Abstract
All-trans retinoid acid (ATRA) is one of the most potent and most thoroughly studied differentiation inducers that induce the differentiation and apoptosis of glioma cells. However, the effect of ATRA on angiogenesis of glioma remains poorly understood. We examined the effect of ATRA on the expression of vascular endothelial growth factor (VEGF) in different glioma cell lines and investigated the underlying mechanism, intending to partially reveal the effects of ATRA on angiogenesis of glioma. Glioma cells were treated by ATRA at 5 and 10 µmol/L. The VEGF mRNA transcript levels were determined by real-time RT-PCR and the protein levels of VEGF in glioma cells were evaluated by Western blotting assays. Moreover, hypoxia-inducible factor-1α (HIF-1α) mRNA expression was analyzed by using real-time RT-PCR. After treatment with 5 and 10 µmol/L ATRA, the VEGF mRNA transcript levels in glioma cells increased remarkably, compared with that in the control group, and the relative protein expression of VEGF was also up-regulated. Meanwhile, the HIF-1α mRNA expression also increased. ATRA increases the expression of VEGF in glioma cells at both transcriptional and translational levels.
Keywords: All-trans retinoic acid (ATRA), vascular endothelial growth factor (VEGF), glioma, hypoxia-inducible factor-1α (HIF-1α), angiogenesis
INTRODUCTION
Despite the use of the best therapeutic regimens available, the prognosis of malignant glioma, the most common form of primary brain tumors, remains unsatisfactory. All-trans retinoic acid (ATRA) is one of the strongest and most thoroughly studied differentiation inducers. Several studies have shown that ATRA can induce the differentiation and apoptosis of a variety of glioma cells[1], including glioma stem cells (GSCs)[2],[3], a highly tumorigenic and therapy-resistant tumor subpopulation. These results showed that ATRA has the therapeutic potential for patients with glioma. However, as one of the most important influencing factors, angiogenesis plays a crucial role in the formation, development and recurrence of glioma, but the effects of ATRA on angiogenesis of glioma remain unknown. In this study, we examined ATRA effects on angiogenesis by detecting the expression of the vascular endothelial growth factor (VEGF) gene in two different glioma cell lines U87 and SHG44. The results provided a better understanding of the mechanisms involved in the therapeutic effects of ATRA for malignant glioma.
MATERIAIS AND METHODS
Reagents
ATRA (Sigma-Aldrich Corp., St. Louis, MO, USA) was dissolved in DMSO at 0.01mol/L and stored in light protected vials at –20°C as stock solution. This stock solution was diluted in media to obtain the desired concentrations before use. All experiments were performed under low-light conditions to minimize ATRA photoisomerization. Anti-human VEGF antibody was purchased from Epitomics (Burlingame, CA, USA).
Cell Culture
Human glioma cell lines U-87MG and SHG44 were purchased from the Cell Resource Center, Chinese Academy of Sciences (Shanghai, China). U87 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone, Logan, UT, USA) supplemented with 5% fetal bovine serum (FBS, Hyclone) in 5% CO2 at 37°C. SHG44 cells were cultured in RPMI1640 (Hyclone) and supplemented with 5% FBS (Hyclone) in 5% CO2 at 37°C.
Real-time PCR
U87MG and SHG44 cells (3×105 cells/well) seeded in 6-well plates were incubated in media containing different concentrations of ATRA (0, 5 µmol/L and10 µmol/L) for 24 hours. Cells were then lysed and the total RNA was isolated by using RNA fast 200 Kit (Fastage) according to the manufacturer's instructions. RNA was reverse-transcribed by PrimeScript RT Master Mix (TaKaRa, Otsu, Japan). Real-time PCR was performed using SYBR Premix Ex TaqTMII (TaKaRa) and analyzed with Bio-Rad iQ5 software version 2.0. Gene expression was compared using the ∆cycle threshold (∆Ct=CtTarget-Ctβ-actin) method, where β-actin was taken as the endogenous reference gene. Change in gene expression was evaluated by the 2−∆∆Ct method[4].All primers were designed and synthesized by TaKaRa Co. and the sequences are listed in Table 1.
Table 1. The sequences of primers used for real-time PCR.
| Gene | Primer sequence | Product length |
| HIF-1α | Forward 5′-TCTGGGTTGAAACTCAAGCAACTG- 3′ | 150 bp |
| Reverse 5′- CAACCGGTTTAAGGACACATTCTG-3′ | ||
| VEGF | Forward 5′-TCACAGGTACAGGGATGAGGACAC- 3′ | 72 bp |
| Reverse 5′-CAAAGCACAGCAATGTCCTGAAG-3′ | ||
| β-actin | Reverse 5′-CAAAGCACAGCAATGTCCTGAAG-3′ | 186 bp |
| Reverse 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ |
Western blotting assays
Logarithmically growing cells cultured in 25 mL cell culture flasks with ATRA at different concentrations (0, 5 µmol/L and 10 µmol/L) were harvested for assays. The cells were washed twice with phosphate-buffered saline (PBS) and then scraped on ice in 300 µL RIPA lysis buffer with 1 mmol/L PMSF. Lysates were clarified by centrifugation, and samples were boiled in 1×sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, resolved by SDS-PAGE, and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were probed with anti-VEGF antibody (Epitomics Bio, Burlingame, CA, USA) diluted in Tris-buffered saline containing 0.02% Tween 20. After wash in Tris-buffered saline containing 0.02% Tween 20, the membranes were incubated with a secondary polyclonal anti-rabbit IgG antibody conjugated to horseradish peroxidase (Bioworld Technology, OH, USA). Membranes were developed with Super Signal West Pico chemiluminescence reagent (Thermo Scientific, USA).
Statistical analysis
Values were calculated as mean±SD and data were analyzed by the SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). One-way ANOVA was used to compare the groups, and LSD test was conducted for comparison between the groups thereafter. P < 0.05 was considered statistically significant.
RESULTS
ATRA induces VEGF mRNA transcription in glioma cells
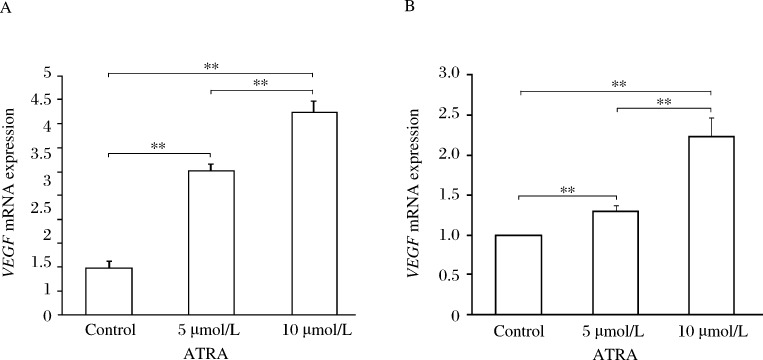
We treated the glioma cells with different concentrations of ATRA for 24 hours and then examined the VEGF mRNA transcript levels by real-time RT-PCR. We found that compared to the control cells, the VEGF mRNA transcript levels were significantly upregulated in the two glioma cell lines treated with ATRA. At 5 µmol/L ATRA, the levels of VEGF mRNA transcripts increased 3.02 and 1.30 fold in U87 and SHG44 glioma cells, respectively. At 10 µmol/L ATRA, the VEGF mRNA transcripts increased 4.25 fold in U87 glioma cells and 2.24 fold in SHG44 glioma cells (Fig.1).
Fig.1. Effects of ATRA on VEGF mRNA expression in different glioma cell lines.
A: VEGF mRNA expression in U87 glioma cells. B: VEGF mRNA expression in SHG44 glioma cells. Significant upregulation of VEGF mRNA expression was found in the two different glioma cell lines treated with ATRA after 24 hours in different concentrations as compared with that in the control group, **P < 0.01.
ATRA induces VEGF protein expression in glioma cells
Consistent with the increase in mRNA transcription levels, the level of VEGF protein expression was also upregulated in glioma cells treated with ATRA. The ratios of VEGF level to that of β-actin were 1.07±0.14 and 0.57±0.06, respectively, in U87 glioma and SHG44 cells treated with 5 µmol/L ATRA. These ratios were even higher in the cells treated with 10 µmol/L ATRA (1.44±0.11 µmol/L in U87 and 0.87±0.01 µmol/L in SHG44) (Fig.2).
Fig.2. VEGF protein expression in different glioma cell lines.
A: VEGF protein expression in U87 glioma cells. B: VEGF protein expression in SHG44 glioma cells. Lane 1: the control group; lane 2: the 5 µmol/L ATRA group; lane 3: the 10 µmol/L ATRA group. VEGF protein expression in glioma cells was also up-regulated by ATRA (*P < 0.05, **P < 0.01).
ATRA induces HIF-1α mRNA transcription in glioma cells
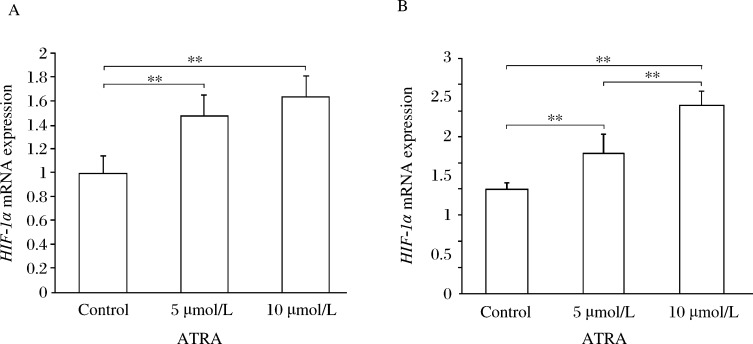
Since VEGF is well known to be regulated by HIF-1α, the effect of ATRA on HIF-1α mRNA transcription in glioma cells was examined by real-time PCR. As shown in Fig 3, ATRA significantly increased the level of HIF-1α mRNA in glioma cells. There was a significant difference in the HIF-1α mRNA transcript levels between the 5 µmol/L ATRA group and the 10 µmol/L ATRA group in SHG44 glioma cells (P < 0.05). But there was no significant difference between the 5 µmol/L ATRA Group and the 10 µmol/L ATRA group in U87 glioma cells (P > 0.05).
Fig. 3. Induction of HIF-1α mRNA expressions in U87MG cells by ATRA.
A: HIF-1α mRNA expression in U87 glioma cells. B: HIF-1α mRNA expression in SHG44 glioma cells. ATRA can significantly increase the expression of HIF-1α mRNA in glioma cells. The difference between the 5 µmol/L ATRA group and the 10 µmol/L ATRA group in SHG44 glioma cells was statistically significant. But there was no significant difference between the 5 µmol/L ATRA group and the 10 µmol/L ATRA group in U87 glioma cells (**P < 0.01).
DISCUSSION
VEGF is a key regulator of angiogenesis and plays an important role in the oncogenesis, progression and recurrence of glioma[5]. Although several studies have reported that the effects of ATRA on the expression of VEGF, the conclusion remains controversial. Some reports have demonstrated that ATRA can inhibit VEGF expression in human gastric cancer cells[6], whereas stimulatory effects of ATRA on the expression of VEGF have been reported in many other studies[7]-[9]. Discrepancies among these studies may be due to the differences in methodology, cell types, experimental conditions and ATRA concentrations used. However, the effects of ATRA on VEGF expression of glioma remain unknown. In this study, we found that ATRA significantly increased VEGF expression in glioma cells at both the transcriptional and translational levels. Moreover, we also found a dose-dependent relationship between the expression level of VEGF and ATRA concentration. Our study thus first demonstrated the stimulatory effects of ATRA on VEGF mRNA and protein expression in glioma cells.
As HIF-1α is well known as a key regulator of VEGF expression[10], we examined the effects of ATRA on HIF-1α expression in glioma cells. We observed that ATRA significantly induced HIF-1α mRNA expression in U87 and SHG44 glioma cell lines. This result was similar to the previous results[11], so we speculate that the upregulation of HIF-1α expression may play a role in the upregulation of VEGF expression in glioma cells. However, the speculation still needs more experimental evidence to support.
The mechanisms of the ATRA-induced HIF-1α expression in glioma cells are still not entirely clear. Recently, researchers have reported that some influencing factors can regulate HIF-1α expression via the MAPK or PI3K signal pathway[12],[13]. Meanwhile, it has also been reported that ATRA can regulate the expression of downstream genes via the MAPK-related rapid phosphorylation pathway[14]. It is therefore speculated that HIF-1α induced by ATRA in U87 cells is partly related to the MAPK pathway. In addition to upregulating the expression of HIF-1α, ATRA may also act through some other pathways to regulate VEGF expression in glioma cells. Some researchers have reported that ATRA can directly induce VEGF expression via the retinoc acid receptor (RAR) signaling pathway[7]-[9], which may be involved in the regulation of VEGF expression of ATRA in treated glioma cells.
As VEGF plays an important role in angiogenesis, the upregulation of VEGF expression in glioma cells induced by ATRA may stimulate angiogenesis of glioma and further promote tumor growth, which requires further investigations. As a therapeutic strategy, ATRA still has some disadvantages in this respect. In conclusion, ATRA can upregulate the expression of VEGF in U87 glioma cells. ATRA may therefore have some disadvantages in the treatment of glioma and the anti-angiogenesis combined therapy may have some more beneficial effects in glioma treatment.
References
- 1.Zang CB, Wachter M, Liu HY, Posch MG, Fenner MH, Stadelmann C, et al. Ligands for PPAR gamma and RAR cause induction of growth inhibition and apoptosis in human glioblastomas. J Neurooncol. 2003;65:107–1. doi: 10.1023/b:neon.0000003728.80052.a8. [DOI] [PubMed] [Google Scholar]
- 2.Karsy M, Albert L, Tobias ME, Murali R, Jhanwar-Uniyal Ml. All-trans Retinoic Acid Modulates Cancer Stem Cells of Glioblastoma Multiforme in an MAPK-dependent Manner. Anticancer Res. 2010;30:4915–20. [PubMed] [Google Scholar]
- 3.Campos B, Wan F, Farhadi M, Ernst A, Zeppernick F, Tagscherer KE, et al. Differentiation Therapy Exerts Antitumor Effects on Stem-like Glioma Cells. Clin Cancer Res. 2010;16:2715–28. doi: 10.1158/1078-0432.CCR-09-1800. [DOI] [PubMed] [Google Scholar]
- 4.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Zhou Y, Zhang XS, Shen BZ. Expression of VEGF and MMP-9 and MRI imaging changes in cerebral glioma. Oncol Lett. 2011;2:1171–5. doi: 10.3892/ol.2011.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li JS, Zhang JP, Chen XY. Effects of all-trans-retinoic on human gastric cancer cells BGC-823. J Dig Dis. 2007;8:29–34. doi: 10.1111/j.1443-9573.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- 7.Sugawara A, Saito A, Uruno A, Kudo M, Kagechika H, Sato Y, et al. All-trans retinoic acid induces in vitro angiogenesis via retinoic acid receptor: Possible involvement of paracrine effects of endogenous vascular endothelial growth factor signaling. Endocrinology. 2007;148:1412–23. doi: 10.1210/en.2006-0900. [DOI] [PubMed] [Google Scholar]
- 8.Chen JT, Liang JB, Chou CL, Shyu RC, Lu DW. Retinoic acid induces VEGF gene expression in human retinal pigment epithelial cells (ARPE-19) J Ocul Pharmaco Ther. 2005;21:413–9. doi: 10.1089/jop.2005.21.413. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi K, Goodison S, Urquidi V, Tarin D, Lotan R, Tahara E. Differential effects of retinoic acid on the growth of isogenic metastatic and non-metastatic breast cancer cell lines and their association with distinct expression of retinoic acid receptor beta isoforms 2 and 4. Int J Oncol. 2003;22:623–9. [PubMed] [Google Scholar]
- 10.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, et al. Role of HIF-1 alpha in hypoxia-mediated apoptosis, cell proliferation and tumor angiogenesis. Nature. 1998;394:485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y, Yang Z, Han YY, You C. Screening of differentially expressed genes related to differentiation and proliferation by gene expression profiling of different grade astrocytoma cell lines. Neural Reg Res. 2008;3:245–9. [Google Scholar]
- 12.Haan C, Vollmer S, Kappler V, Kaczor J, Flugel D, Rolvering C, et al. Hypoxia-Inducible Factor 1 alpha Is Up-Regulated by Oncostatin M and Participates in Oncostatin M Signaling. Hepatology. 2009;50:253–60. doi: 10.1002/hep.22928. [DOI] [PubMed] [Google Scholar]
- 13.Li JX, Davidson G, Huang Y, Jiang BH, Shi XL, Costa M, et al. Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer Res. 2004;64:94–101. doi: 10.1158/0008-5472.can-03-0737. [DOI] [PubMed] [Google Scholar]
- 14.Zheng XY, Liu JG, Zhou R, He QR, Li WI, Zhang T, et al. Calmodulin kinase II activation of mitogen-activated protein kinase in PC12 cell following all-trans retinoic acid treatment. Neurotoxicology. 2009;30:599–604. doi: 10.1016/j.neuro.2009.03.006. [DOI] [PubMed] [Google Scholar]