Abstract
Ribosomal protein L1 from Saccharomyces cerevisiae binds 5S rRNA and can be released from intact 60S ribosomal subunits as an L1-5S ribonucleoprotein (RNP) particle. To understand the nature of the interaction between L1 and 5S rRNA and to assess the role of L1 in ribosome assembly and function, we cloned the RPL1 gene encoding L1. We have shown that RPL1 is an essential single-copy gene. A conditional null mutant in which the only copy of RPL1 is under control of the repressible GAL1 promoter was constructed. Depletion of L1 causes instability of newly synthesized 5S rRNA in vivo. Cells depleted of L1 no longer assemble 60S ribosomal subunits, indicating that L1 is required for assembly of stable 60S ribosomal subunits but not 40S ribosomal subunits. An L1-5S RNP particle not associated with ribosomal particles was detected by coimmunoprecipitation of L1 and 5S rRNA. This pool of L1-5S RNP remained stable even upon cessation of 60S ribosomal subunit assembly by depletion of another ribosomal protein, L16. Preliminary results suggest that transcription of RPL1 is not autogenously regulated by L1.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archambault J., Milne C. A., Schappert K. T., Baum B., Friesen J. D., Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J Biol Chem. 1992 Feb 15;267(5):3282–3288. [PubMed] [Google Scholar]
- Baim S. B., Pietras D. F., Eustice D. C., Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Pringle J. R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Mar;11(3):1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Isolation of a 5S RNA-protein complex from mammalian ribosomes. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1881–1885. doi: 10.1073/pnas.68.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Riggs D. L., Kassavetis G. A., Geiduschek E. P. Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2530–2534. doi: 10.1073/pnas.86.8.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D. A., Geiduschek E. P. Modulation of yeast 5 S rRNA synthesis in vitro by ribosomal protein YL3. A possible regulatory loop. J Biol Chem. 1987 Oct 15;262(29):13953–13958. [PubMed] [Google Scholar]
- Chan Y. L., Lin A., McNally J., Wool I. G. The primary structure of rat ribosomal protein L5. A comparison of the sequence of amino acids in the proteins that interact with 5 S rRNA. J Biol Chem. 1987 Sep 15;262(26):12879–12886. [PubMed] [Google Scholar]
- Chen-Schmeisser U., Garrett R. A. A new method for the isolation of a 5 S RNA complex with proteins L5, L18 and L25 from Escherichia coli ribosomes. FEBS Lett. 1977 Mar 1;74(2):287–291. doi: 10.1016/0014-5793(77)80866-1. [DOI] [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991 May 31;65(5):797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fragapane P., Caffarelli E., Lener M., Prislei S., Santoro B., Bozzoni I. Identification of the sequences responsible for the splicing phenotype of the regulatory intron of the L1 ribosomal protein gene of Xenopus laevis. Mol Cell Biol. 1992 Mar;12(3):1117–1125. doi: 10.1128/mcb.12.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Guddat U., Bakken A. H., Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in xenopus oocytes. Cell. 1990 Feb 23;60(4):619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- Helser T. L., Baan R. A., Dahlberg A. E. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol. 1981 Jan;1(1):51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991 Jun;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992 Jul;11(7):2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maicas E., Pluthero F. G., Friesen J. D. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol Cell Biol. 1988 Jan;8(1):169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Paulovich A. G., Tsay Y. F., Woolford J. L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990 Dec;111(6 Pt 1):2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Pulaski B. A., Woolford J. L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991 Nov;11(11):5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H. G., Fried H. M. Effects of progressive depletion of TCM1 or CYH2 mRNA on Saccharomyces cerevisiae ribosomal protein accumulation. Mol Cell Biol. 1986 May;6(5):1535–1544. doi: 10.1128/mcb.6.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Yaguchi M., Willick G. E., Rollin C. F., Roy C. The 5-S RNA binding protein from yeast (Saccharomyces cerevisiae) ribosomes. Evolution of the eukaryotic 5-S RNA binding protein. Eur J Biochem. 1979 Dec 17;102(2):573–582. doi: 10.1111/j.1432-1033.1979.tb04274.x. [DOI] [PubMed] [Google Scholar]
- Neigeborn L., Warner J. R. Expression of yeast 5S RNA is independent of the rDNA enhancer region. Nucleic Acids Res. 1990 Jul 25;18(14):4179–4184. doi: 10.1093/nar/18.14.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., Wegnez M. Isolation of a 7S particle from Xenopus laevis oocytes: a 5S RNA-protein complex. Proc Natl Acad Sci U S A. 1979 Jan;76(1):241–245. doi: 10.1073/pnas.76.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., le Maire M., Wegnez M., Denis H. Biochemical Research on oogenesis. Composition of the 42-S storage particles of Xenopus laevix oocytes. Eur J Biochem. 1980 Aug;109(2):359–368. doi: 10.1111/j.1432-1033.1980.tb04802.x. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rotenberg M. O., Moritz M., Woolford J. L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988 Feb;2(2):160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N., Matheson A. T., Yaguchi M., Willick G. E., Nazar R. N. The 5-S RNA . protein complex from an extreme halophile, Halobacterium cutirubrum. Purification and characterization. Eur J Biochem. 1978 Sep 1;89(2):501–509. doi: 10.1111/j.1432-1033.1978.tb12554.x. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Berg C., Hendrick J. P., La Branche-Chabot H., Metspalu A., Rinke J., Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol. 1988 Mar;106(3):545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B. Z., Nazar R. N. Structure of the yeast ribosomal 5 S RNA-binding protein YL3. J Biol Chem. 1991 Apr 5;266(10):6120–6123. [PubMed] [Google Scholar]
- Tao J., Frankel A. D. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991 Mar;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
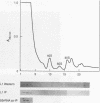
- Tsay Y. F., Thompson J. R., Rotenberg M. O., Larkin J. C., Woolford J. L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988 Jun;2(6):664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Udem S. A., Kaufman K., Warner J. R. Small ribosomal ribonucleic acid species of Saccharomyces cerevisiae. J Bacteriol. 1971 Jan;105(1):101–106. doi: 10.1128/jb.105.1.101-106.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972 Mar 28;65(2):227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- Vioque A., Hernández F., Palacián E. Involvement of lysine and arginine residues in the binding of yeast ribosomal protein YL3 to 5S RNA. Mol Cell Biochem. 1987 Aug;76(2):141–146. doi: 10.1007/BF00223479. [DOI] [PubMed] [Google Scholar]
- Wang C. K., Weil P. A. Purification and characterization of Saccharomyces cerevisiae transcription factor IIIA. J Biol Chem. 1989 Jan 15;264(2):1092–1099. [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989 Jun;53(2):256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- Wormington W. M. Developmental expression and 5S rRNA-binding activity of Xenopus laevis ribosomal protein L5. Mol Cell Biol. 1989 Dec;9(12):5281–5288. doi: 10.1128/mcb.9.12.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi M., Rollin C. F., Roy C., Nazar R. N. The 5S RNA binding protein from yeast (Saccharomyces cerevisiae) ribosomes. An RNA binding sequence in the carboxyl-terminal region. Eur J Biochem. 1984 Mar 15;139(3):451–457. doi: 10.1111/j.1432-1033.1984.tb08026.x. [DOI] [PubMed] [Google Scholar]
- Yu R. S., Wittmann H. G. The sequence of steps in the attachment of 5-S RNA to cores of Escherichia coli ribosomes. Biochim Biophys Acta. 1973 Oct 26;324(3):375–385. doi: 10.1016/0005-2787(73)90282-7. [DOI] [PubMed] [Google Scholar]
- elBaradi T. T., van der Sande C. A., Mager W. H., Raué H. A., Planta R. J. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr Genet. 1986;10(10):733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]