD3 (clone UCHT1; eBioscience, Vienna, Austria), PE-Cy5-conjugated anti-CD4 (clone RPA-T4; eBioscience), allophycocyanin (APC)-conjugated anti-CD8 (clone RPA-T8; eBioscience), APC-conjugated anti-CD44 (clone IM-7; eBioscience), PE-Cy5-conjugated anti-CD62L (clone DREG-56; eBioscience) and polyclonal goat-anti-CCX-CKR (CCR11; Capralogics, Hardwick, MA, USA). hCCX-CKR was detected using PE-conjugated anti-goat IgG (Jackson ImmunoResearch, Newmarket, UK) as a secondary antibody. Isotype-matched primary antibodies of irrelevant specificity served as negative controls. 20.000–30.000 events were measured using a FACSCalibur flow cytometer (BD Biosciences, Breda, The Netherlands) and analysed by WinMDI software.
Chemotaxis assay
Cell migration in response to chemokines was assessed using a 48-well chemotaxis microchamber (NeuroProbe, Gaithersburg, MD, USA). 10 μM chemokine stock solutions (containing recombinant human CCL4, CCL13, CCL17, CCL20, CCL21, CCL25, CCL27, CXCL9, CXCL10 or CXCL12; R&D Systems) were prepared in sterile PBS and further diluted in serum-free DMEM for use in the assay. Serum-free DMEM without chemokines served as control. 27 μL of the chemoattractant solution or control medium was applied to the lower well of the chamber. Upper and lower chamber were separated by a polyvinylpyrrolidone-free polycarbonate filter (5-μm pore size for T cells and 8-μm pore size for HEK293/HEK293T cells). In the upper wells of the chamber, 50-μL cell suspension containing 5 × 104 cells was applied. The chamber was incubated in a humidified atmosphere (5% CO2) at 37°C for 2 h. Determinations were done in hexaplicate for each group. After incubation the filter was washed, fixed in methanol and stained with toluidine blue. Migrated cells per group were counted by a double-blinded person with a scored eyepiece [three fields (1 mm2) per well].
Reverse transcription and quantitative real-time PCR (QPCR)
T cells were lysed in guanidinium isothiocyanate/mercaptoethanol buffer and total RNA was extracted according to Chomczynski and Sacchi (1987) with slight modifications. One μg of total RNA was transcribed into cDNA as described (Biber et al., 1997). Primers used for RT-PCR are described in the Supporting Information Table S1. Real-time PCR, using an iCyclerR (Bio-Rad, Veenendaal, The Netherlands) and iQ SYBR Green supermixR (Bio-Rad), was performed on 4 ng cDNA from T cells. Primers (Supporting Information Table S1) for QPCR, yielding PCR products of approximately 100 base pairs long, were designed by ‘Primer Designer’ (Version 3.0, Scientific and Educational Software, Cary, NC, USA). PCR reactions with primers for hCCX-CKR and hCXCR3 were run in parallel with primers for the housekeeping genes GAPDH and hypoxanthine guanine phosphoribosyl transferase. All primers were purchased from Biolegio (Nijmegen, The Netherlands). Melt-curve analysis was performed immediately following amplification in order to check primer specificity. For analysis, the comparative Ct method was used. In each experiment samples were run in duplicate. Results are the averaged data of three independent experiments and are given as mean ± SEM.
Immunocytochemistry
HEK293 cells were plated on poly-L-lysine (PLL) coated glass coverslips and fixed in 4% paraformaldehyde (PFA) solution. Cells were pre-incubated in PBS containing 10% FCS and 0.3% Triton-X100 for 30 min at room temperature. Incubation with primary antibodies against hCXCR3 (R&D, 1:100) and hCCX-CKR (Capralogics, 1:400) was performed O/N at 4°C. The next day, cells were incubated with secondary antibodies conjugated with fluorescent labels (Jackson Immuno Research) for 2 h at room temperature. Cells were washed, stained with Hoechst nuclear dye and finally mounted in Vectashield Mounting Media (Vector, Peterborough, UK). Stained cells were analysed with a Zeiss Axioskop 2 microscope and confocal fluorescence microscopy (see FRET section).
Ligand-binding experiments
The CXCR3 antagonist VUF10085 was first described in literature as AMG 487 (Johnson et al., 2007) and re-synthesized in our department. [125I]-CXCL10 and Na125I were purchased from PerkinElmer Life and Analytical Sciences (Boston, MA, USA). CCL19 was purchased from Peprotech (Rocky Hill, NJ, USA) and labelled with Na125I using iodogen pre-coated tubes (Thermo Fischer Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Twenty-four hours post-transfection, HEK293T cells were transferred to poly-L-lysine (Sigma-Aldrich) coated 48-well plates. The next day, cells were incubated for 4 h at 4°C with 0.5–1 nM [125I]-CCL19 or 50–100 pM [125I]-CXCL10 in binding buffer (50 mM HEPES, 100 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, pH 7.4, 0.5% BSA) containing unlabeled displacer. After incubation, cells were washed three times with ice-cold wash buffer (50 mM HEPES, 0.5 M NaCl, 1 mM CaCl2, 5 mM MgCl2, pH 7.4) and subsequently lysed. Cell lysates were counted in a Wallac Compugamma counter (PerkinElmer Life and Analytical Sciences). For ligand binding cooperativity assays, percentages of CXCR3-specific [125I]-CXCL10, [125I]-CXCL11 and CCX-CKR-specific [125I]-CCL19 binding were calculated by normalizing data to binding in the absence of unlabeled displacer (100%) and binding in the presence of 10 μM VUF10085, 100 nM CXCL11 and 100 nM CCL19, respectively (0%). For homologous radioligand displacement curves, CXCR3-specific [125I]-CXCL10, [125I]-CXCL11 and CCX-CKR-specific [125I]-CCL19 binding were calculated by normalizing data to radioligand binding in the absence of unlabeled displacer (100%) and in the presence of 10 μM VUF10085, 100 nM CXCL11 and 100 nM CCL19, respectively (0%), in cells expressing only CXCR3 ( [125I]-CXCL10 and [125I]-CXCL11) or only CCX-CKR ([125I]-CCL19).
ELISA measurements
Cells were plated in 48-well plates, fixed 30 min with 4% PFA in PBS 48 h after plating and washed two times with TBS. For the permeabilized samples, cells were incubated 30 min with 0.5% NP-40 (Roche Molecular Biochemicals) in TBS after fixation. Samples were blocked for 4 h at RT with 1% fat-free milk in 0.1 M NaHCO pH 8.6. Primary antibody (R&D Systems, MAB160) was diluted 1:1000 in 0.1% BSA in TBS and incubated O/N at 4°C and washed three times with TBS. Secondary antibody (Bio-rad 170–6516 goat-anti-mouse/HRP) was diluted 1:2500 in 1% fat free milk in 0.1 M NaHCO pH 8.6 and incubated 3 h at RT. o-Phenylenediamine substrate solution [2.2 mM ophenylenediamine (Sigma-Aldrich P-1526), 35 mM citric acid, 66 mM Na2HPO4, 0.015% H2O2, pH 5.6) was added. The reaction was stopped by addition of 1 M H2SO4 and the absorbance at 490 nm was measured with a Wallac Victor plate reader.
FRET experiments
HEK 293T cells were transfected with plasmids encoding Venus- or CFP-tagged versions of hCXCR3, hCCX-CKR or both. For control experiments Venus-tagged versions of hGABAB-R1 and hCCR5 were used. Imaging of the expression of the various fusion proteins was performed on a Leica AOBS_TCS SP2 confocal laser scanning microscope using an 63x NA 1.4 oil-immersion objective (Leica Microsystems) and the 458 nm line of an AR/Kr laser. The microscope was set-up and corrections were applied according to Van Rheenen et al. (2004). FRET efficiency was determined using the method by Jalink and Van Rheenen (2009). Shortly, images were corrected by substracting the background and correcting for bleedtrough by using CFP- and Venus-transfected cells followed by correction of intensity. Values were measured by scaling all samples to the same level of the hCXCR3-CFP/hCXCR3-Venus homomer followed by measuring the intensity at different region of interests at the membrane.
Results
CCX-CKR co-expression inhibits chemotactic signalling of various chemokine receptors
In order to investigate if co-expression of hCCX-CKR influences the signalling of other chemokine receptors that don't share ligands with CCX-CKR, chemotaxis experiments were performed. We observed that the migration of HEK293 cells transfected with the chemokine receptors hCCR6, hCCR10, hCXCR3 or hCXCR4 was significantly reduced when hCCX-CKR was co-expressed (Table 2). Co-expression together with hCCR2 and hCCR4 showed weak, non-consistent reduction in the chemotactic response. Chemotaxis towards receptor hCCR5 was not inhibited by the presence of hCCX-CKR. Finally, hCCR7 and hCCR9 were also investigated, which both share common ligands with CCX-CKR. The chemotactic response of these receptors was also blunted when co-expressed with CCX-CKR (Table 2). In order to analyse the effects of co-expression in more detail, we generated HEK293 cells stably co-expressing CCX-CKR and CXCR3, an important receptor for immune cell trafficking.
Table 2.
Chemokine receptors tested for chemotaxis inhibition by hCCX-CKR
| Receptor | Ligand (1 nM) | % of migration (control is set to 100%) | n | P-value |
|---|---|---|---|---|
| CCR2 CCR2+CCX-CKR | CCL13 | 125 ± 13 92 ± 6 | 3 | 0.069 |
| CCR4 CCR4+CCX-CKR | CCL17 | 139 ± 5 111 ± 11 | 3 | 0.069 |
| CCR5 CCR5+CCX-CKR | CCL4 | 143 ± 13 121 ± 6 | 3 | 0.15 |
| CCR6 CCR6+CCX-CKR | CCL20 | 135 ± 10 96 ± 9 | 3 | 0.02 |
| CCR7 CCR7+CCX-CKR | CCL21 | 125 ± 15 75 ± 9 | 3 | 0.023 |
| CCR9 CCR9+CCX-CKR | CCL25 | 168 ± 15 94 ± 5 | 3 | <0.001 |
| CCR10 CCR10+CCX-CKR | CCL27 | 158 ± 15 84 ± 11 | 3 | 0.003 |
| CXCR3 CXCR3+CCX-CKR | CXCL10 | 148 ± 9 82 ± 7 | 3 | <0.001 |
| CXCR4 CXCR4+CCX-CKR | CXCL12 | 154 ± 16 83 ± 10 | 3 | 0.009 |
HEK293T cells were either transfected with a chemokine receptor alone or with a combination of a chemokine receptor and CCX-CKR. Cells were then subjected to a chemotaxis experiment using the chemokine specific for the transfected receptor diluted in medium at a concentration of 1 nM. Control migration was done in presence of medium alone. In all control experiments, between 25 and 45 cells migrated towards the medium. Using these numbers, control migration was set to 100%. Data represent the mean and SEM of 3 independent experiments. ANOVA was performed to illustrate statistical differences in migration. P < 0.05 indicates that the chemotaxis mediated by the chemokine receptor towards its ligand was inhibited by the presence of CCX-CKR.
Expression of hCCX-CKR does not affect expression of CXCR3
First the presence of chemokine receptors expression was examined by immunocytochemistry. Non-transfected HEK293 cells did not show staining for hCXCR3 (green signal) (Figure 1A) and hCCX-CKR (red signal) (Figure 1B). Staining for hCXCR3 was observed in both hCXCR3-transfected (Figure 1D) and double transfected HEK293 cells (Figure 1H) and hCCX-CKR expression was observed in hCCX-CKR-transfected (Figure 1F) and double-transfected HEK293 cells (Figure 1H). Omission of primary antibodies generated no signal (Figure 1C,E,G). These findings were also confirmed at mRNA level (Supporting InformationFigure S1; Supporting Information Table S2). To exclude the possibility that the presence of hCCX-CKR might hamper the normal expression of hCXCR3, expression levels of hCXCR3 in these cells were further investigated by ELISA measurements (Figure 1I). We show that comparable levels of hCXCR3 expression were found in hCXCR3-transfected cells and hCXCR3 + hCCX-CKR-transfected cells. Although there is a decrease in hCXCR3 expression in presence of hCCX-CKR, which is probably due to the cytomegalovirus promoter driving the protein machinery to its limit, it is not sufficient to explain the complete inhibition of chemotaxis towards CXCL10 that we observed. Thus the co-expression of hCCX-CKR did not affect protein expression of hCXCR3 in HEK293 cells.
Figure 1.
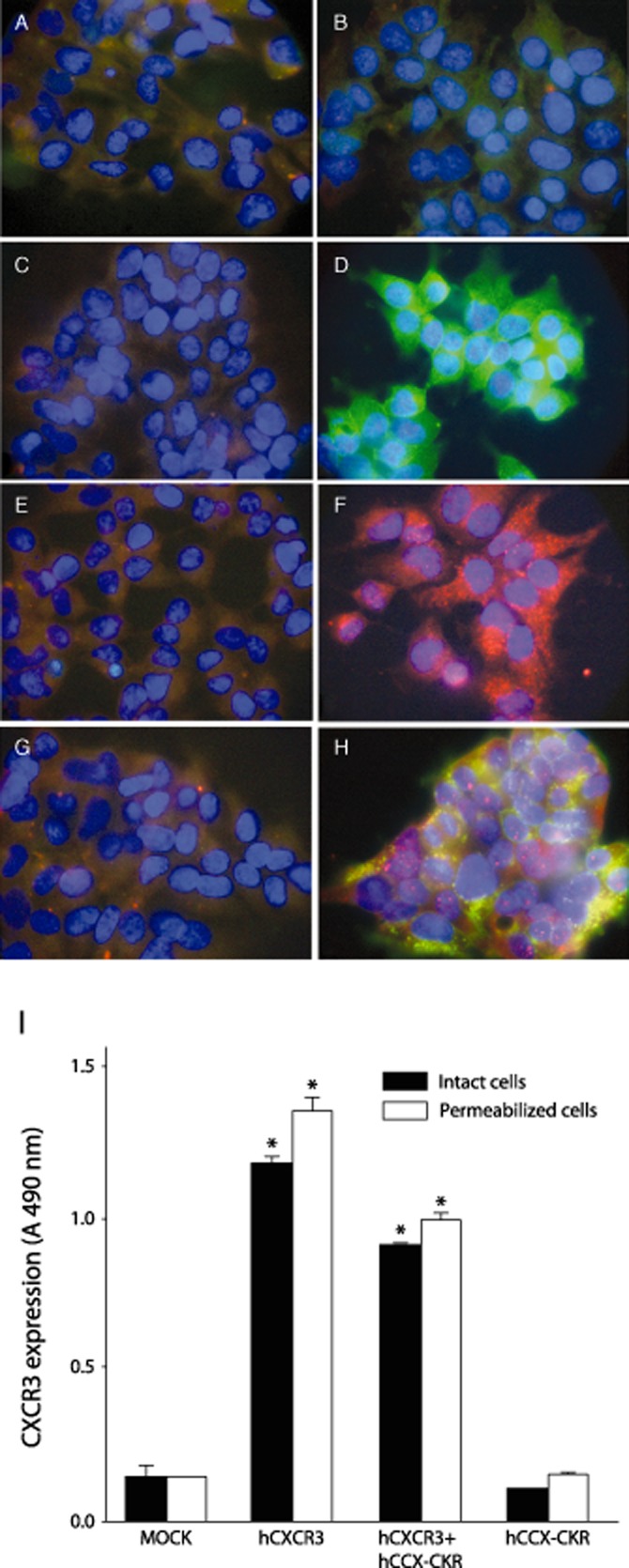
Expression of hCCX-CKR does not influence expression of hCXCR3. Immunocytochemical stainings revealed that non-transfected HEK293 cells did not express hCXCR3 (A) nor hCCX-CKR (B), whereas positive labelling was observed in hCXCR3 (D), hCCX-CKR (F), and hCXCR3/hCCX-CKR (H) stably transfected HEK293 cells. Cells stained in the absence of primary antibodies or in presence of an isotype control antibody with irrelevant specificity were devoid of signals (C,E,G). The magnification used in panels A-H was 40X. (I) ELISA experiments showed no influence of hCCX-CKR co-expression on hCXCR3 protein levels when compared with single hCXCR3-expressing cells. No hCXCR3 protein expression was found in mock- or hCCX-CKR-transfected cells. Data represent the means ± SEM of three independent experiments. Statistical significance against mock transfected cells was tested by means of multiple comparison anova and Tukey's post hoc analysis. Asterisks indicate P ≤ 0.05.
Effect of CCX-CKR on CXCR3-mediated chemotaxis
Having confirmed the expression levels of our stable cell lines, we tested the effect of hCCX-CKR expression on hCXCR3-mediated migration. Whereas HEK293 cells expressing hCXCR3 cells displayed typical chemotaxis responses to CXCL9 and CXCL10, HEK293 cells expressing both hCXCR3 and hCCX-CKR failed to migrate in response to these two chemokines at all concentrations tested (Figure 2A,B). The lack of migration in double-transfected cells could have been due to an artefact as a result of the transfection procedure. To exclude this possibility, we transfected hCXCR3-expressing HEK293 cells with another chemokine receptor subtype, hCX3CR1 (also known as fractalkine receptor), and examined migration in response to CXCL10 as well as to the CX3CR1 ligand CX3CL1. After transfection, these HEK293 cells responded significantly to both CXCL10 and CX3CL1 (Figure 2C) with no difference compared with single transfected cells (data not shown).
Figure 2.
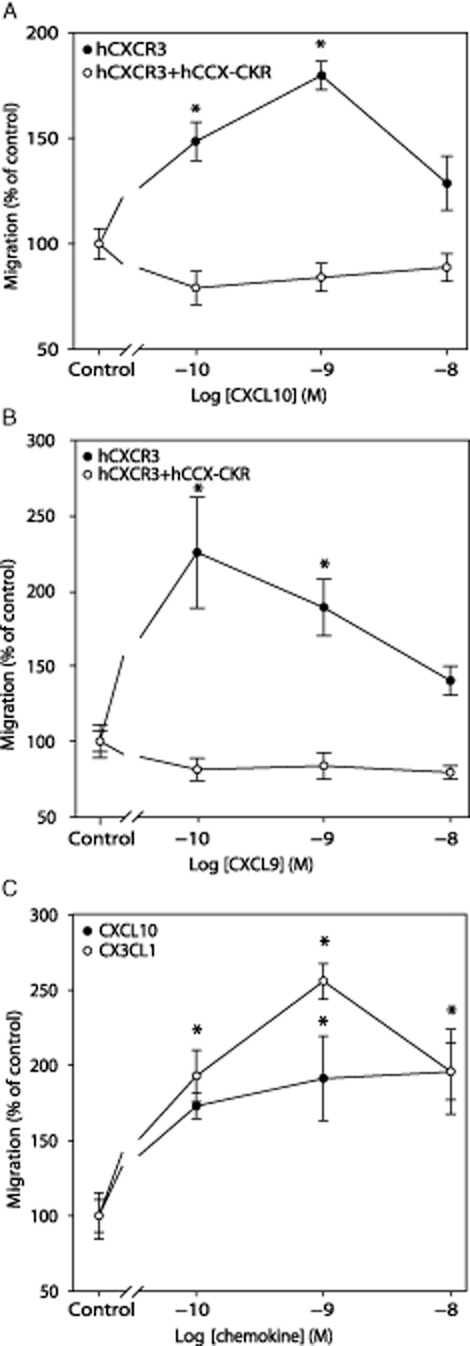
Expression of CCX-CKR impairs CXCR3-dependent migrationHEK293 expressing stable hCXCR3 migrated towards various concentration of CXCL10 whereas migration of HEK293 expressing hCXCR3+hCCX-CKR was abolished (A). The same migratory behavior was observed with various concentrations of CXCL9, another CXCR3 ligand (B). HEK293 stably expressing hCXCR3+hCX3CR1 show migratory behaviour towards various concentration of CXCL10 or CX3CL1 (C). Data represent the means ± SEM of three independent experiments. Statistical significance was tested by means of multiple comparison anova and Tukey's post hoc analysis. Asterisks indicate P ≤ 0.05.
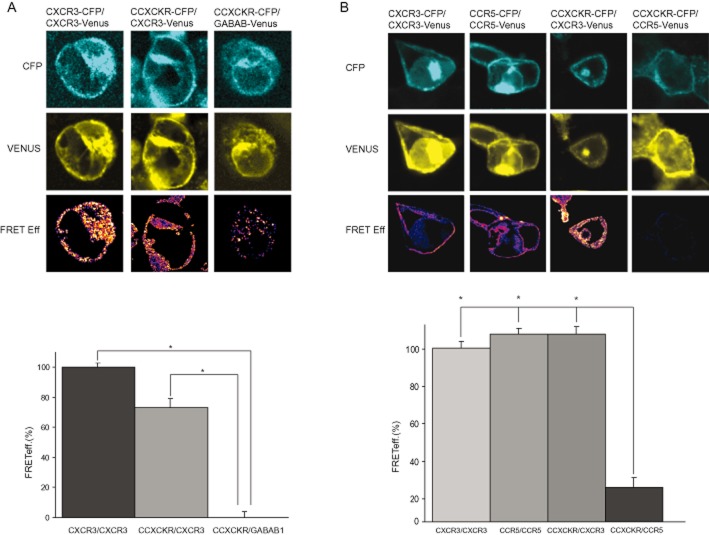
hCCX-CKR heteromerizes with hCXCR3 but not with hCCR5
To investigate whether hCCX-CKR might form heteromers with hCXCR3 we performed FRET experiments in HEK293T cells. Co-transfection of hCXCR3-CFP and hCXCR3-Venus revealed high level of FRET efficiency, which was set to 100%, indicating that CXCR3 per se shows homomerization (Figure 3A) When the hCCX-CKR-CFP construct was co-expressed with hCXCR3-Venus, a strong FRET signal was observed (75 ± 2.81%; Figure 3A, indicating heteromerization of hCCX-CKR with hCXCR3. On the contrary, when hCCX-CKR was co-expressed with CCR5 or a completely unrelated transmembrane receptor (hGABAB1-CFP) no FRET signal was observed (0 ± 3.92%; Figure 3A,B). These data indicate that hCCX-CKR is able to heteromerize with hCXCR3, but not with CCR5 or GABAb1, and thus inhibition of hCXCR3-mediated chemotaxis probably occurs through a direct interaction between both receptors.
Figure 3.
hCCX-CKR forms heteromers with hCXCR3 but not with hCCR5. (A) HEK293T cells transfected with both hCXCR3-CFP and hCXCR3-Venus constructs showed high FRET efficiency, indicating that both proteins are forming abundant homomeric complexes. When transfected with hCXCR3-CFP and hCCX-CKR-Venus, HEK293T also displayed a relatively strong FRET signal, demonstrating that both proteins are interact and form heteromers. HEK293T cells transfected with hGABABR1-CFP and hCCX-CKR-Venus failed to show any FRET signal indicating that these receptors do not interact with each other. (B) FRET experiments were performed after co-expression of hCCR5 and hCCX-CKR. Strong FRET signals were found for hCXCR3 and hCCR5 homomers. Whereas the heteromer hCXCR3/hCCX-CKR showed a strong FRET signal, no signal was observed after co-expression of contructs containing hCCX-CKR-venus and hCCR5-CFP. FRET efficiencyis expressed in % and was scaled on the signal of hCXCR3-CFP and hCXCR3-Venus homomers.
Presence of hCCX-CKR affects binding properties of hCXCR3
Using homologous radioligand displacement assays, we determined the affinities of CXCL10, CXCL11 for hCXCR3 and the affinity of CCL19 for hCCX-CKR in cells expressing the receptors alone or co-expressing CXCR3 and CCX-CKR (Figure 4). Binding affinities of CXCL10 and CXCL11 for hCXCR3 (pKi of 9.7 ± 0.1 and 9.2 ± 0.1, respectively) were not significantly affected by the presence of hCCX-CKR (pKi of 10.0 ± 0.3 and 9.4 ± 0.1, respectively). However, total binding of [125I]-CXCL10 was decreased by 47% on co-expression of hCCX-CKR, whereas total binding of [125I]-CXCL11 was decreased by 20% (Figure 4A, B). Similarly, the binding affinity of CCL19 for hCCX-CKR (pKi of 8.7 ± 0.1) was not significantly affected by the presence of hCXCR3 (pKi of 8.9 ± 0.3). Total [125I]-CCL19 binding was decreased by 65% on co-expression of CCX-CKR with CXCR3 (Figure 4C). As negative ligand binding cooperativity has been observed for chemokine receptor heterodimers (Springael et al., 2006; Sohy et al., 2007), we investigated whether this allosteric ligand interactions also occur within the CXCR3-CCX-CKR heterodimer (Figure 5). Next, [125I]-CXCL10, [125I]-CXCL11 and [125I]-CCL19 equilibrium binding was performed in HEK293T cells expressing hCXCR3 alone, hCCX-CKR alone or co-expressing hCXCR3 and hCCX-CKR, in the absence or presence of 100 nM of the unlabeled chemokine receptor ligands CXCL9, CXCL10, CXCL11 (CXCR3) and CCL19, CCL21, CCL25 and CXCL13 (CCX-CKR). Whereas in hCXCR3-expressing cells [125I]-CXCL10 binding was displaced only by the CXCR3 chemokines, CXCL9, CXCL10 and CXCL11 (Figure 5A), in cells co-expressing hCXCR3and hCCX-CKR, negative [125I]-CXCL10 binding cooperativity was found for CCL19, CCL21 and CCL25, but not CXCL13 (Figure 5B). CXCL10 did not decrease [125I]-CXCL10 binding in cells co-expressing hCXCR3 and hCCX-CKR (Figure 5B). [125I]-CXCL11 was displaced by CXCL10 and CXCL11 in cells expressing hCXCR3 (Figure 5C). On co-expression of hCCX-CKR with CXCR3, CCL19 also significantly decreased [125I]-CXCL11 binding (Figure 5D). Conversely, in cells expressing hCCX-CKR (Figure 5E), the binding of [125I]-CCL19 was only displaced by CCL19 and CCL21, whereas in cells expressing hCXCR3+hCCX-CKR (Figure 5F) a negative cooperative effect of CXCL11, but not CXCL9 or CXCL10, was observed.
Figure 4.
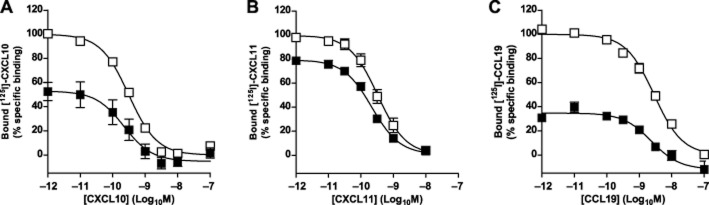
hCXCR3 and hCCX-CKR co-expression decreases the number of chemokine binding sites, but not chemokine affinities. Homologous displacement curves were obtained for [125I]-CXCL10 (A), [125I]-CXCL11 (B) and [125I]-CCL19 (C) in cells expressing hCXCR3 alone (A and B, empty symbols), hCCX-CKR alone (C, empty symbols) or cells co-expressing hCXCR3 and hCCX-CKR (A-C, filled symbols). Data were normalized to specific binding in cells expressing CXCR3 or CCX-CKR alone. Data are given as averages ± SEM of specific binding from three independent experiments performed in triplicate.
Figure 5.
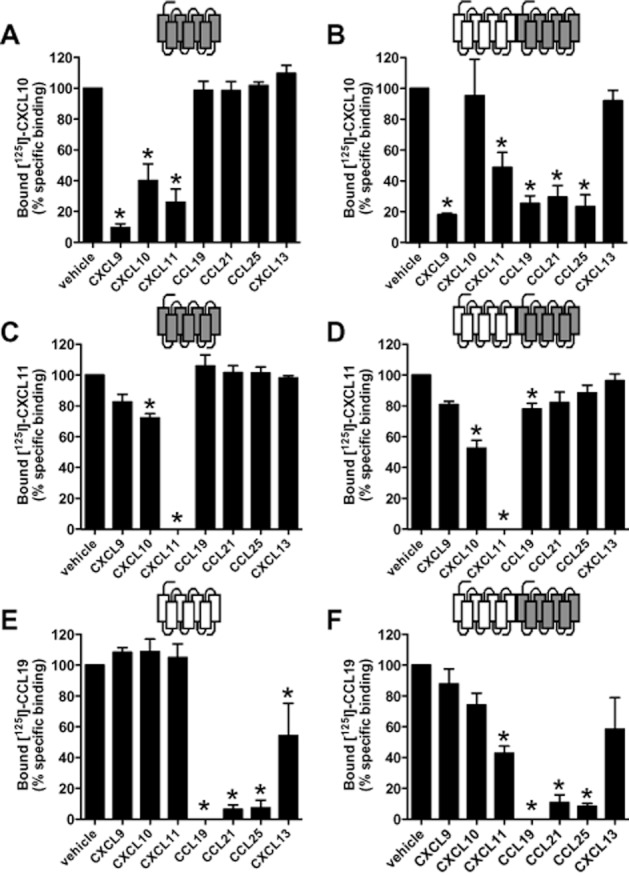
Negative ligand binding cooperativity occurs in cells expressing both hCCX-CKR and hCXCR3. [125I]-CXCL10 (A-B), [125I]-CXCL11 (C-D) and [125I]-CCL19 (E-F) equilibrium binding was performed with HEK293T cells expressing hCXCR3 alone (A and C), hCCX-CKR alone (E) or both hCXCR3 and hCCX-CKR (B, D, F), in the absence or presence of unlabeled chemokines (100 nM) as indicated. Data are given as averages ± SEM of normalized specific binding from 2–5 independent experiments performed in triplicate. Statistical significance against vehicle samples was tested by means of multiple comparison ANOVA and Tukey's post hoc analysis. Asterisks indicate P ≤ 0.05.
CXCR3 and CCX-CKR expression in human T cells
Because CXCR3 and CCX-CKR are among the chemokine receptors present in T cells (Loetscher et al., 1996; Gosling et al., 2000; Thomsen et al., 2003), the inhibition of hCXCR3-medited migration by hCCX-CKR could be of relevance for the migratory behaviour of T cells. Therefore, expression of hCCX-CKR and hCXCR3 mRNA as well as protein was investigated in freshly isolated T cells and T cells activated by phytohemagglutinin (PHA) and IL-2 treatment (Figure 6A). RT-PCR and QPCR analysis showed that both mRNAs were present in freshly isolated T cells (Figure 6B). Whereas PHA/IL-2 treatment did not significantly influence hCXCR3 mRNA expression, a profound down-regulation of hCCX-CKR mRNA expression was observed (Figure 6B). hCCX-CKR and hCXCR3 protein expression was investigated by flow cytometry analysis. It is shown (Figure 6C) that 54.4% of the freshly isolated T cells expressed hCCX-CKR. This percentage dropped to 43.2% positive cells in PHA/IL-2-treated cells (Figure 6C). In contrast a clear increase in expression of hCXCR3 expression was found after PHA/IL-2 treatment, from 18.3 to more that 50.3% CXCR3 positive cells after PHA/IL-2 treatment was observed (Figure 6C). Thus, the treatment with PHA/IL-2 induced a small downregulation of hCCX-CKR and an up-regulation of hCXCR3 expression in human T cells. Similar data were obtained in primary mouse microglia and primary mouse macrophages (Supporting Information Figure S2). Indeed, QPCR analysis indicated the presence of both CCX-CKR and CXCR3 receptors mRNA in these cells. Moreover, treatment with a combination of 50 ng·mL−1 INF and 100 ng·mL−1 LPS in the case of primary macrophages and with 100 ng·mL−1 LPS in the case of microglia provoked a pronounced down-regulation of CCX-CKR mRNA in both cell types (Figure S2A,B). Up-regulation of CXCR3 after activation was observed only in primary microglia (Supporting Information Figure S2A).
Figure 6.
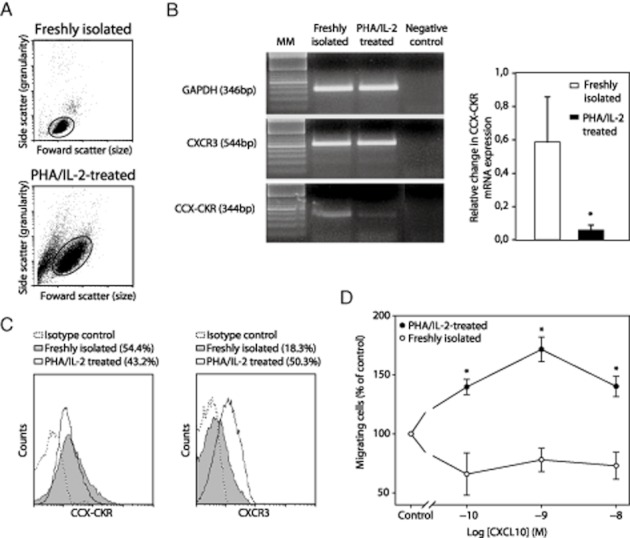
Expression of hCCX-CKR in freshly isolated and in activated T cells. (A) Freshly isolated T cells and PHA/IL-2-activated T cells were sorted by FACS. (B) RT-PCR analysis show that freshly isolated T cells express both hCXCR3 and hCCX-CKR mRNA. Activated T cells display similar levels of hCXCR3 mRNA as freshly isolated T cells but levels of hCCX-CKR mRNA were considerably lower. * = P < 0.05. (C) FACS analysis revealed that 54.4% and 18.3% of freshly isolated T cells expressed hCCX-CKR and hCXCR3, respectively. hCCX-CKR expression dropped to 43.2% in activated T cells whereas a major up-regulation (50.3%) of hCXCR3-expressing-T cells was observed following activation. (D) Chemotaxis experiments show that freshly isolated T cells failed to migrate towards CXCL10 whereas PHA/IL-2-activated T cells demonstrated a prominent migratory behaviour towards the chemokine. * = P < 0.05 compared with control.
Effect of hCCX-CKR expression on CXCR3 mediated migration in human T cells
Next, chemotaxis of freshly isolated and PHA/IL-2-treated T cells in response to CXCL10 was examined. Whereas freshly isolated T cells did not show migration in response to CXCL10 at all concentrations tested, significant migration was observed in PHA/IL-2-treated T cells, with a peak migratory response at 1 nM CXCL10 (Figure 6D). Thus, similar to HEK293 cells, downregulation of hCCX-CKR was corelated to the migration of human T cells towards CXCR3 ligands. However, since the treatment with PHA/IL-2 caused a robust up-regulation of CXCR3 and does most likely induce various other cellular reactions that could account for the observed change in migratory behavior, we aimed at modifying CCX-CKR levels in freshly isolated and PHA/IL-2-treated T cells. We therefore transfected freshly isolated T cells with ShRNA for CCX-CKR to mimic the down-regulation observed after treatment with PHA/IL-2. This treatment indeed caused a clear down-regulation of hCCX-CKR (Figure 7A). Whereas the negative control ShRNA did not affect the chemotactic response of T cells to 1 nM CXCL10, treatment with the ShRNA targeting hCCX-CKR clearly enhanced the migration of T cells to CXCL10 (Figure 7B). In addition, hCCX-CKR was over-expressed in PHA/IL-2-treated T cells as demonstrated by flow cytometry (Figure 7C). Whereas a significant migration in response to CXCL10 was observed in GFP-transfected cells (control transfection), no migratory response was observed in PHA/IL-2-treated T cells that had been transfected with hCCX-CKR (Figure 7D). Similar data were obtained with primary mouse microglia. Since we observed that treatment with LPS provoked a down-regulation of CCX-CKR mRNA and an up-regulation of CXCR3, we hypothesized that activated microglia would migrate more towards CXCR3 ligands. Indeed, we observed a significant increase in cell migration towards both CXCL9 and CXCL10 when microglia were activated with LPS (Supporting Information Figure S2C), showing that the down-regulation of CCX-CKR is correlated to an increased chemotactic response.
Figure 7.
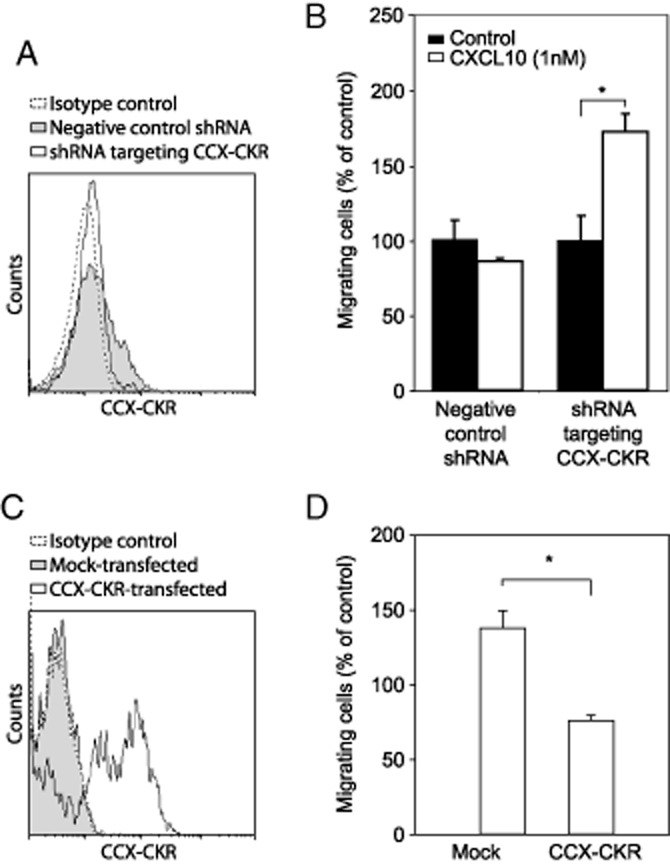
Presence of hCCX-CKR is associated with suppression of chemotaxis towards CXCL10 in T cells (A) Silencing hCCX-CKR using vectors containing DNA encoding for short hairpin RNA targeting hCCX-CKR resulted in a down-regulation of hCCX-CKR expression by freshly isolated T cells as determined by flow cytometry. Data are representative for 4 different human donors. (B) Down-regulating hCCX-CKR rescued chemotaxis towards CXCL10. Data are representative for five different human donors and presented as mean ± SEM. *P < 0.05 (Mann-Whitney U-test). (C) Flow cytometric analysis showed successful transfection of PHA/IL-2-treated T cells with hCCX-CKR. The data represent one out of five separate experiments with different human donors. (D) Re-expressing hCCX-CKR in PHA/IL-2-treated T cells inhibited CXCR3-mediated chemotaxis towards CXCL10. The data are representative for 6 separate experiments using different human donors and are presented as mean ± SEM. *P < 0.05 (Mann-Whitney U-test).
Discussion
Atypical chemokine receptors including DARC, D6 have been shown to efficiently internalize their cognate ligands and act as chemokine scavengers (Hansell et al., 2006). Also CCX-CKR binds and scavenges its ligands CCL21, CCL19, CCL25 and CXCL13 (Gosling et al., 2000; Townson and Nibbs, 2002; Comerford et al., 2006). Here, we show that co-expression of hCCX-CKR abolished hCXCR3-mediated migratory responses towards CXCL9 and CXCL10, which is not due to scavenging of hCXCR3 ligands, as CXCL9 and CXCL10 have no affinity for hCCX-CKR (Gosling et al., 2000). It has been demonstrated that chemokines can signal through monomeric and heteromeric receptor complexes (Salanga et al., 2009). Well-known chemokine receptor heteromers are CCR2/CCR5 (Mellado et al., 2001; El-Asmar et al., 2005; Sohy et al., 2009), CXCR1/CXCR2 (Wilson et al., 2005), CXCR4/CCR2 (Percherancier et al., 2005; Sohy et al., 2007; 2009), CXCR4/CXCR7 (Sierro et al., 2007), CXCR4/CCR5 (Contento et al., 2008; Sohy et al., 2009) and DARC/CCR5 (Chakera et al., 2008). In some of these cases, heteromerization had essential implications for agonist-induced chemotaxis and calcium signalling (Sohy et al., 2007; 2009; Chakera et al., 2008). We therefore decided to investigate possible heteromerization of hCCX-CKR and hCXCR3 by FRET. Expressed in HEKT293 cells, hCXCR3 formed homomers and also displayed pronounced heteromerization with hCCX-CKR. In contrast, hCCX-CKR did not heteromerize with hCCR5 or hGABAB1 receptor, thus confirming the specificity of the FRET assay. Because CCR5-induced chemotaxis is not affected by co-expression of CCX-CKR, this suggests that heteromerization of hCCX-CKR with hCXCR3 plays an essential role in its inhibitory effect on chemotaxis. Heteromerization could induce changes in receptor conformation that possibly would reduce the receptor-ligand binding properties leading to chemotaxis inhibition. Accordingly, the loss of CXCL10 binding sites observed in binding experiments in cell co-expressing hCXCR3 and hCCX-CKR as compared with cells expressing only hCXCR3, is larger than the loss of CXCL11 binding sites and hCXCR3 cell surface expression as measured by ELISA. Cox et al. (2001) have shown that CXCL10 and CXCL11 occupy distinct binding sites at hCXCR3. In contrast to CXCL11 binding, the interaction of CXCL10 with CXCR3 is highly dependent on an active conformation of the receptor. The loss of CXCL10 binding sites in the absence of decreased CXCL10 affinity for hCXCR3 observed in the presence of hCCX-CKR may be explained by a relative increase in the population of CXCR3 in an inactive conformation. Additionally, negative binding cooperativity, induced by ligands both for hCXCR3 and hCCX-CKR was observed. Thus, chemokines for hCCX-CKR suppressed chemokine binding to hCXCR3 and vice versa. In cells co-expressing hCXCR3 and hCCX-CKR, CCL19 could displace both radiolabeled CXCL10 and CXCL11, whereas CCL21 and CCL25 caused significant displacement of [125I]-CXCL10 but not [125I]-CXCL11. The lack of cooperativity between [125I]-CXCL11 and CCL21 or CCL25 may be due to the lower affinity of these two chemokines than that of CCL19 for hCCX-CKR as previously reported by Gosling et al. 2000 (i.e. CCL19, CCL21, CCL25, and CXCL13 displaced [125I]-CCL19 with IC50 values of 6, 12, 7 and 140 nM, respectively). The lack of negative binding cooperativity between [125I]-CCL19 and CXCL10 may be due to the loss of CXCL10 high affinity binding sites on co-expression of hCCX-CKR with hCXCR3. Additionally, the lack of negative binding cooperativity between hCXCR3 radioligands and CXCL13 is likely due to the moderate affinity of this chemokine for hCCX-CKR. In cells co-expressing hCXCR3 and hCCX-CKR, CXCL10 actually increased [125I]-CXCL10 binding, possibly promoting non-specific binding of the radioligand. This effect was also observed in cells that did not express CXCR3 (data not shown). In order to study the potential influence of hCCX-CKR co-expression on hCXCR3 dependent chemotaxis in primary cells experiments with human T cells and mouse microglia and macrophages were performed. Interestingly, in all cells it was observed that the presence of hCCX-CKR significantly inhibited the chemotactic response of hCXCR3 ligands. These results thus suggest that co-expression of hCCX-CKR also in a more physiological setting negatively influences the chemotactic response of hCXCR3. In all these cells it was found that an inflammatory activation down-regulated hCCX-CKR expression and thus increased the migratory capacity of these cells to hCXCR3 ligands. It therefore is tempting to speculate that the loss of hCCX-CKR expression is part of the activation programme of immune cells that enable them to migrate towards sites of inflammation. Interestingly, Gosling et al. (2000) described a similar decrease in CCX-CKR mRNA expression for maturing dendritic cells. Whether CCX-CKR might also influence the migration of dendritic cells remains to be established.
Little is yet known about the physiological function of hCCX-CKR. Since no signalling effects of this receptor-like protein have yet been described, hCCX-CKR is currently considered to function as a chemokine scavenger (Comerford and Nibbs, 2005), similar to the chemokine receptor-like proteins D6 and DARC (Jamieson et al., 2005; Martinez de la Torre et al., 2005). Our current data suggest that hCCX-CKR forms complexes with hCXCR3. This heteromerization might underlie the inhibition of hCXCR3-induced chemotaxis. Negative binding cooperativity upon co-expression of hCXCR3 and hCCX-CKR may also serve as inhibitory mechanism. Similar inhibitory functions have been reported for the atypical CRAM receptor and CCR7 (Catusse et al., 2010). However, contrary to CCX-CKR, which do not display affinity for CXCR3 ligands, the inhibitory effect of CRAM on CCR7 was a combination of ligand competition as well as other unknown factors that might include negative binding cooperativity or heterodimerization. Recently, a possible role of hCCX-CKR as a regulator of growth and metastasis in breast cancer has been reported (Feng et al., 2009). In this study, over-expression of hCCX-CKR inhibited cancer cell proliferation in vitro, and a significant correlation between hCCX-CKR expression and survival rate in breast cancer patients was found. Whether or not heteromerization of hCCX-CKR with other chemokine receptors [as suggested by our chemotaxis experiments (Table 2)] is also involved in inhibition of chemotaxis and proliferation of breast cancer remains to be established.
Conclusion
Here, we show that hCCX-CKR co-expression has a negative influence on the chemotactic response of the chemokine receptor CXCR3. FRET studies and ligand displacement experiments in hCCX-CKR and hCXCR3 co-expressing cells showed heteromerization and negative binding cooperativity, both of which may influence the migratory capacity of CXCR3. Moreover, we demonstrate that CCX-CKR co-expression also inhibits CXCR3-mediated chemotaxis in human T cells, and mouse myeloid cells. The loss of CCX-CKR expression in these primary cells by activation caused a prominent increase in chemotaxis towards CXCR3 ligands. The inhibition of CXCR3-mediated chemotaxis by CCX-CKR co-expression may therefore be part of the downregulatory programme of non-primed immune cells. Thus CCX-CKR apart from being a scavenger for chemokines may have direct influence on the activity status and chemotactic properties of immune cells.
Glossary
Abbreviations
- FCS
fetal calf serum
- Pen/Strep
penicillin/streptomycin
- PHA
phytohemagglutinin
Acknowlegements
Financial support: This study was performed within the framework of the Dutch Top Institute Pharma project: number D1-105. The work was supported by Dutch Foundation for Scientific Research (NWO) Vidi grant (K.B.). J.V. was supported by a training (research) fellowship FISM – Fondazione Italiana Sclerosi Multipla – cod. 2011/B/7.
Conflict of interest
None.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Figure S1 In situ hybridization visualizing hCXCR3 and hCCX-CKR mRNA expression in HEK293 cells stably transfected with hCXCR3 + hCCX-CKR. Stably transfected HEK293 cells were hybridized with sense hCXCR3 (A, 200× magnification) or hCCX-CKR (B, 200× magnification) probes to determine background staining. Antisense probes for both hCXCR3 (C, 200× magnification and E, 400× magnification) and hCCX-CKR (D, 200× magnification and F, 400× magnification) revealed perinuclear mRNA accumulation.
Figure S2 Primary mouse microglia and macrophages behave similarly as human T cells. Treatment of primary mouse microglia with LPS induced a significant reduction of CCX-CKR mRNA expression, whereas CXCR3 mRNA levels were increased (A). mRNA levels of CCXCKR were also downregulated in mouse macrophages when they were treated with LPS and interferon gamma (B). Strnagely, levels of CXCR3 also were greatly reduced. Chemotaxis experiments with primary mouse microglia show that there migration significantly increase towards ATP, CXCL10 and CXCL9 when treated with LPS (C).
Table S1 Primers used for quantitative real-time PCR.
Table S2 Threshold cycle number for hCXCR3, hCCX-CKR and the housekeeping gene GAPDH for all HEK293 cell lines.
References
- Biber K, Klotz KN, Berger M, Gebicke-Harter PJ, van Calker D. Adenosine A1 receptor-mediated activation of phospholipase C in cultured astrocytes depends on the level of receptor expression. J Neurosci. 1997;17:4956–4964. doi: 10.1523/JNEUROSCI.17-13-04956.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer N, Zuurman MW, Wei T, Ransohoff RM, Bodekke HW, Biber K. Induction of glial L-CCR mRNA expression in spinal cord and brain in experimental autoimmune encephalomyelitis. Glia. 2004;46:84–94. doi: 10.1002/glia.10352. [DOI] [PubMed] [Google Scholar]
- Catusse J, Leick M, Groch M, Clark DJ, Buchner MV, Zirlik K, et al. Role of the atypical chemoattractant receptor CRAM in regulating CCL19 induced CCR7 responses in B-cell chronic lymphocytic leukemia. Mol Cancer. 2010;9:297. doi: 10.1186/1476-4598-9-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakera A, Seeber RM, John AE, Eidne KA, Greaves DR. The duffy antigen/receptor for chemokines exists in an oligomeric form in living cells and functionally antagonizes CCR5 signaling through hetero-oligomerization. Mol Pharmacol. 2008;73:1362–1370. doi: 10.1124/mol.107.040915. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Combadiere C, Ahuja SK, Murphy PM. Cloning, chromosomal localization, and RNA expression of a human beta chemokine receptor-like gene. DNA Cell Biol. 1995;14:673–680. doi: 10.1089/dna.1995.14.673. [DOI] [PubMed] [Google Scholar]
- Comerford I, Nibbs RJ. Post-translational control of chemokines: a role for decoy receptors? Immunol Lett. 2005;96:163–174. doi: 10.1016/j.imlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Comerford I, Milasta S, Morrow V, Milligan G, Nibbs R. The chemokine receptor CCX-CKR mediates effective scavenging of CCL19 in vitro. Eur J Immunol. 2006;36:1904–1916. doi: 10.1002/eji.200535716. [DOI] [PubMed] [Google Scholar]
- Contento RL, Molon B, Boularan C, Pozzan T, Manes S, Marullo S, et al. CXCR4-CCR5: a couple modulating T cell functions. Proc Natl Acad Sci USA. 2008;105:10101–10106. doi: 10.1073/pnas.0804286105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MA, Jenh CH, Gonsiorek W, Fine J, Narula SK, Zavodny PJ, et al. Human interferon-inducible 10-kDa protein and human interferon-inducible T cell alpha chemoattractant are allotopic ligands for human CXCR3: differential binding to receptor states. Mol Pharmacol. 2001;59:707–715. doi: 10.1124/mol.59.4.707. [DOI] [PubMed] [Google Scholar]
- El-Asmar L, Springael JY, Ballet S, Andrieu EU, Vassart G, Parmentier M. Evidence for negativebinding cooperativity within CCR5-CCR2b heterodimers. Mol Pharmacol. 2005;67:460–469. doi: 10.1124/mol.104.003624. [DOI] [PubMed] [Google Scholar]
- Feng LY, Ou ZL, Wu FY, Shen ZZ, Shao ZM. Involvement of a novel chemokine decoy receptor CCX-CKR in breast cancer growth, metastasis and patient survival. Clin Cancer Res. 2009;15:2962–2970. doi: 10.1158/1078-0432.CCR-08-2495. [DOI] [PubMed] [Google Scholar]
- Gosling J, Dairaghi DJ, Wang Y, Hanley M, Talbot D, Miao Z, et al. Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J Immunol. 2000;164:2851–2856. doi: 10.4049/jimmunol.164.6.2851. [DOI] [PubMed] [Google Scholar]
- Hansell CA, Simpson CV, Nibbs R. Chemokine sequestration by atypical chemokine receptors. Biochem Soc Trans. 2006;34:1009–1013. doi: 10.1042/BST0341009. [DOI] [PubMed] [Google Scholar]
- Hensbergen PJ, van der Raaij-Helmer EM, Dijkman R, van der Schors RC, Werner-Felmayer G, Boorsma DM, et al. Processing of natural and recombinant CXCR3-targeting chemokines and implications for biological activity. Eur J Biochem. 2001;268:4992–4999. doi: 10.1046/j.0014-2956.2001.02433.x. [DOI] [PubMed] [Google Scholar]
- Horuk R. Chemokine receptors. Cytokine Growth Factor Rev. 2001;12:313–335. doi: 10.1016/s1359-6101(01)00014-4. [DOI] [PubMed] [Google Scholar]
- Jalink K, Van Rheenen J. Quantitative imaging of sensitized emission FRET and FLIM techniques. In: Gadella TWJ, editor. Laboratory Techniques in Biochemistry and Molecular Biology. Vol. 33. Burlington, MA: Academic Press; 2009. pp. 289–349. [Google Scholar]
- Jamieson T, Cook DN, Nibbs RJ, Rot A, Nixon C, McLean P, et al. The chemokine receptor D6 limits the inflammatory response in vivo. Nat Immunol. 2005;6:403–411. doi: 10.1038/ni1182. [DOI] [PubMed] [Google Scholar]
- Johnson M, Li AR, Liu J, Fu Z, Zhu L, Miao S, et al. Discovery and optimization of a series of quinazolinone-derived antagonists of CXCR3. Bioorg Med Chem Lett. 2007;17:3339–3343. doi: 10.1016/j.bmcl.2007.03.106. [DOI] [PubMed] [Google Scholar]
- Loetscher M, Gerber B, Loetscher P, Jones SA, Piali L, Clark-Lewis I, et al. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J Exp Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de la Torre Y, Locati M, Buracchi C, Dupor J, Cook DN, Bonecchi R, et al. Increased inflammation in mice deficient for the chemokine decoy receptor D6. Eur J Immunol. 2005;35:1342–1346. doi: 10.1002/eji.200526114. [DOI] [PubMed] [Google Scholar]
- Mellado M, Rodriguez-Frade JM, Vila-Coro AJ, Fernandez S, Martin de Ana A, Jones DR, et al. Chemokine receptor homo- or heterodimerization activates distinct signaling pathways. EMBO J. 2001;20:2497–2507. doi: 10.1093/emboj/20.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy PM. International Union of Pharmacology. XXX. Update on Chemokine Receptor Nomenclature. Pharmacol Rev. 2002;54:227–229. doi: 10.1124/pr.54.2.227. [DOI] [PubMed] [Google Scholar]
- Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- Percherancier Y, Berchiche YA, Slight I, Volkmer-Engert R, Tamamura H, Fujii N, et al. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J Biol Chem. 2005;280:9895–9903. doi: 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- Rios CD, Jordan BA, Gomes I, Devi LA. G-protein-coupled receptor dimerization: modulation of receptor function. Pharmacol Ther. 2001;92:71–87. doi: 10.1016/s0163-7258(01)00160-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Frade JM, Vila-Coro AJ, de Ana AM, Albar JP, Martinez AC, Mellado M. The chemokine monocyte chemoattractant protein-1 induces functional responses through dimerization of its receptor CCR2. Proc Natl Acad Sci USA. 1999;96:3628–3633. doi: 10.1073/pnas.96.7.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- Salanga CR, O'Hayre M, Handel T. Modulation of chemokine receptor activity through dimerization and cross talk. Cell Mol Life Sci. 2009;66:1307–1386. doi: 10.1007/s00018-008-8666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Madrid F, del Pozo MA. Leukocyte polarization in cell migration and immune interactions. EMBO J. 1999;18:501–511. doi: 10.1093/emboj/18.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro F, Biben C, Martinez-Munoz L, Mellado M, Ransohoff RM, Li M, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci USA. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohy D, Parmentier M, Springael JY. Allosteric transinhibition by specific antagonists in CCR2/CXCR4 heterodimers. J Biol Chem. 2007;282:30062–30069. doi: 10.1074/jbc.M705302200. [DOI] [PubMed] [Google Scholar]
- Sohy D, Yano H, de Nadai P, Urizar E, Guillabert A, Javitch JA, et al. Hetero-oligomerization of CCR2, CCR5, and CXCR4 and the protean effects of ‘selective’ antagonists. J Biol Chem. 2009;284:31270–31279. doi: 10.1074/jbc.M109.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael JY, Urizar E, Parmentier M. Dimerization of chemokine receptors and its functional consequences. Cytokine Growth Factor Rev. 2005;16:611–623. doi: 10.1016/j.cytogfr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Springael JY, Le Minh PN, Urizar E, Costagliola S, Vassart G, Parmentier M. Allosteric modulation of binding properties between units of chemokine receptor homo- and hetero-oligomers. Mol Pharmacol. 2006;69:1652–1661. doi: 10.1124/mol.105.019414. [DOI] [PubMed] [Google Scholar]
- Thelen M. Dancing to the tune of chemokines. Nat Immunol. 2001;2:129–134. doi: 10.1038/84224. [DOI] [PubMed] [Google Scholar]
- Thomsen AR, Nansen A, Madsen AN, Bartholdy C, Christensen JP. Regulation of T cell migration during viral infection: role of adhesion molecules and chemokines. Immunol Lett. 2003;85:119–127. doi: 10.1016/s0165-2478(02)00236-5. [DOI] [PubMed] [Google Scholar]
- Townson JR, Nibbs RJ. Characterization of mouse CCX-CKR, a receptor for the lymphocyte-attracting chemokines TECK/mCCL25, SLC/mCCL21 and MIP-3beta/mCCL19: comparison to human CCX-CKR. Eur J Immunol. 2002;32:1230–1241. doi: 10.1002/1521-4141(200205)32:5<1230::AID-IMMU1230>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Van Rheenen J, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophysical J. 2004;86:2517–2529. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijl D, Storelli S, Scholten DJ, Bosch L, Reinhart TA, Streblow DN, et al. Noncompetitive antagonism and inverse agonism as mechanism of action of nonpeptidergic antagonists at primate and rodent CXCR3 chemokine receptors. J Pharmacol Exp Ther. 2008;325:544–555. doi: 10.1124/jpet.107.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Wilkinson G, Milligan G. The CXCR1and CXCR2 receptors form constitutive homo- and heterodimers selectively and with equal apparent affinities. J Biol Chem. 2005;280:28663–28674. doi: 10.1074/jbc.M413475200. [DOI] [PubMed] [Google Scholar]
- Zuurman MW, Heeroma J, Brouwer N, Boddeke HW, Biber K. LPS-induced expression of a novel chemokine receptor (L-CCR) in mouse glial cells in vitro and in vivo. Glia. 2003;41:327–336. doi: 10.1002/glia.10156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.