Abstract
Hearing loss is the most frequent sensory disorder involving a multitude of factors, and at least 50% of cases are due to genetic etiology. To further characterize the molecular etiology of hearing loss in the Chinese population, we recruited a total of 135 unrelated patients with nonsyndromic sensorineural hearing loss (NSHL) for mutational screening of GJB2, GJB3, GJB6, SLC26A4, SLC26A5 IVS2-2A>G and mitochondrial 12SrRNA, tRNASer(UCN) by PCR amplification and direct DNA sequencing. The carrier frequencies of deafness-causing mutations in these patients were 35.55% in GJB2, 3.70% in GJB6, 15.56% in SLC26A4 and 8.14% in mitochondrial 12SrRNA, respectively. The results indicate the necessity of genetic screening for mutations of these causative genes in Chinese population with nonsyndromic hearing loss.
Keywords: nonsyndromic hearing loss, GJB2, GJB3, GJB6, SLC26A4, SLC26A5, mitochondrial DNA, gene mutation
INTRODUCTION
Hearing loss is the most common sensory disorder in humans. Profound hearing loss affects approximately one in 1,000 live births in the general population, and 50%-60% of these cases have genetic etiologies[1]. Genetic hearing loss can follow a pattern of autosomal dominant, autosomal or X-linked recessive, or mitochondrial inheritance. About 70% are classified as nonsyndromic since hearing loss is the only symptom, while 30% are classified as syndromic and are associated with other clinical features[2].
There are more than 150 genetic loci that have been described for nonsyndromic sensorineural hearing loss (NSHL) in humans, and about 60 of them were cloned (Hereditary Hearing Loss Homepage: http://hereditaryhearingloss.org). It is believed that alterations in several members of the connexin protein family, mutations in the solute carrier 26 (SLC26) family and in the mitochondrial DNA (mtDNA) contribute to the development of the majority of genetic hearing losses[2]–[8].
It is estimated that there are approximately 20 million babies born every year in China, of whom about 30,000 are expected to have congenital hearing loss[9]. Carrier frequencies of some mutational hot spots associated with NSHL such as GJB2 235delC and mtDNA A1555G have been reported, but the molecular etiology of NSHL in Chinese population has not been investigated completely in most areas. In this study, we screened the GJB2, GJB3, GJB6, SLC26A4, SLC26A5 IVS2-2A>G and mitochondrial genes to determine the etiology of hearing loss in eastern China.
MATERIALS AND METHODS
Subjects
All the individuals were recruited to participate in the study by signing a written informed consent, and the study protocol was approved by the Ethical Committee of the Nanjing Medical University for Human Studies, and the participants were asked to donate a blood sample. The genetic studies were conducted on two groups: a case group with moderate to profound and nonsyndromic sensorineural hearing impairment (n = 135) aged 7-12 (11.3±2.6) years, and a control group (n = 162) with normal hearing aged 8-14 (11.5±2.9) years. The female-male ratio of these groups are 74/61 and 75/87, respectively. One hundred thirty-five blood samples were obtained from a panel of sporadic hearing-impaired individuals from Nanjing City School for Deaf Children, and 162 control blood samples were gotten from a panel of unaffected individuals in Jiangsu province. All subjects were Han Chinese in origin, and were evaluated through otological examination and audiological evaluations including pure-tone audiometry (Madsen Orbiter 922), immittance (Madsen Zodiac 901), auditory brainstem response (ABR, Interacoustic EP25), and transient evoked otoacoustic emissions (Madsen Celesta 503). Hearing impairment was defined by the level of hearing loss in the better ear for pure-tone threshold average in the speech frequencies 0.5, 1, 2, and 4k Hz. Hearing loss of 26-40 dB was considered mild; 41-60 dB, moderate; 61-80 dB, severe, and more than 80 dB, profound.
Molecular screening
The mutations on the GJB2, GJB3, GJB6, SLC26A4, SLC26A5 IVS2-2A>G and mitochondrial genes were selected for molecular screening. Genomic DNA was isolated from 2 mL of peripheral leukocytes of all participants using Puregene DNA Isolation Kits (DNA fast 2000, Shanghai China). The DNA fragments spanning the entire coding region of those genes were PCR amplified by using reference primers[8],[9]. The quality and quantity of purified genomic DNA were determined by running a 0.8% agarose gel and spectrophotometry. Each fragment was analyzed by direct sequencing in an Applied Biosystems 3730 automated DNA sequencer. The resultant sequence data were compared with the wildtype GJB2 (GenBank Accession No. GI62999485), GJB3 (GenBank Accession No.NM_024009), GJB6 (GenBank Accession No. NT_009799.12), SLC26A4 (GenBank Accession No. NM_000441.1), SLC26A5 (GenBank Accession No. AC 005064), and mtDNA (GenBank accession No. NC_001807.4) gene sequences to identify the mutations.
Data compilation
To determine the most common gene mutations in China, many data in several typical areas of China were reviewed. To examine the possible role of the nuclear genes in patients with non-syndromic hearing loss, we compared the carrier frequencies of GJB2, GJB3, GJB6 and SLC26 family mutations, which are common deafness-associated nucleotide changes, from different areas of the world.
RESULTS
Molecular analysis
All the results were compared with the standard sequences of GJB2, GJB3, GJB6, SLC26A4, SLC26A5 and mtDNA 12SrRNA, tRNASer(UCN). A series of sequence variations were detected in these genes as shown in Table 1 and Table 2.
Table 1. Variations in mtDNA identified in the subjects.
| Genetype | Nucleotide change | Amino acid change | Mutation type | Category | Patients (n) |
Controls (n) |
||
| Homo-plasmy | Heteio-plasmy | Homo-plasmy | Heteio-plasmy | |||||
| Mitochondrial | 709G>A | 12SrRNA | Missense | undefined | 3 | 0 | 2 | 0 |
| 752C>T | 12SrRNA | Missense | polymorphism | 4 | 0 | 1 | 0 | |
| 827A>G | 12SrRNA | Missense | pathogenic | 6 | 0 | 0 | 0 | |
| 961T>C | 12SrRNA | Missense | pathogenic | 2 | 0 | 0 | 0 | |
| 1005T>C | 12SrRNA | Missense | polymorphism | 6 | 0 | 3 | 0 | |
| 1048C>T | 12SrRNA | Missense | polymorphism | 2 | 0 | 0 | 0 | |
| 1095T>C | 12SrRNA | Missense | pathogenic | 1 | 0 | 0 | 0 | |
| 1119T>C | 12SrRNA | Missense | polymorphism | 3 | 0 | 0 | 0 | |
| 1382A>G | 12SrRNA | Missense | polymorphism | 4 | 0 | 3 | 0 | |
| 1438A>G | 12SrRNA | Missense | polymorphism | 125 | 0 | 112 | 0 | |
| 1555A>G | 12SrRNA | Missense | pathogenic | 2 | 0 | 0 | 0 | |
| 7445A>G | tRNASer(UCN) | Missense | pathogenic | 0 | 0 | 0 | 0 | |
| 7472insC | tRNASer(UCN) | Missense | pathogenic | 0 | 0 | 0 | 0 | |
| 7511T>C | tRNASer(UCN) | Missense | pathogenic | 0 | 0 | 0 | 0 | |
| Subtotal | 158 | 0 | 121 | 0 | ||||
| DM subtotal | 11 | 0 | ||||||
DM: definite deafening mutations.
Table 2. Variations in GJB2, GJB3, GJB6, SLC26A4 and SLC26A5 genes identified in the subjects.
| Gene | Nucleotide change | Amino acid change | Domain | Mutation type | Category | Patients (n) |
Controls (n) |
||||
| Homo | Hetero | Allele frequency(%) | Homo | Hetero | Allele frequency(%) | ||||||
| GJB2 | 79G>A | V27I | TM1 | Missense | Polymorphism | 4 | 10 | 6.67 | 13 | 20 | 17.04 |
| 79G>A+341A>G | V27I+E114G | TM1+IC2 | Missense | Undefined | 3 | 2 | 2.96 | 1 | 1 | 1.11 | |
| 79G>A+109G>A | V27I+V37I | TM1 | Missense | Undefined | 0 | 1 | 0.37 | 0 | 0 | 0.00 | |
| 109G>A | V37I | TM1 | Missense | Undefined | 2 | 5 | 3.33 | 0 | 3 | 1.11 | |
| 101T>C | M34T | TM1 | Missense | Polymorphism | 0 | 1 | 0.37 | 0 | 0 | 0.00 | |
| 176-191del16 | 176-191del16 | EC1 | Deletion | Pathogenic | 0 | 1 | 0.37 | 0 | 1 | 0.37 | |
| 235delC | 235delC | TM2 | Deletion | Pathogenic | 18 | 19 | 20.37 | 0 | 2 | 0.74 | |
| 299-300delAT | 299-300delAT | IC2 | Deletion | Pathogenic | 0 | 6 | 2.22 | 0 | 1 | 0.37 | |
| 341A>G | E114G | IC2 | Missense | Polymorphism | 3 | 7 | 4.81 | 1 | 5 | 2.59 | |
| 368C>A | T123N | IC2 | Missense | Pathogenic | 1 | 1 | 1.11 | 0 | 1 | 0.37 | |
| 504insAAGG | 504insAAGG | EC2 | Insertion | Pathogenic | 0 | 2 | 0.74 | 0 | 0 | 0.00 | |
| 608TC>AA | I203K | TM4 | Missense | Polymorphism | 0 | 2 | 0.74 | 0 | 8 | 2.96 | |
| GJB3 | 357C>T | N119N | IC2 | Samesense | Polymorphism | 0 | 2 | 0.74 | 0 | 0 | 0.00 |
| 866G>A | 3′UTR | Missense | Polymorphism | 0 | 6 | 2.22 | 0 | 1 | 0.37 | ||
| GJB6 | 232 kb del | Frameshift | TM3 | Deletion | Pathogenic | 4 | 1 | 3.33 | 0 | 0 | 0.00 |
| SLC26A4 | IVS7-2A>G | Splice site | intravening | Missense | Pathogenic | 7 | 11 | 9.26 | 0 | 0 | 0.00 |
| IVS7-2A>G+ | Splice site + H723R | sequence7 | Missense | Pathogenic | 0 | 2 | 0.74 | 0 | 0 | 0.00 | |
| 2168A>G | intravening | ||||||||||
| 2167 C>G | H723D | sequence7 STAS | Missense | Pathogenic | 0 | 1 | 0.37 | 0 | 0 | 0.00 | |
| SLC26A5 | IVS2-2A>G | Splice site | intravening sequence2 | Missense | Pathogenic | 0 | 0 | 0.00 | 0 | 0 | 0.00 |
UTR: untranslated region; IC: intracellular; TM: transmembrane; EC: extracellular; STAS: sulfate transporter and anti-sigma antagonist
In the case group, there were totally eleven mitochondrial 12SrRNA sequence variations detected. Of those, four variants are known deafness-associated mutations (Fig. 1) with variable frequencies of 1.48% in A1555G, 4.44% in A827G, 1.48% in T961C and 0.74% in T1095C (cosegregate with A1555G mutation). All of these mutations were not found in the control group. One variant, G709A, which was reported to be a polymorphism in some reports, was detected both in the case group (2.22%) and the control group (1.23%). Other variants of mtDNA 12SrRNA, such as T1005C, C1048T, T1119C, C752T, A1382G and A1438G, seem to be polymorphisms rather than causes of disease. On the other hand, we did not find C1494T mutation in the 12SrRNA and any of the known deafness-associated mutations in tRNASer(UCN), such as A7445G, 7472insC, T7510C, T7511C, T7512C and G7444A, in all individuals.
Fig. 1. Partial sequence chromatograms of mtDNA 12SrRNA from the patients.
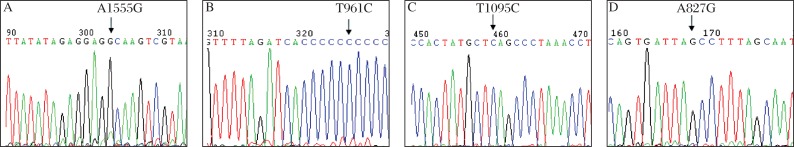
Arrows indicate the location of the base changes. A: A to G transition at position 1555. B: T to C transition at position 961. C: T to C transition at position 1095. D: A to G transition at position 827.
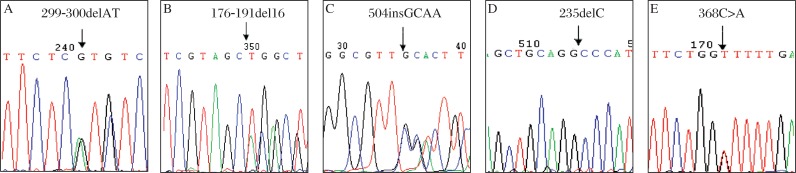
Compared to the standard sequence of GJB2, we identified twelve types of sequence changes in 88 patients, of which 5 types of variations (299-300delAT, 176-191del16, 504insGCAA, 235delC, and 368C>A) (Fig. 2) in 48 patients were thought to be pathogenic, and 4 belonged to benign variant (79G>A, 101T>C, 341A>G and 608TC>AA). The carrier frequencies of deafness-causing GJB2 mutations in our case group were 0.74% for 176-191del16, 27.41% for 235delC, 4.44% for 299-300delAT, 1.48% for T123N and 1.48% for 504insGCAA, respectively. In addition, the pathogenicity of the other 3 kinds of variants (79G>A+341A>G, 79G>A+109G>A, and 109G>A) have not come to an agreement. Thus, the total carrier frequency of deafness-causing GJB2 mutations in our case group was 35.55% (48/135) at least. The 235delC appeared to be the most common deafness-related GJB2 mutation (37/135, 27.41%) with the highest allele frequency of 20.37% (Table 2 and Table 3). In the control group, only five subjects were found to carry the heterozygous deafness-causing mutation.
Fig. 2. Partial sequence chromatograms of GJB2 from the patients.
Arrows indicate the location of base changes. A: AT deletion at position 299-300. B: 16-base deletion at position 176-191. C: GCAA insert at 504. D: C deletion at position 235. E: C to A transversion at position 368.
Table 3. Classification of variants in GJB2, GJB3, GJB6, SLC26A4 and SLC26A5 genes identified in all subjects.
| Variant classification | Patients (n= 135) |
Controls (n =162) |
|||||
| Homo | Hetero | Total | Homo | Hetero | Total | ||
| AR | GJB2 | 19 | 29 | 48 | 0 | 5 | 5 |
| GJB3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| GJB6 | 4 | 1 | 5 | 0 | 0 | 0 | |
| SLC26A4 | 7 | 14 | 21 | 0 | 0 | 0 | |
| SLC26A5 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Polymorphism and AR and undefined | GJB2 | 31 | 57 | 88 | 15 | 42 | 57 |
| GJB3 | 0 | 8 | 8 | 0 | 1 | 1 | |
| GJB6 | 4 | 1 | 5 | 0 | 0 | 0 | |
| SLC26A4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| SLC26A5 | 0 | 0 | 0 | 0 | 0 | 0 | |
AR: autosomal recessive.
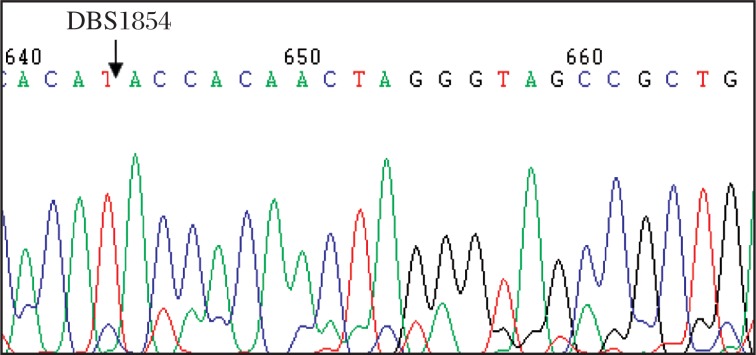
Apart from GJB2 mutations, one kind of deafness-causing GJB6 mutation (⊿GJB6-D13S1854 or 232 kb del, Fig. 3) was identified in 5 of 135 hearing-impaired individuals. The heterozygous gene variation frequency of GJB6 and GJB2 was 5/135 (3.70%), GJB2 and mtDNA was 3/135 (2.22%). All of the patients with⊿GJB6-D13S1854 were also found to have the 235delC mutation in GJB2. None of the normal-hearing person in the control group carries the GJB6 mutation.
Fig. 3. Partial sequence chromatograms of GJB6 from the patient.
The arrow indicates the location of 232 kb deletion (GJB6-D13S1854 mutation).
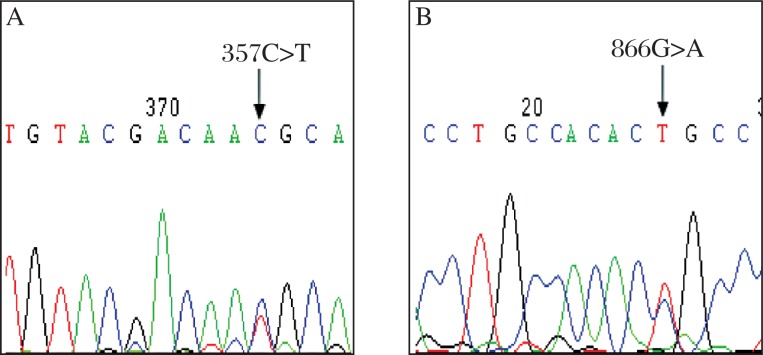
In this observation, none of the deafness-causing GJB3 mutation was detected in both the case and control groups by sequence analysis. Alternatively, we identified two polymorphic variants of the GJB3 gene, 866G>A (2.22%) and 357C>T (0.74%), as shown in Fig. 4 and Table 2, which were, however, absent in the 162 matched normal-hearing controls.
Fig. 4. Partial sequence chromatograms of GJB3 from the patients.
Arrows indicate the location of base changes. A: C to T transition at position 357. B: G to A transition at position 866.
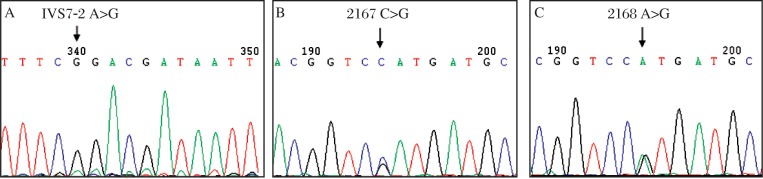
There were 3 different kinds of SLC26A4 mutations (Fig. 5) with variable frequencies of 13.33% in IVS7-2A>G, 1.48% in IVS7-2A>G+2168A>G, and 0.74% in 2167C>G, which was reported for the first time. The total carrier frequency of deafness causing SLC26A4 mutations was 15.55% (21/135) in the case group and no SLC26A4 mutation was found in the control group. The SLC26A5 IVS2-2A>G variant, however, was not found in a total of 297 individuals with either impaired or normal hearing by sequence analysis.
Fig. 5. Partial sequence chromatograms of SLC26A4 from the patients.
Arrows indicate the location of base changes. A: A to G transition at position IVS7-2. B: C to G transversion at position 2167. C: A to G transition at position 2168.
Data congregate
Based on the widely and systematically collected information about NSHL from several typical areas of China, we summarize the existing reports on molecular epidemiology of deafness. The prevalence of the GJB2, GJB3, GJB6, SLC26A4 and mtDNA genes in Chinese populations is listed in Table 4. As for genetic factors of nonsyndromic deafness in China, the average value of variant in GJB2 seems to be the most common (23.37%), followed by SLC26A4 (14.74%), mtDNA 12SrRNA A1555G ( 2.44%), GJB3 (1.97%) and GJB6 (1.33%) genes.
Table 4. Prevalence of GJB2, GJB3, GJB6, and SLC26A4 genes and mtDNA 12SrRNA A1555G mutation in the Chinese population.
| Different areas of China | Patients (n) | Frequency of pathologic variants(%) |
References | ||||
| 12SrRNA A1555G | GJB2 | GJB3 | GJB6 | SLC26A4 | |||
| Northern China | 743 | 2.92 | 18.12 | - | - | 18.15 | Yu Fei et al.2006[25] |
| Liu Xin et al.2006[26] | |||||||
| Song RD et al.2007[27] | |||||||
| Zhu QW et al.2007[28] | |||||||
| Zhu YH et al.2008[29] | |||||||
| Northeastern China | 377 | 1.00 | 20.80 | - | - | - | Yu Fei et al.2006[25] |
| Liu Xin et al.2006[26] | |||||||
| Chen JX et al.2007[30] | |||||||
| Wang Ping et al.2001[31] | |||||||
| Eastern China | 135 | 1.50 | 35.55 | 0 | 3.70 | 15.56 | This study |
| Southern China | 404 | 2.70 | 10.80 | - | - | - | Yu Fei et al.2006[25] |
| Liu Xin et al.2006[26] | |||||||
| Northwestern China | 1639 | 4.10 | 17.61 | 2.20 | 0 | 12.85 | Dai Pu et al.2006[32] |
| Guan Jing et al.2006[33] | |||||||
| Yu Fei et al.2006[25] | |||||||
| Liu Xin et al.2006[26] | |||||||
| Yuan YY et al.2008[34] | |||||||
| Guo YF et al.2008[35] | |||||||
| Du RL et al.2009[36] | |||||||
| Southeastern China | 140 | - | 40.00 | - | - | - | Wang SH et al.2009[37] |
| Others | 533 | - | 20.70 | 3.70 | 0.30 | 12.38 | Li QZ et al.2005[38] |
| Han DY et al.2006[39] | |||||||
| Li Qi et al.2007[40] | |||||||
| Yuan YY et al.2007[41] | |||||||
| Sun Qing et al.2008[18] | |||||||
| Average value | 2.44 | 23.37 | 1.97 | 1.33 | 14.74 | 43.57% (total) | |
After the systematic comparison of various genes in different countries or ethnic differences between the mutation frequencies, we calculated the average frequency of GJB2, GJB3, GJB6, SLC26A4 and mtDNA mutations in patients with nonsyndromic hearing loss. As shown in Table 5, for hearing-impaired people in the world, the GJB2 mutation is also the most common causative factor. The variant accounts for about 29.03%, followed by the SLC26A4 (15.16%) and GJB6 (5.58%) mutations. The GJB3 mutation and mtDNA 12SrRNA A1555G, however, seem not to play an important part in the causative effects of deafness in Western countries as compared with the Chinese people.
Table 5. Prevalence of GJB2, GJB3, GJB6, and SLC26A4 genes and mtDNA 12SrRNA A1555G mutation in different countries.
| Different areas | Samples(n) | Frequency of pathogenic mutations |
References | ||||
| 12SrRNA A1555G | GJB2 | GJB3 | GJB6 | SLC26A4 | |||
| Argentina | 252 | 0.00 | 41.27 | - | 1.59 | - | Viviana Dalamón et al. 2010[42] |
| Austria | 122 | 0.00 | 65.57 | 0.00 | 16.39 | - | RENÉ UTRERA et al. 2007[43]; Reinhard Ramsebner et al. 2007[20] |
| Brazil | 300 | - | 13.67 | 0.00 | 1.00 | - | Ana Carla Batissoco et al. 2009[44] |
| Croatia | 121 | - | 38.84 | - | 0.00 | - | Ivona Sansovic et al. 2009[14]; Igor Medica et al. 2005[45] |
| Poland | 1313 | - | 17.75 | - | - | - | Agnieszka Pollak et al. 2007[46] |
| Germany | 164 | 0.60 | 1.22 | - | - | - | Li R et al. 2004[47] |
| Greek | 30 | - | 33.33 | - | 0.00 | - | Vassos Neocleous et al. 2006[24] |
| India | 484 | - | 6.82 | - | - | 5.40 | Park HJ et al. 2003[7]; Padma G et al. 2009[48] |
| Korea | 176 | - | 37.50 | - | 0.00 | 40.00 | KY Lee et al. 2008[13]; Park HJ et al. 2003[7] |
| Morocco | 116 | - | 24.14 | 0.00 | - | - | RENÉ UTRERA et al. 2007[43] |
| Hungary | 410 | - | 54.88 | 0.24 | 0.49 | - | TÍMEA TÓTH et al. 2007[19] |
| Spain | 152 | - | 23.40 | - | 27.66 | 27.00 | Del Castillo F J et al. 2010[22]; Pera A et al. 2008[49] |
| Turkey | 418 | - | 24.80 | - | 0.00 | 1.70 | Sirmaci A et al. 2006[23]; Duman D et al. 2010[5] |
| United Kingdom | 160 | - | 22.22 | - | 22.22 | 3.50 | Del Castillo F J et al. 2005[22]; Hutchin T et al. 2008[16] |
| USA | 464 | 0.00 | 28.44 | - | 0.00 | 12.55 | Joy Samanich et al. 2007[50]; Dai Pu et al. 2009[9] |
| Venezuela | 40 | - | 27.50 | - | 2.50 | - | RENÉ UTRERA et al. 2007[43] |
| Average value | 0.15 | 28.83 | 0.06 | 5.99 | 15.03 | 50.06% (total) | |
DISCUSSION
GJB2 (OMIM No.121011) encodes the gap junction protein connexin 26 (CX26), which is expressed in the cochlea and may play a role in K+ circulation between different partitions in the cochlea. Both in China and many other countries, GJB2 mutations are responsible for a large proportion of NSHL. For example, the 35delG, 167delT, 235delC and R143W alleles are the most common GJB2 mutations in Europeans, Americans, Africans and Asians[10]–[17]. In the Asian nonsyndromic hearing-impaired populations, the 235delC of GJB2 are 34.0% and 14.3% in two Japanese reports[3],[10], 8.52% and 6.70% in two studies in Taiwan[11],[12], 5.10% in one Korean study[13], and 27.41%(37/135) in our study. In our observation on patients with NSHL from eastern China, 235delC mutation of GJB2 was higher than that in the Caucasians. None of 35delG was detected in both the case and control groups of our country; however, the 35delG of GJB2 occupied the main cause of deafness in the Caucasian populations. Therefore, in different ethnic groups, there was a significant difference in genetic mutation between Mongolians and Caucasians.
The Chinese population is made up of several major ethnicities, such as Han, Man, Mon, Hui, Zhuang and Miao. All of the subjects included in our study were Han Chinese in origin, which is the predominant ethnicity (85.56%) of Chinese. Previous studies demonstrated that there was no significant difference of GJB2 mutations between Han and other minorities through the comparative analysis of reports from six Chinese typical areas (Table 4).
GJB3 (OMIM No. 603324) encodes the gap junction protein connexin 31(Cx31), which is also thought to be a good candidate for hereditary hearing impairment. Mutations of GJB3 cause three different disorders: nonsyndromic deafness, syndromic deafness, and a genodermatosis. More than 10 mutations in GJB3 have been found in patients with deafness from China[18], Hungary[19], and Brazil[6]. In contrast, several studies demonstrated that the variations in GJB3 with no or low genetic relevance in Moroco and in Austrialia[15],[20]. In our study, two kinds of variants in GJB3 were detected from 135 hearing-impaired subjects: the 866G>A in three and the 357C>T in one patient. Both of the GJB3 variations were heterozygotes, which had previously been reported as polymorphisms. However, we did not find these changes in the 162 matched normal-hearing controls. This led us to the assumption that these variants may be associated with autosomal-dominantly nonsyndromic hearing loss in some patients.
GJB6 (OMIM No. 604418) encoding connexin 30 (Cx30) was an obvious candidate gene for deafness owing to its chromosomal location at 13q12, and because connexin 26 and connexin 30 are expressed in the same inner ear structures and share 77% homology in amino acid sequence[21]. Despite the high prevalence of GJB2 and GJB6 mutations in some Western populations, for example, two large deletions of the GJB6 (one of 309 kb,⊿GJB6-D13S1830 and another of 232 kb,⊿GJB6-D13S1854) upstream the GJB2 are frequently found among individuals who are deaf in Spain[22], these mutations seem to account for a smaller percentage of hereditary hearing loss in Turkey[23], Greek Cyprus[24] and Austria[20], and few data have been reported on the presence of these mutations in the Chinese population[8]. In the present study, we assessed the prevalence of GJB6 mutations in both hearing-impaired individuals and normal-hearing controls from Nanjing city. In five of the 135 patients (3.70%), the 232 kb del (⊿GJB6-D13S1854) was detected, one was heterozygote and others were homozygotes. All of them were also found to have the 235delC mutation in GJB2. None of the patients negative for GJB2 mutations carried this mutation. The result illustrates the complexity of genetic epidemiology of deafness, and a possible interaction between GJB2 235delC and⊿GJB6-D13S1854 in nonsyndromic hearing impairment. Furthermore, 162 controls exhibited only wild-type alleles of GJB6, and none of the cases and controls screened showed ⊿GJB6-D13S1830 mutation.
Mutations in several SLC family 26 genes are responsible for some distinct recessive disorders. SLC26A4 (OMIM No. 600791) at 7q31 encodes a chloride-iodide transport protein expressed in the thyroid, kidney and inner ear. Its different mutations can lead to either syndromic deafness (Pendred syndrome) or non-syndromic recessive deafness. As shown in Table 4 and Table 5, SLC26A4 mutation accounts for almost fifteen percent causative factors of hearing loss. In addition, mutations of SLC26A4 usually cause enlarged vestibular aqueduct or cochlear deformity, which can be diagnosed with high-resolution CT scan of the temporal bone. In our study, deafness-associated mutations in SLC26A4 were screened. There are 3 kinds of mutations in the SLC26A4 gene (IVS7-2A>G, IVS7-2A>G+2168A>G, and 2167C>G) in the case group. A novel mutation, 2167C>G in exon 19, leading to a His–to-Asp substitution, was found.
Cochlear outer hair cells change their length in response to variations in membrane potential. This capability is believed to enable the sensitivity and frequency selectivity of the mammalian cochlea. Prestin is a transmembrane protein required for cochlear electromotility. This makes SLC26A5 (OMIM No. 604943), the restricted expression of prestin in the outer hair cells of the cochlea, a strong candidate for human deafness[8]. Indeed, a single nucleotide change, IVS2-2A>G (NM_198999.1:c.-53-2A>G), in the second intron of SLC26A5 has been reported in association with NSHL[9]. It was, however, observed only in the Caucasian probands with the carrier frequency of about 4.10%. The SLC26A5 IVS2-2A>G sequence variation was not detected in Asians or African Americans according to previous reports. In this study, the IVS2-2A>G variant was either not found in a total of 297 Chinese Han people with either impaired or normal hearing by sequence analysis. The results indicated a special ethnic background between this sequence variation and human hearing loss.
Although the majority of cases with hereditary hearing loss are caused by nuclear gene defects, it has become clear that mtDNA (OMIM No. 561000) mutations can also cause deafness. Among the identified non-syndromic deafness-causing mtDNA mutations are A1555G, C1494T, T1095C, A827G and mutations at position 961 in the 12SrRNA, and A7445G, 7472insC, T7510C, T7511C, T7512C and G7444A in the tRNASer(UCN). Currently, it is estimated that these mutations are present in about 3.10% of patients with NSHL, but it is expected that this number will increase as genetic testing becomes more readily available[4],[9].
Similar to that of the SLC26A5 IVS2-2A>G mutation, the association between mtDNA mutation and hearing loss has also a special ethnic difference. In the Asian nonsyndromic hearing-impaired populations, the incidence of the mtDNA mutation appears to be higher than in Caucasians as indicated by previous reports and as shown in Table 4 and Table 5. In our observation on 135 hearing-impaired subjects, the deafness-causing mtDNA mutations were detected in 8.14% (11/135) of the patients, a relatively high rate of incidence. Interestingly, these mitochondrial mutations were only found in the 12SrRNA, and no deafness-associated mutations in tRNASer(UCN) were identified.
In summary, current data revealed that near half of the patients with NSHL carry deafness-causing mutation in GJB2, GJB3, GJB6, SLC26A4, or mtDNA 12SrRNA genes: 43.57% in China (Table 4) and 50.06% in other countries (Table 5). Although the racial background is different, the results come to converge. As for causative factors, mutation in GJB2 is the most common, followed by the SLC26A4 variation. Other genes, such as GJB3, GJB6 and mtDNA 12SrRNA, may also play an important part in the pathogenesis of hearing loss in different countries or areas. These results indicate the necessity of genetic screening for mutations of these genes in patients with nonsyndromic deafness.
Footnotes
This work was supported by the Research Grant Award from the National Natural Science Foundation of China (No.31171217), the Open Research Grant of Medical Key Department (No.XF200719) from Jiangsu Province (No. KF200910) and Technology Developmental Program from Nanjing Medical University (No.09NJMUM005).
References
- 1.Cohen MM, Gorlin RJ. Epidermiology, etiology and genetic patterns. In: Gorlin RJ, Toriello HV, Cohen MM, editors. Hereditary Hearing Loss and Its Syndromes. New York and Oxford: Oxford University Press; 1995. pp. 9–21. [Google Scholar]
- 2.Bitner-Glindzicz M. Hereditary deafness and phenotyping in humans. Br Med Bull. 2002;63:73–94. doi: 10.1093/bmb/63.1.73. [DOI] [PubMed] [Google Scholar]
- 3.Abe S, Usami S, Shinkawa H, Kelley PM, Kimberling WJ. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet. 2000;37:41–3. doi: 10.1136/jmg.37.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischel-Ghodsian N. Mitochondrial deafness. Ear Hear. 2003;24:303–13. doi: 10.1097/01.AUD.0000079802.82344.B5. [DOI] [PubMed] [Google Scholar]
- 5.Duman D, Sirmaci A, Cengiz FB, Ozdag H, Tekin M. Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet Test Mol Biomarkers. 2011;15(1-2):29–33. doi: 10.1089/gtmb.2010.0120. [DOI] [PubMed] [Google Scholar]
- 6.Alexandrino F, Oliveira CA, Reis FC, Maciel-Guerra AT, Sartorato EL. Screening for mutations in the GJB3 gene in Brazilian patients with nonsyndromic deafness. J Appl Genet. 2004;45:249–54. [PubMed] [Google Scholar]
- 7.Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4(PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40:242–8. doi: 10.1136/jmg.40.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu XZ, Ouyang XM, Xia XJ, Zheng J, Pandya A, Li F, et al. Prestin, a cochlear motor protein, is defective in non-syndromic hearing loss. Hum Mol Genet. 2003;12:1155–62. doi: 10.1093/hmg/ddg127. [DOI] [PubMed] [Google Scholar]
- 9.Dai P, Liu X, Yu F, Zhu Q, Yuan Y, Yang S, et al. Molecular etiology of patients with nonsyndromic hearing loss from deaf-mute schools in 18 provinces of China. Chin J Otol (in Chinese) 2006;4:1–5. [Google Scholar]
- 10.Ohtsuka A, Yuge I, Kimura S, Namba A, Abe S, Van Laer L, et al. GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet. 2003;112:329–33. doi: 10.1007/s00439-002-0889-x. [DOI] [PubMed] [Google Scholar]
- 11.Hwa HL, Ko TM, Hsu CJ, Huang CH, Chiang YL, Oong JL, et al. Mutation spectrum of the connexin 26 (GJB2) gene in Taiwanese patients with prelingual deafness. Genet Med. 2003;5:161–5. doi: 10.1097/01.GIM.0000066796.11916.94. [DOI] [PubMed] [Google Scholar]
- 12.Wang YC, Kung CY, Su MC, Su CC, Hsu HM, Tsai CC, et al. Mutations of Cx26 gene (GJB2) for prelingual deafness in Taiwan. Eur J Hum Genet. 2002;10:495–8. doi: 10.1038/sj.ejhg.5200838. [DOI] [PubMed] [Google Scholar]
- 13.Leea KY, Choib SY, Baeb JW, Kimc S, Chungd KW, Draynae D, et al. Molecular analysis of the GJB2, GJB6 and SLC26A4 genes in Korean Deafness Patients. Int J Pediatr Otorhinolaryngol. 2008;72:1301–9. doi: 10.1016/j.ijporl.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sansović I, Knezević J, Musani V, Seeman P, Barisić I, Pavelić J. GJB2 mutations in patients with nonsyndromic hearing loss from Croatia. Genet Test Mol Biomarkers. 2009;13:693–9. doi: 10.1089/gtmb.2009.0073. [DOI] [PubMed] [Google Scholar]
- 15.Omar A, Redouane B, Halima N, Khadija B, Lahcen W, Hassan R, et al. Carrier frequencies of mutations/polymorphisms in the Connexin 26 Gene (GJB2) in the Moroccan population. Genetic Testing. 2008;4:569–74. doi: 10.1089/gte.2008.0063. [DOI] [PubMed] [Google Scholar]
- 16.Hutchin T, Coy NN, Conlon H, Telford E, Bromelow K, Blaydon D, et al. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK - implications for genetic testing. Clin Genet. 2005;68:506–12. doi: 10.1111/j.1399-0004.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- 17.Brobby GW, Muller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N Engl J Med. 1998;338:548–50. doi: 10.1056/NEJM199802193380813. [DOI] [PubMed] [Google Scholar]
- 18.Sun Q, Yuan HJ, Liu X, Yu, Kang DY, Zhang X, Dai P, et al. Mutation analysis of GJB3 in Chinese population with DFNA. Chin Arch Otolaryngol Head Neck Surg. 2008;11:625–7. [Google Scholar]
- 19.Tóth T, Kupka S, Haack B, Fazakas F, Muszbek L, Blin N, et al. Coincidence of mutations in different connexin genes in Hungarian patients. Int J Mol Med. 2007;20:315–21. [PubMed] [Google Scholar]
- 20.Reinhard R, Trevor L, Christian S, Martin L, Wolf-Dieter B, Franz J. Wachtler, Karin Kirschhofer, and Klemens Frei. Relevance of the A1555G mutation in the 12S rRNA gene for hearing impairment in Austria. Otol Neurotol. 2007;28:884–6. [PubMed] [Google Scholar]
- 21.Kelley PM, Abe S, Askew JW, Smith SD, Usami S, Kimberling WJ. Human connexin 30 (GJB6), a candidate gene for nonsyndromic hearing loss: molecular cloning, tissue-specific expression, and assignment to chromosome 13q12. Genomics. 1999;62:172–6. doi: 10.1006/geno.1999.6002. [DOI] [PubMed] [Google Scholar]
- 22.Del Castillo FJ, Rodríguez-Ballesteros M, Alvarez A, Hutchin T, Leonardi E, de Oliveira CA, et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J Med Genet. 2005;42:588–94. doi: 10.1136/jmg.2004.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirmaci A, Akcayoz-Duman D, Tekin M. The c. IVS1+1G>A mutation in the GJB2 gene is prevalent and large deletions involving the GJB6 gene are not present in the Turkish population. J Genet. 2006;85:213–6. doi: 10.1007/BF02935334. [DOI] [PubMed] [Google Scholar]
- 24.Neocleous V, Aspris A, Shahpenterian V, Nicolaou V, Panagi C, Ioannis Ioannou I, et al. High frequency of 35delG mutation and absence of del(GJB6-D13S1830) in Greek Cypriot patients with nonsyndromic hearing loss. Genet Test. 2006;10:285–9. doi: 10.1089/gte.2006.10.285. [DOI] [PubMed] [Google Scholar]
- 25.Yu F, Dai P, Han DY, Cao JY, Kang DY, Liu X, et al. 233-235delC mutation analysis of GJB2 gene in nonsyndromic hearing impairment patients fromChina. Chin Arch Otolaryngol Head Neck Su (in Chinese) 2006;4:223–6. [Google Scholar]
- 26.Liu X. Molecular epidemiologyand mechanism study of mitochondrial DNA A1555G mutation in hearing imPairment:[Doctoral Dissertation]BeiJing. Chin PLA Med Ins (in Chinese) 2006 [Google Scholar]
- 27.Song RD, Yuan YY, Dai P, Lin YJ, Yu F, Liu X, et al. Screening of GJB2 235delC mutation, SLC26A4 IVS7-2A>G mutation and mtDNA 12SrRNA A1555G mutation in severe-profound hearing loss patients in Zhuozhou and Gaobeidian, Hebei province. Chin J Otol (in Chinese) 2007;1:64–7. [Google Scholar]
- 28.Zhu QW, Dai P, Han DY, Ling HY, Yuan YY, Yu F, et al. Analysis of SLC26A4 mutation in severe in the deafness patients from the city of Anyang. J Audiol Speech Pathol (in Chinese) 2007;3:181–3. [Google Scholar]
- 29.Zhu YH, Neng LL, Li MS, Dong MM. Molecular genetic analysis of SLC26A4 2168A>G mutations in sensorineural hearing loss with unknown reason in Henan province. J Chin Otorhinolarvnol Head Neck Su (in Chinese) 2008;22:1026–31. [PubMed] [Google Scholar]
- 30.Chen JX, Zhang GR. Molecular genetic analysis of the mitochondrial DNA 1555 mutation gene among nonsyndromic hearing impairment patients from Jilin province. Chin J Lab Diagn (in Chinese) 2007;6:782–5. [Google Scholar]
- 31.Wang P, Wang XM, An XF, Wang YS, Du B, Du BD. High frequency mutation of the 233delC in connexin26 gene in Chinese deafness populations. Chin Arch Otolaryngol Head Neck Su (in Chinese) 2001;1:24–6. [Google Scholar]
- 32.Dai P, Zhu XH, Yuan YY, Zhu QW, Teng GC, Zhang X, et al. Patients suffered from enlarged vestibular aqueduct syndrome in Chifeng deaf and dumb school detected by Pendred's syndrome gene hot spot mutation screening. J Chin Otorhinolaryngol Head Neck Su (in Chinese) 2006;7:497–500. [PubMed] [Google Scholar]
- 33.Guan J, Guo YF, Xu BC, Zhao YL, Li QZ, Wang QJ. Molecular epidemiological study of GJB2 mutations in the prelingual deafness in northwestern China. Med J Chin PLA (in Chinese) 2006;4:303–5. [Google Scholar]
- 34.Yuan YY, Huang DL, Dai P, Zhu XH, Yu F, Zhang X, et al. Mutation analysis of GJB2, GJB3 and GJB6 gene in deaf population from special educational school of Chifeng city. J Clin Otorhinolaryngol Head Neck Su (in Chinese) 2008;1:14–21. [PubMed] [Google Scholar]
- 35.Guo YF, Bao XL, Wang QJ, Liu XW, Guan J, Xu BC, et al. An Investigation of GJB2,SLC26A4 Gene mutations in nonsyndromic hearing loss in the northwest of China. J Audiol Speech Patho l (in Chinese) 2008;4:263–6. [Google Scholar]
- 36.Du RL, Li HW, Zhao XX, Li YH. Mutation analysis of GJB3 between Chinese and Uighur population with nonsyndromic hearing impairment. MMJC (in Chinese) 2009;6:1–4. [Google Scholar]
- 37.Wang SH, Hu ZM, Xiao Z, Tang QL, Xia K, Yang XM. GJB2 (connex in26) gene mutation screen in patients with nonsydromic hearing loss in Hunan. J Cent South Univ (in Chinese) 2009;34:498–503. [PubMed] [Google Scholar]
- 38.Li QZ, Wang QJ, Zhao LD, Yuan H, Li L, Liu Q, et al. Mutation analysis of GJB3 in Chinese population with non-syndromic hearing impairment. J Audiol Speech Pathol (in Chinese) 2005;3:145–8. [Google Scholar]
- 39.Han DY, Li QZ, Lan L, Zhao YL, Yuan H, Li L, et al. A novel mutation of GJB6 in Chineses Poradic nonsyndromic hearing impairment. Chin Arch Otolaryngol Head Neck Su (in Chinese) 2006;10:670–2. [Google Scholar]
- 40.Li Q, Dai P, Huang DL, Yuan YY, Zhu QW, Han B, et al. Frequency of SLC26A4 IVS7-2A>G mutation in patients with severe to profound hearing loss from diferent area and ethnic group in China. J Chin Otorhinolarvnol Head Neck Su (in Chinese) 2007;12:893–7. [PubMed] [Google Scholar]
- 41.Yuan YY, Huang DL, Dai P, Zhu QW, Liu X, Wang GJ, et al. GJB6 gene mutation analysis in Chinese nonsyndromic deaf population. J Clin Otorhinolaryngol Head Neck Su (in Chinese) 2007;1:3–6. [PubMed] [Google Scholar]
- 42.Dalamón V, Lotersztein V, Béhèran A, Lipovsek M, Diamante F, Pallares N, et al. GJB2 and GJB6 genes: molecular study and identification of novel GJB2 mutations in the hearing-impaired Argentinean population. Audiol Neurootol. 2010;15:194–202. doi: 10.1159/000254487. [DOI] [PubMed] [Google Scholar]
- 43.René U, Vanessa R, Yuryanni R, Maria J. R, Leomig M, Simón A, et al. Detection of the 35delG/GJB2 and del(GJB6-D13S1830) mutations in Venezuelan patients with autosomal recessive nonsyndromic hearing loss. Genet Test. 2007;4:347–52. doi: 10.1089/gte.2006.0526. [DOI] [PubMed] [Google Scholar]
- 44.Batissoco AC, Abreu-Silva RS, Braga MC, Lezirovitz K, Della-Rosa V, Alfredo T, et al. Prevalence of GJB2 (connexin-26) and GJB6 (connexin-30) mutations in a cohort of 300 Brazilian hearing-impaired individuals: implications for diagnosis and genetic counseling. Ear Hear. 2009;30:1–7. doi: 10.1097/AUD.0b013e31819144ad. [DOI] [PubMed] [Google Scholar]
- 45.Igor M, Gorazd R, Manuela B, Borut P. C. 35delG/GJB2 and del(GJB6-D13S1830) mutations in Croatians with prelingual non-syndromic hearing impairment. BMC Ear, Nose Throat Disord. 2005;5:11. doi: 10.1186/1472-6815-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollak A, Skórka A, Mueller-Malesińska M, Kostrzewa G, Kisiel B, Waligóra J, et al. M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A. 2007;143(A 21):2534–43. doi: 10.1002/ajmg.a.31982. [DOI] [PubMed] [Google Scholar]
- 47.Li R, Greinwald JH, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J Med Genet. 2004;41:615–20. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padma G, Ramchander PV, Nandur UV, Padma T. GJB2 and GJB6 gene mutations found in Indian probands with congenital hearing impairment. J Genet. 2009;88:267–72. doi: 10.1007/s12041-009-0039-5. [DOI] [PubMed] [Google Scholar]
- 49.Pera A, Villamar M, Viñuela A, Gandía M, Medà C, Moreno F, et al. A mutational analysis of the SLC26A4 gene in Spanish hearing-impaired families provides new insights into the genetic causes of Pendred syndrome and DFNB4 hearing loss. Eur J Hum Genet. 2008;16(8):888–96. doi: 10.1038/ejhg.2008.30. [DOI] [PubMed] [Google Scholar]
- 50.Samanich J, Lowes C, Burk R. Mutations in GJB2, GJB6, and mitochondrial DNA are rare in African American and Caribbean Hispanic individuals with hearing impairment. Am J Med Genet A. 2007;143(A 8):830–8. doi: 10.1002/ajmg.a.31668. [DOI] [PubMed] [Google Scholar]