Abstract
Microfluidics is a platform technology that has been used for genomics, proteomics, chemical synthesis, environment monitoring, cellular studies, and other applications. The fabrication materials of microfluidic devices have traditionally included silicon and glass, but plastics have gained increasing attention in the past few years. We focus this review on thermoplastic microfluidic devices and their applications in protein and DNA analysis. We outline the device design and fabrication methods, followed by discussion on the strategies of surface treatment. We then concentrate on several significant advancements in applying thermoplastic microfluidic devices to protein separation, immunoassays, and DNA analysis. Comparison among numerous efforts, as well as the discussion on the challenges and innovation associated with detection, is presented.
1. Introduction
Microfluidics is a platform technology that has been used for genomics, proteomics, chemical synthesis, environment monitoring, cellular studies, and other biomedical applications.1–3 Benefits of conducting such a study in a microfluidic device may include low sample and reagent consumption, large surface-to-volume ratio, high-throughput, portability, and potential for point-of-care uses, making the device an alternative to a traditional bench-top apparatus.1–3 One of the promises of the field is that multiple functions such as sample preparation may be integrated in the device, achieving the goal of “lab on a chip” (LOC). Although there have been significant advances in the field through a large number of research groups and efforts in the past two decades, the most of earlier promises including proliferation of commercial LOC systems “have not yet happened”.4
One of the reasons of the slow commercialization could be the device material used. Most pioneering works were carried out in silicon and glass devices5–8 while many efforts and exciting development have been achieved in devices made from poly-(dimethylsiloxane) (PDMS).9–12 Although each of these materials has been used in commercial products, they are generally considered as not ideal for the manufacturable process. For instance, PDMS is a great material for prototyping, especially in an academic setting, but it is “not readily or economically formed in high-throughput production.”13
Thermoplastics, including polycarbonate, poly(methyl methacrylate) (PMMA), cyclic olefin copolymer (COC), polystyrene, and poly(ethylene terephthalate), have been emerging as commercially viable fabrication materials, since they are amenable for industrial manufacturing and yet possess excellent optical properties such as low intrinsic fluorescence and broad visible transmittance. The process of manufacturing low-cost, high-volume plastic parts with micro-scale features is well-developed in other fields, exemplified by the ubiquitous compact disc (CD) that consists of microgrooves embossed on the back side of a thermoplastic.14 Additionally, their biocompatibility (evidenced from plastic labwares), wide range of mechanical stiffness, and convenient prototyping methods outside clean-room environment allow rapid implementation of numerous research interests and thereby accelerate the advancement of microfluidics.
As a result, we chose to review the recent development of thermoplastic microfluidic devices, excluding those made from other polymers/plastics. We focused on protein separation for proteomics studies, immunoassays, and DNA analysis. For readers who are new to field, we presented the device design and fabrication methods so that they will understand the pros and cons of different methods and the efforts required before getting hold of a device for the endeavors they might pursue. For other readers, we discussed in length on the strategies of surface treatment and reviewed innovative detection approaches enabled by thermoplastics, both of which are important to their applications and to commercialization of microfluidics.
2. Device fabrication
Numerous merchant devices associated with various disciplines including biology, chemistry, medicine, and therapy, are made from thermoplastics. Commercially available thermoplastics, specifically PMMA,15–26 COC,27–39 and polycarbonate,40–44 have also been utilized as the fabrication materials of a number of microfluidic devices over the past few years due to their optical properties, manufacturability, and other attributes mentioned above. The overall fabrication process and methods14,45–47 are delineated below and illustrated in Fig. 1.
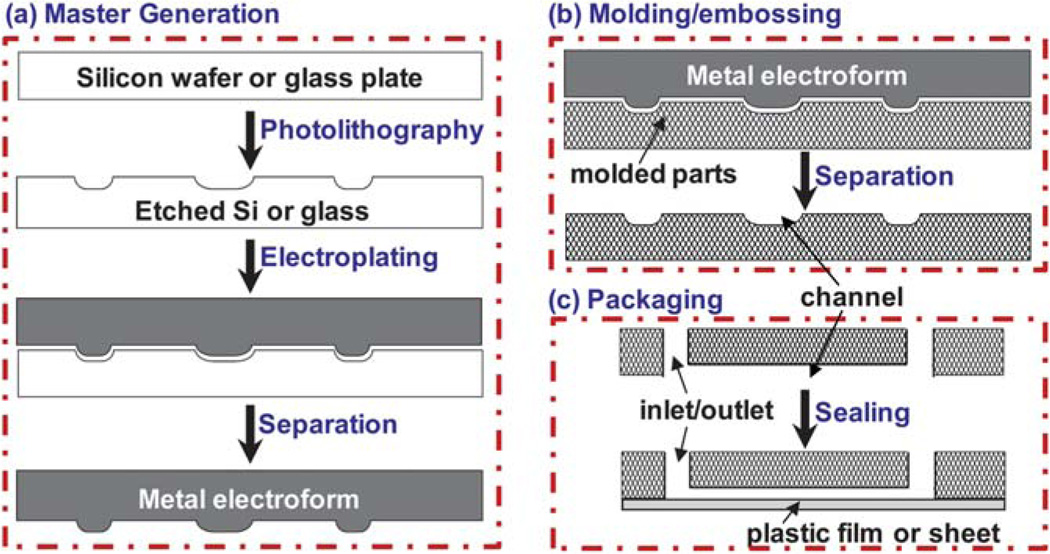
Fig. 1.
(a) The process of generating a master or molding die. A pattern of microfeatures (e.g. channels or chambers) is created in a silicon or glass substrate using photolithography. A metal mold (called electroform) is produced by electroplating on the patterned substrate. Note that the topology of the mold is exactly the opposite of the patterned silicon or glass substrate; a channel in the substrate becomes a ridge in the mold. (b) Plastic parts are fabricated using molding or embossing. The channels are in the direction out of the paper. Note that the topology of the plastic part is a negative image of the mold. Therefore, the plastic part has the exactly same shape as the patterned silicon or glass substrate. Repetitive molding would produce a large number of plastic parts without going through the photolithography steps for each individual device. (c) Packaging steps include drilling the inlet/outlet at the ends of each channel, followed by sealing the part with a plastic film or sheet. Shown in (c) is the molded part in (b) that is flipped vertically and then rotated 90° so that the channel is in the direction along the paper to show the inlet/outlet.
To start the fabrication process, a master or molding die must be generated first (Fig. 1a). One way is to use conventional photolithography and chemical etching to make a pattern in a glass, silicon, or other substrate, followed by electroplating to form a molding master.14,48 This process can be classified into the LiGA (Lithographie-Galvanoformung-Abformung, German for lithography-electroplating-molding) process, though LiGA was originally associated with using X-rays produced by a synchrotron to create high-aspect ratio structures (UV-LiGA now also exists).49 An alternative method of generating a molding master is to directly fabricate microstructures in a metal, including computer-numerically-controlled (CNC) precision milling, laser ablation, or other approaches.50 After obtaining the molding master, a large number of high fidelity replicates can be made from the master in a relatively rapid manner as shown in Fig. 1b. Injection molding, compression molding, embossing are popular among the molding methods. The master can be repeatedly used with faithful replication.
For prototyping and low-volume device fabrication, direct patterning onto a thermoplastic substrate may be used without generating an expensive molding master and undertaking the time-consuming process. The direct fabrication methods include CNC milling, laser ablation and direct lithography,23,51–54 all of which are suitable for producing high-aspect-ratio channels on a wide range of polymer surfaces regardless of their hardness. However, it should be noted that the direct fabrication techniques have the drawbacks of rough channel surfaces, curvature-shaped corners, and possible changes in the surface chemistry of the polymer.39,54 In addition, methods like lamination-based prototyping, wire-imprinting, and in situ photo-initiated polymerization have also been reported in the literature.36,55,56
To complete a device, a substrate with patterned microstructures must be sealed with a film or sheet to form channels or other closed architectures as shown in Fig. 1c. Before bonding, inlets and outlets can be created by drilling holes at the ends of channels. The bonding methods include thermal diffusion bonding,26,30 plasma or ozone-activated bonding,36 lamination,14,48,56 solvent-assisted sealing,43,51,57–60 and use of an adhesive layer.56,61 The bonding process may often be viewed to be trivial, but it is a critical step to have a fully assembled, functional device. Using different bonding methods will affect the device quality and performance as discussed in the literature.62 We compared these methods for the compatibility of a device with chemical and biological reagents to be used, the sustainable pressure for pumping, possible distortion of microstructures due to a high pressure/temperature or an additional adhesive layer, possibility to compensate the surface topology by filling micro-voids or smoothing surfaces, surface property changes due to exposure to solvent or others, and applicability of the bonding method to bonding two completely different types of thermoplastic sheets, as summarized in Table 1.
Table 1.
Comparison of bonding methods for thermoplastic microfluidic devices
| Thermal bonding | Plasma or O3 activated | Lamination | Solvent bonding | Adhesive bonding | |
|---|---|---|---|---|---|
| Chemical compatibility | High | High | High | Low | Low |
| Pressure sustainability | Medium | Low | Medium-low | High | High-medium |
| Feature distortion | Yes | No | Depends | Depends | Yes |
| Topology compensation | Yes | No | No | Yes | Yes |
| Surface property change | No | Yes | No | Yes | Yes |
| Applicable to hybrid devices | No | Yes | No | Yes | Yes |
3. Surface treatment
Thermoplastics are generally hydrophobic. As a result, surface adsorptions exist when a sample contains proteins, peptides, cells, or other reagents with hydrophobic moieties. The interactions between these reagents and channel surfaces not only broaden the peaks when electrophoretic separation takes place, but also change the surface property and thus electroosmotic mobility; all of them degrade the separation resolution, efficiency and reproducibility. In addition, surface absorption may cause appreciable sample loss. Therefore, a variety of surface treatment methods have been explored by a number of groups to diminish the effects of surface absorption.
As in capillary electrophoresis, there are two major categories for surface treatment of thermoplastic channels: dynamic coating and permanent chemical modification. Dynamic coatings include the use of ionic or nonionic small molecules, as well as linear polymers, in the separation buffer. They have been used to prevent protein adsorption and suppress electroosmotic flow in a plastic microfluidic device. We have employed hydroxyethylcellulose to dynamically coat COC channels and found out that the electroosmotic mobility decreased by 72%.63 Isoelectric focusing of proteins was carried out in the device. Shadpour and Soper employed the dynamic coating of methyl hydroxyethylcellulose for protein separation in a PMMA device.26 The electroosmotic mobility was determined to be reduced from (1.56 ± 0.05) × 10−4 cm2 V−1 s−1 to (1.20 ± 0.07) × 10−5 cm2 V−1 s−1. The peak capacity and theoretical plate height for a 10 mm separation distance were 59 and 0.87 µm, respectively. Although the dynamic coating addresses the surface absorption and is convenient to carry out, it requires the sample and buffer solution containing the reagents used for dynamic coating so that coating can be continuously replenished. For those applications coupled with a mass spectrometer in the downstream of the analysis, the use of dynamic coating becomes problematic since it could complicate the peak identification in the mass spectrum.
An alternative surface treatment method is permanent chemical modification by irreversibly attaching a moiety to polymer surfaces. For instance, Soper’s research group reported formation of a layer of linear polyacrylamide on the channel surfaces of a PMMA device via an amide bond and they achieved high-resolution electrophoretic separation of single-stranded DNA with a single base pair resolution.64 Polyacrylamide can also be coated on the surfaces of PMMA, COC and polycarbonate via photografting.32,65 Other materials such as poly(ethylene glycol) (PEG) have also been grafted onto the channel surfaces of a PMMA device for electrophoresis of proteins and peptides.17 Separation efficiencies up to 5.2 × 104 plates were obtained for a 3.5 cm long separation channel. The channel surfaces were often activated using ultraviolet/ozone or oxygen plasma before grafting.17,32,65 COC surfaces may also be coated with silanes and silica.66,67
Permanent surface treatment also possesses shortcomings. One is that the coatings are often not sustainable in a harsh chemical environment and some covalent bonds (e.g. Si–O–C) are prone to hydrolyze in an aqueous environment. The other drawback is that the modification procedures are typically time-consuming and require skilled personnel. The fundamental approach to address the concerns is to create plastic materials that require no surface treatment. For instance, Lee’s research group used monomers containing PEG moiety to form a polymer backbone that has desired properties without the requirement of surface treatment.55,68 Three types of acrylate monomers were mixed at an appropriate ratio to form a copolymer substrate and the device was successfully demonstrated for separation of proteins and peptides. High theoretical plate numbers (4.7 × 104 plates per 3.5 cm) and the symmetrical shape of protein peaks indicate negligible interactions between the channel surface and proteins.55,68 Along the same line, the authors also synthesized a monomer to create a PMMA derivative, namely poly(glycidyl methacrylate-co-methyl methacrylate).69,70 A device made from this polymer was found to be superior to traditional PMMA in terms of surface treatment owing to the presence of epoxy functional groups in the backbone. Electrophoretic separation of multiple proteins and peptides was carried out in the devices with high separation efficiency and reproducibility.
Although the discussion above has been focused on the surface treatment for electrophoretic separation, these methods and/or their variations are generally applicable to other applications (since the goal is the same, for preventing molecules from being absorbed on thermoplastic surfaces). For instance, Zhao et al. oxidized the surfaces of COC using ozone to obtain carboxylic acid groups, followed by covalent binding for DNA microarray hybridization.71 Similarly, Liu et al. modified the channel surface in a PMMA device using poly(ethyleneimine) to obtain amine groups, followed by glutaraldehyde treatment for antibody binding.72
In the light of the work discussed above, in particular the efforts by Lee’s group,55,68–70 the design and synthesis of new or modified polymer substrates specifically for microfluidic applications could be a very fruitful direction. They likely lead to materials with desirable physical and chemical properties for a wide range of applications targeted by the microfluidics community.
4. Protein analysis
Two-dimensional protein separation
Protein separation and identification are important in both medical diagnostics and laboratory research. Among the techniques used for proteomics studies, two-dimensional gel electrophoresis (2DGE) remains “one of the most potent methods”.73 Conventional 2DGE consists of isoelectric focusing (IEF) as the first dimension and polyacrylamide gel electrophoresis (PAGE) as the second dimension. In analogy of DNA sequencing that has shifted from slab gel electrophoresis to capillary array electrophoresis,74,75 2DGE has been implemented in parallel microchannels in a device by a number of research groups.76,77 The potential advantages of microfluidics-enabled 2DGE include faster electrophoresis and higher separation resolution as a result of higher electric fields applied to a microdevice, enhanced reproducibility due to elimination of gel-warping and other features associated with microfluidics.
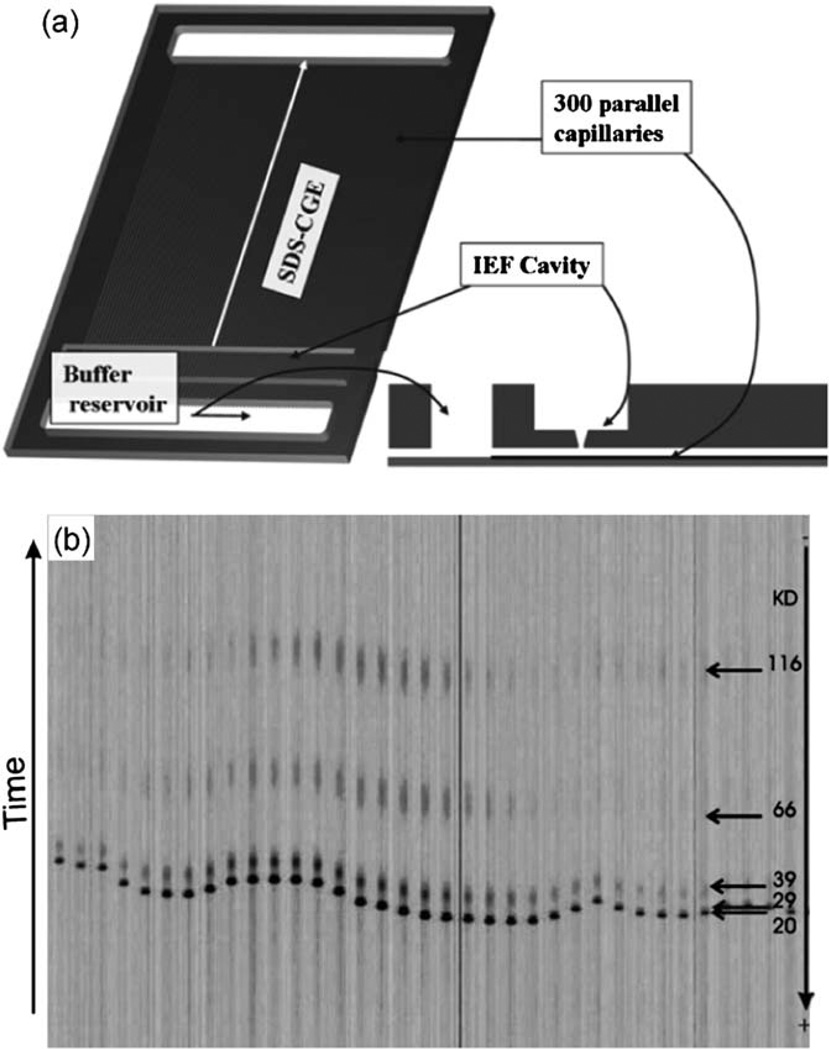
Due to the aforementioned advantages of thermoplastics, efforts have been made in implementing 2DGE in thermoplastic devices.26,53,77–83 Table 2 lists these efforts in the chronological order of the publications. An early example by Griebel et al. was a credit-card-sized PMMA device, which was comprised of an IEF slot in the top layer and 300 parallel PAGE channels in the bottom layer (Fig. 2a).79 IEF was carried out using an immobilized pH gradient strip as in conventional 2DGE. The transfer of proteins from IEF to PAGE was carried out by a sequence of voltage controls on three electrodes (in the IEF slot and two buffer reservoirs); only a fraction of proteins were transferred due to the difference between macroscale IEF and microscale PAGE. The protein separation in parallel PAGE channels (Fig. 2b) is similar to the earlier work of DNA sequencing in 96 capillaries.84 Although authors characterized the protein separation in each dimension as well as sample transfers between two dimensions, a 2D map of multiple proteins was not reported presumably due to the difficulty in the protein transfer between different scales.
Table 2.
List of efforts in two-dimensional protein separation in plastic microfluidic devicesa
| Device materials | Separation mechanisms | Separation time | Peak capacity | References |
|---|---|---|---|---|
| Polycarbonate | IEF-PAGE | 10 min | 1700 | 78 |
| PMMA | IEF-PAGE | 90 min | n/r | 79 |
| PMMA | PAGE-MEKC | 12 min | 1000 | 26 |
| PMMA | IEF-PAGE | n/r | n/r | 80 |
| COC | IEF-PAGE | 10 min | n/r | 81 |
| PMMA | IEF-PAGE | 10 min | n/r | 53 |
| PMMA | PAGE-MEKC | 30 min | 2600 | 82 |
| COC | IEF-RPLC | 25 min | n/r | 83 |
Note: the efforts are listed in the chronological order. RPLC and n/r stand for “reverse phase liquid chromatography” and “not reported” while other abbreviations can be found in the text.
Fig. 2.
(a) Schematic representation of the PMMA device for 2DGE. The credit-card sized device (85 mm × 55 mm × 2 mm) contains a cavity for IEF and two buffer reservoirs that are connected by 300 parallel channels (50 µm × 50 µm × 64 mm) for PAGE. The IEF cavity and the PAGE channels are connected to each other through a 50 µm wide opening. (b) Electropherograms of six proteins in parallel PAGE channels after 1 cm separation. Adapted from ref. 79.
Both IEF and PAGE were carried out in the microscale in several efforts later on. The key challenge to implement 2DGE in a microdevice is the interface design, which must (1) allow the introduction of two separation media into the respective dimension without cross-contamination and (2) fully transfer proteins from the first to the second dimension with negligible disturbance on protein bands. To address the challenge, different methods have been explored, including the use of viscous separation medium,78 pseudo-valve arrays,81 gel plugs in appropriate locations,53,83 and narrower interface channels.85 All of these interface designs demonstrated various degrees of success in the introduction of the separation media and the transfer of protein samples from the first to the second dimension.76
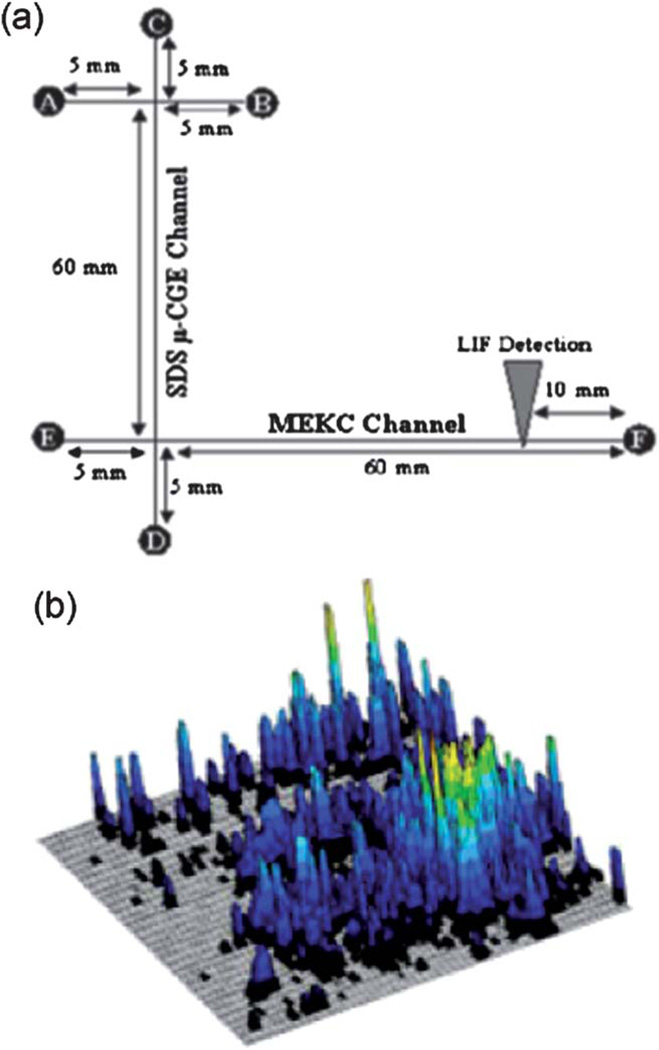
Rather than integrating one channel (first dimension) with an array of channels (second dimension), work has also been carried out by coupling two orthogonal channels, in which analytes eluted from one channel (as the first dimension) are consecutively transferred into the other channel (as the second dimension). The consecutive, sequential transfers of analytes between two dimensions require the waiting time, but the published results exemplified by the work of Soper and his colleagues are very impressive.82 Fig. 3a shows their PMMA device consisting of a channel for PAGE and an orthogonal channel for micellar electrokinetic chromatography (MEKC). The effluents from the first dimension (PAGE) were transferred into the second dimension periodically, at a sampling frequency of 1 Hz, followed by MEKC separation. A 2D map of fetal calf serum proteins was developed within 30 min (Fig. 3b), yielding an average peak capacity of 2600. The peak capacity was three times larger than that obtained from the same sample using conventional 2DGE.
Fig. 3.
(a) Layout of a device to carry out PAGE and MEKC. The channels were 50 mm deep and 20 mm wide. (b) 2D map of proteins from fetal calf serum. The sample was placed into reservoir A and electro-kinetically injected into the separation channel at 200 V cm−1. PAGE and MEKC were performed at 300 and 400 V cm−1, respectively. A 10 s separation time was utilized in the first dimension prior to performing the serial 10 s MEKC cycles. A total of 159 MEKC cycles were carried out. Adapted from ref. 82 with permission from John Wiley & Sons, Inc.
While the performance of some microfluidics-enabled devices is reaching to (or slightly better than) the level of conventional slab gels, it should be noted that one of the key advantages is its rapid analysis. The analysis time in the most work listed in Table 2 is 30 minutes or less. In contrast, conventional 2DGE is often completed in 2 days.
Immunoassays
Immunoassays measure the concentration of analytes in a sample using specific immunological reaction of antibodies to their target antigens. They are extensively used in biomedical research, clinical diagnostics, food/water safety testing, and biological warfare defense. Among several modes of immunoassays, one common format is enzyme-linked immunosorbent assay (ELISA).
Efforts to implement ELISA in microfluidic devices have been made by several research groups.72,86–95 Among them, thermoplastic devices have also been employed.72,92–95 For instance, Liu et al. developed a PMMA device to determine α-fetoprotein, a hepatocellular carcinoma biomarker from human serum.72 The channel surface was modified with amine groups, followed by covalent immobilization of α-fetoprotein antibody. After antigen capture, a horseradish peroxidase-conjugated secondary antibody was used, followed by detection. Woolley’s research group integrated an immunoaffinity column with electrophoretic separation in a PMMA device for detection of the same biomarker.95 The purification step through the immunoaffinity column and the separation step via electrophoresis enable multiple biomarkers to be used for potential prognosis and diagnosis.
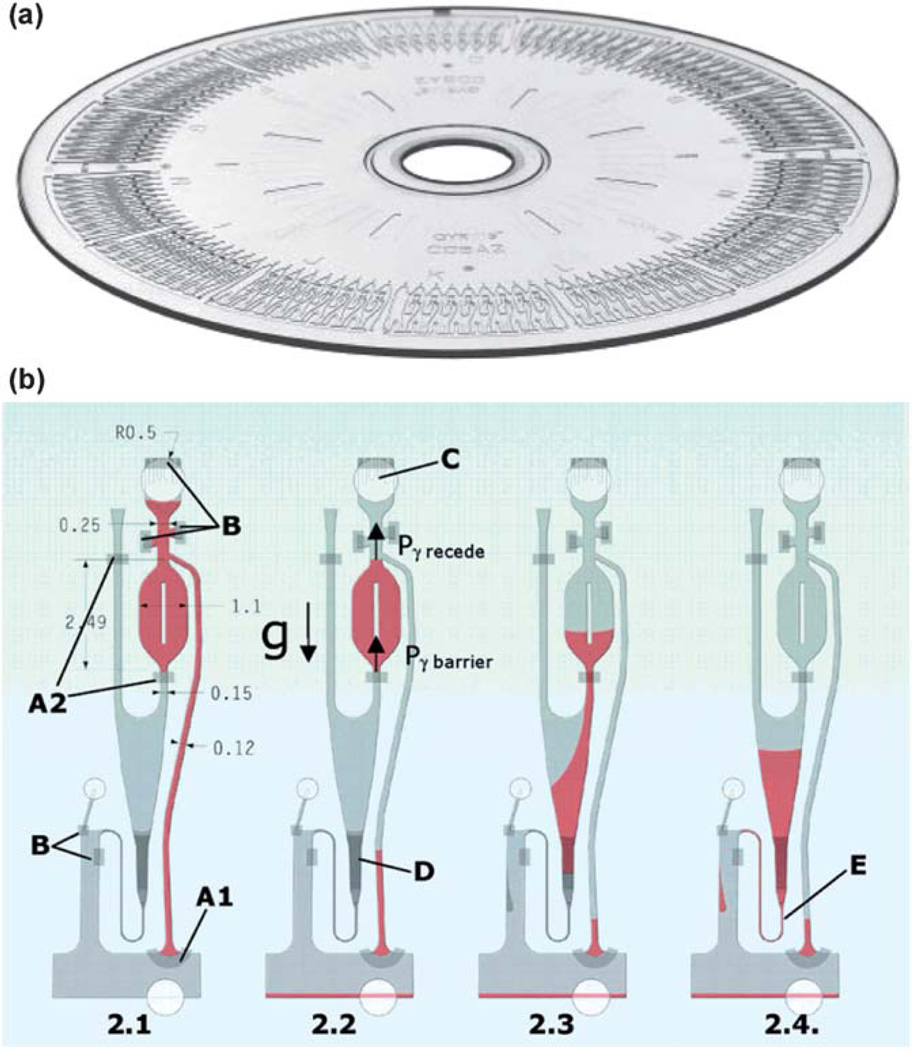
Detection of α-fetoprotein using ELISA has also been implemented in a compact disc (CD)-shaped microdevice. Honda et al. exploited commercially available Gyrolab CD devices as an immunoassay platform for detection of several biomarkers in 200 nL of samples.96 The 12 cm-diameter CD devices were made from polycarbonate and they were manufactured using injection molding.97 As shown in Fig. 4a, there are 112 parallel analysis units in one CD. Each unit consists of various functional elements based on capillary action, centrifugal force, and hydrophobic barriers as illustrated in Fig. 4b. These elements enabled reagent/sample introduction, precise metering, and flow controls. The precision of 112 parallel volume definition operations was determined to be 0.75% CV (coefficient of variation) at 200 nL.
Fig. 4.
(a) Photograph of a Gyrolab CD device with 112 analysis units. (b) Exploded view of an analysis unit with hydrophobic barriers (A1 and A2), hydrophobic patches (B), capillary posts (C), and packed column (D). Part 2.1 shows liquid drawn in by capillary action. In part 2.2, the overflow channel was activated at 1000 rpm, removing excess liquid. Part 2.3 shows the volume (200 nL) defined within the chamber when a short pulse of a higher speed forced the liquid through the barrier. In part 2.4, a defined volume moved through the packed column at a constant flow rate by a spin speed program. Adapted from ref. 96 and 97.
Compact disc is a unique microfluidic platform for running immunoassays. The rotation speed of a CD dictates the magnitude of centrifugal force, thus it can be adjusted to control the passage of flows in microstructures with different channel resistance and/or hydrophobic properties as exemplified in Fig. 4b. Moreover, it is possible to use the data-reading optics and electronics already incorporated in a conventional CD player. Yu’s research group reported using a standard optical drive of an ordinary computer, without any modification of hardware or software, to demonstrate three different types of biochemical recognition reactions in a CD device.98 Lee et al. described a PMMA-based CD device to perform ELISA for hepatitis detection in whole blood samples.93 In addition to those mentioned above, a number of research groups as well as commercial entities have explored this platform for a variety of applications.99–102
5. DNA analysis
Polymerase chain reaction (PCR) is an important tool for genomics studies, forensic analysis, and medical diagnostics. Significant efforts have been made in carrying out PCR in a microfluidic device as reviewed by Landers and colleagues.103 Some of these works were implemented in thermoplastic devices. For instance, Sundberg et al. fabricated a three-layer thermoplastic CD that consists of 1000 compartments for PCR.104 Sample introduction to an individual compartment was enabled by centrifugal forces. A 300 bp plasmid DNA was amplified in each compartment of 33 nL and analyzed in 50 min for whole disc. Klapperich and coworkers described an integrated COC device and the associated instrument for the detection of bacteria (Bacillus subtilis was used).52 The system conducted bacterial lysis, nucleic acid isolation, PCR, and fluorescent detection.
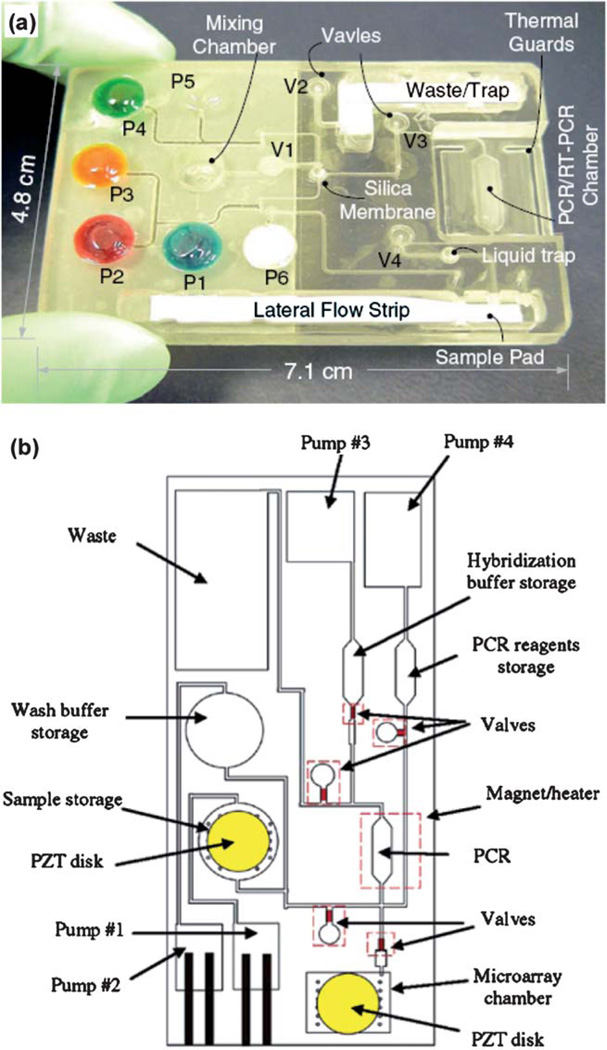
Bau’s research group reported a polycarbonate device for real-time PCR.44 The device allowed pre-storage of paraffin-encapsulated reagents in the PCR chamber and they were reconstituted when thermocycling was initiated after aqueous solutions were introduced. Detection was achieved by using waveguides to couple the device with a fluorescence reader. The authors also integrated PCR with other components in a thermoplastic cassette.105 As shown in Fig. 5a, the cassette contained flexible pouches for reagent storage and valves for flow controls. The process operations included sample introduction, cell lysis, solid-phase extraction of nucleic acids, elution of the nucleic acids into a PCR chamber, and thermal cycling. Afterward, amplicons (PCR products) were transported onto an integrated lateral flow strip, where detection took place. The functionality of the device was demonstrated by detecting the presence of HIV virus in saliva samples.105
Fig. 5.
(a) Photograph of a polycarbonate cassette, in which pouches were filled with dyes for better visualization and they were designed to store various reagents and buffers. Courtesy of Dr. Haim Bau (ref. 105). (b) Schematic of a polycarbonate device containing pumps, valves, PCR chambers and microarrays for detection. Adapted from ref. 106.
A similar effort with a different design and detection scheme was reported by Liu et al.106 As shown in Fig. 5b, a thermoplastic device consists of microfluidic mixers, valves, pumps, channels, chambers, heaters, and DNA microarrays. Cavitation micro-streaming was implemented to enhance mixing while thermally actuated microvalves and electrochemical pumps were integrated to regulate flows. Pathogenic bacteria detection from whole blood samples was demonstrated.106
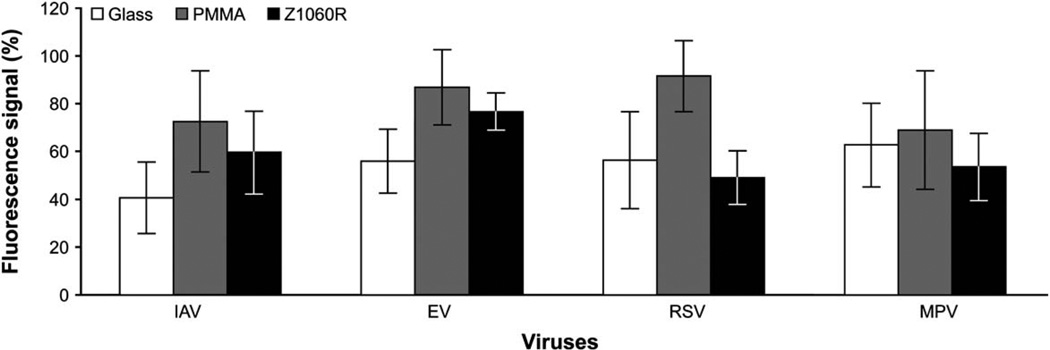
As illustrated in Fig. 5b, DNA microarray is one of the means to analyze amplicons. However, conventional DNA microarrays are fabricated on glass slides and they are “not easily adaptable to integration into microfluidic systems”.71 Therefore Zhao et al. studied polymeric materials and compared PMMA and COC with glass slides for DNA microarray hybridization.71 They extracted RNA from four human respiratory viruses (IAV: influenza A virus, EV: enterovirus, RSV: respiratory syncytial virus, and MPV: metapneumovirus), followed by reverse transcriptase PCR. These samples went through DNA hybridization on both glass and plastic slides printed with DNA probes. Two thermoplastics, PMMA and COC (Zeonor 1060R) were modified to have carboxylic acid groups on the surface before use. As shown in Fig. 6, both materials had comparable performance with a glass slide. Yu’s research group also investigated DNA hybridization on polycarbonate substrates, which were activated using UV ozone treatment.107 Comparable selectivity and efficiency of hybridization were observed relative to other materials.
Fig. 6.
Comparison among glass, PMMA, and COC (Zeonor 1060R) for flow-through microarray hybridizations. Each fluorescence signal represents an average from 36 spots on 3 slides and it is expressed as a percentage of the signal obtained from the corresponding control. The fluorescence signals are from Cy3-labeled amplicons hybridized to DNA microarray. Adapted from ref. 71 with permission from American Society for Microbiology.
It should be noted that all of these efforts have aims to be used in a clinic environment. As a result, potential cross-contamination of sequential samples is of concern if a device is reused. A disposable, low-cost device would be ideal for such applications. High-volume manufacturing capability in plastic parts with micro-scale features (e.g. CD) is one of the driving forces to explore thermoplastic microfluidics for DNA-based diagnostics.
6. Detection
Detection is often a challenge when a tiny amount of a sample is analyzed in a microfluidic device. Moreover, the device often possesses a significantly shortened path length when optical detection is used. According to Beer–Lambert law, A = abc, where A is the absorbance, a is the absorptivity, b is the optical path length, and c is the analyte concentration. The optical path length from mm-scale in a conventional cuvette to µm-scale in a microchannel dramatically reduces the detection sensitive, which is the reason that laser-induced fluorescence detection is often the default detection method for microfluidics. As a result, a variety of innovative detection methods have been explored as reviewed by Myers and Lee.108
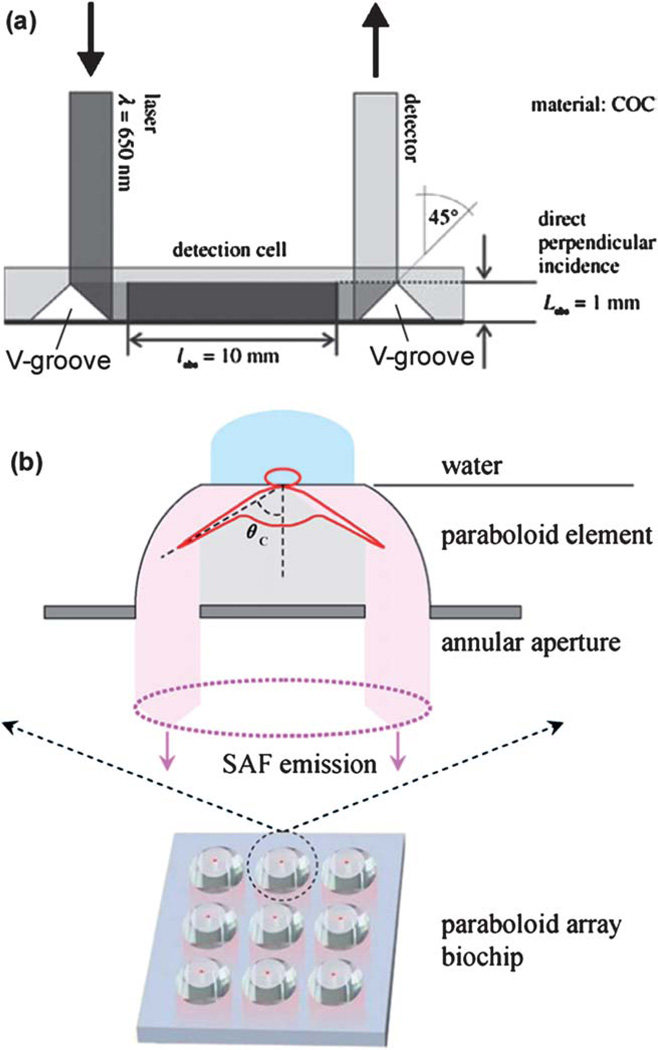
Thermoplastics could offer additional advantage in addressing this challenge. Their facile fabrication procedures enable the device to possess optical-quality surfaces, integrate optic elements, and have specialized channel/well shapes. For instance, Ducree and colleagues fabricated a pair of triangle ridges (i.e., V-grooves from the bottom surface of the substrate in Fig. 7a) in a CD device.109 When a laser beam was directly at an incident angle exceeding the critical angle of the construction material (COC), total internal reflection occurred. As illustrated in Fig. 7a, light was guided to take place along the planar length rather than the vertical height of a channel. The increased optical path length in this integrated detection cell resulted in an improvement factor of 10 when they used the device for detection of glucose in blood. Similarly, Soper’s research group reported embedding a COC waveguide into a multi-channel PMMA device with an orthogonal configuration.110 Light propagated through the COC waveguide and simultaneously illuminated 11 PMMA channels.
Fig. 7.
(a) Detection cell integrated in a CD device with an increased optical path length. The incident optical beam was guided by the V-groove on the left, through the detection cell, and reflected by the V-groove on the right before being detected. V-grooves were created from the bottom surface of the device. Courtesy of Dr. Jens Ducrée (ref. 109). (b) Layout of a 3 × 3 COC array with paraboloid wells. Shown above the array is an exploded view of a well with supercritical angle fluorescence (SAF) emission. Courtesy of Dr. Colette McDonagh (ref. 111).
MacCraith’s research group recently described a COC device consisting of an array of wells.111 As shown in Fig. 7b, the wells were made in a shape of paraboloid, which substantially enhanced the fluorescence collection efficiency due to the phenomenon of supercritical angle fluorescence (SAF, which refers to a fact that substantial amount of fluorescence is emitted into angles above a critical angle). The SAF phenomenon also confined the fluorescence detection volume strictly to the close proximity of the paraboloid surface, thereby discriminating against fluorescence background from the analyte solution. The increased signal-to-noise ratio resulted in 20-fold improvement when they incorporated it with surface plasmon-coupled emission detection.
7. Conclusion
Microfluidics has been significantly advanced since the pioneering work in earlier 1990s.5–8 The recent shift of the bulk of the research activities from silicon/glass to manufacturable thermoplastics could help the commercialization process of microfluidics. As discussed by Becker, a transition “from prototypes to volume manufacturing” must be a part of the development phase.112 Many of the work discussed in this review will assist this transition and help bridge the gap between the research activities and commercialization efforts. The advantageous attributes of thermoplastics, strategies in surface treatment, demonstration of thermoplastic microfluidic devices for a range of applications, and innovative techniques to improve the detection sensitive are a part of the overall endeavors that will help eventually reach the holy grail of LOC.
Acknowledgements
We thank the financial support from National Institute of Health (R21RR024397), Flight Attendant Medical Research Institute (FAMRI-082502), Defense Advanced Research Projects Agency (DARPA) via Micro/Nano Fluidics Fundamentals Focus Center at the University of California at Irvine, and National Science Foundation (OISE-0968313). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Biographies

Dr Ke Liu is a postdoctoral fellow in Microfluidics and BioMEMS Laboratory, Department of Mechanical and Aerospace Engineering, University of Florida, Gainesville, USA. He received his BSc in applied chemistry from Sichuan University, China, in 2002, his MS in chemistry from Louisiana Tech University, Ruston, in 2004, and his PhD in analytical chemistry from Texas Tech University, Lubbock, in 2009. His research interests focus on surface chemistry, sensing techniques, and development of microfluidic platforms for proteomics and cellular studies.

Dr Z. Hugh Fan is an associate professor of Departments of Mechanical and Aerospace Engineering and Biomedical Engineering at University of Florida (UF). Prior to joining UF in 2003, he was Principal Scientist at ACLARA BioSciences Inc. and Member of Technical Staff at Sarnoff Corp. He received his BSc from Yangzhou University in China and PhD from University of Alberta in Canada. His research interests include microfluidics, BioMEMS, sensors, and bioengineering. Dr Fan is the recipient of Fraunhofer–Bessel Award from Alexander von Humboldt Foundation in 2010 and of E.T.S. Walton Award from Science Foundation Ireland in 2009.
References
- 1.Arora A, Simone G, Salieb-Beugelaar GB, Kim JT, Manz A. Anal. Chem. 2010;82:4830–4847. doi: 10.1021/ac100969k. [DOI] [PubMed] [Google Scholar]
- 2.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 3.Melin J, Quake SR. Annu. Rev. Biophys. Biomol. Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 4.Whitesides G. Lab Chip. 2010;10:2317–2318. doi: 10.1039/c0lc90036b. [DOI] [PubMed] [Google Scholar]
- 5.Harrison DJ, Manz A, Fan ZH, Ludi H, Widmer HM. Anal. Chem. 1992;64:1926–1932. [Google Scholar]
- 6.Harrison DJ, Fluri K, Seiler K, Fan ZH, Effenhauser CS, Manz A. Science. 1993;261:895–897. doi: 10.1126/science.261.5123.895. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson SC, Hergenroder R, Koutny LB, Warmack RJ, Ramsey JM. Anal. Chem. 1994;66:1107–1113. [Google Scholar]
- 8.Manz A, Fettinger JC, Verpoorte E, Ludi H, Widmer HM, Harrison DJ. TrAC, Trends Anal. Chem. 1991;10:144–149. [Google Scholar]
- 9.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 10.McDonald JC, Whitesides GM. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 11.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 12.Maerkl SJ, Quake SR. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 13.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 14.Boone TD, Fan ZH, Hooper HH, Ricco AJ, Tan H, Williams SJ. Anal. Chem. 2002;74:78A–86A. doi: 10.1021/ac021943c. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Duan H, Zhang L, Chen G. Electrophoresis. 2008;29:4922–4927. doi: 10.1002/elps.200800093. [DOI] [PubMed] [Google Scholar]
- 16.Kriikku P, Grass B, Hokkanen A, Stuns I, Siren H. Electrophoresis. 2004;25:1687–1694. doi: 10.1002/elps.200305872. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Pan T, Woolley AT, Lee ML. Anal. Chem. 2004;76:6948–6955. doi: 10.1021/ac040094l. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Lee ML. Electrophoresis. 2006;27:3533–3546. doi: 10.1002/elps.200600082. [DOI] [PubMed] [Google Scholar]
- 19.Sun S, Ossandon M, Kostov Y, Rasooly A. Lab Chip. 2009;9:3275–3281. doi: 10.1039/b912097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi H, Meng S, Li Y, Guo K, Chen Y, Kong J, Yang P, Zhong W, Liu B. Lab Chip. 2006;6:769–775. doi: 10.1039/b600326e. [DOI] [PubMed] [Google Scholar]
- 21.Vogt O, Pfister M, Marggraf U, Neyer A, Hergenroder R, Jacob P. Lab Chip. 2005;5:205–211. doi: 10.1039/b411739p. [DOI] [PubMed] [Google Scholar]
- 22.Bi H, Weng X, Qu H, Kong J, Yang P, Liu B. J. Proteome Res. 2005;4:2154–2160. doi: 10.1021/pr050240j. [DOI] [PubMed] [Google Scholar]
- 23.Tsougeni K, Papageorgiou D, Tserepi A, Gogolides E. Lab Chip. 2010;10:462–469. doi: 10.1039/b916566e. [DOI] [PubMed] [Google Scholar]
- 24.Nagata H, Tabuchi M, Hirano K, Baba Y. Electrophoresis. 2005;26:2247–2253. doi: 10.1002/elps.200410395. [DOI] [PubMed] [Google Scholar]
- 25.Mei Q, Xia Z, Xu F, Soper SA, Fan ZH. Anal. Chem. 2008;80:6045–6050. doi: 10.1021/ac800843v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shadpour H, Soper SA. Anal. Chem. 2006;78:3519–3527. doi: 10.1021/ac0600398. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharyya A, Klapperich CM. Anal. Chem. 2006;78:788–792. doi: 10.1021/ac051449j. [DOI] [PubMed] [Google Scholar]
- 28.Castano-Alvarez M, Fernandez-Abedul MT, Costa-Garcia A. J. Chromatogr., A. 2006;1109:291–299. doi: 10.1016/j.chroma.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 29.Gustafsson O, Mogensen KB, Kutter JP. Electrophoresis. 2008;29:3145–3152. doi: 10.1002/elps.200800131. [DOI] [PubMed] [Google Scholar]
- 30.Kim DS, Lee SH, Ahn CH, Lee JY, Kwon TH. Lab Chip. 2006;6:794–802. doi: 10.1039/b516495h. [DOI] [PubMed] [Google Scholar]
- 31.Laib S, MacCraith BD. Anal. Chem. 2007;79:6264–6270. doi: 10.1021/ac062420y. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Yang Y, Craighead HG, Lee KH. Electrophoresis. 2005;26:1800–1806. doi: 10.1002/elps.200410309. [DOI] [PubMed] [Google Scholar]
- 33.Mela P, van den Berg A, Fintschenko Y, Cummings EB, Simmons BA, Kirby BJ. Electrophoresis. 2005;26:1792–1799. doi: 10.1002/elps.200410153. [DOI] [PubMed] [Google Scholar]
- 34.Park J, Lee D, Kim W, Horiike S, Nishimoto T, Lee SH, Ahn CH. Anal. Chem. 2007;79:3214–3219. doi: 10.1021/ac061714g. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Li C, Kameoka J, Lee KH, Craighead HG. Lab Chip. 2005;5:869–876. doi: 10.1039/b503025k. [DOI] [PubMed] [Google Scholar]
- 36.Tsao CW, Hromada L, Liu J, Kumar P, DeVoe DL. Lab Chip. 2007;7:499–505. doi: 10.1039/b618901f. [DOI] [PubMed] [Google Scholar]
- 37.Illa X, De Malsche W, Bomer J, Gardeniers H, Eijkel J, Morante JR, Romano-Rodriguez A, Desmet G. Lab Chip. 2009;9:1511–1516. doi: 10.1039/b818918h. [DOI] [PubMed] [Google Scholar]
- 38.Illa X, Ordeig O, Snakenborg D, Romano-Rodriguez A, Compton RG, Kutter JP. Lab Chip. 2010;10:1254–1261. doi: 10.1039/b926737a. [DOI] [PubMed] [Google Scholar]
- 39.Nunes PS, Ohlsson PD, Ordeig O, Kutter JP. Microfluid. Nanofluid. 2010;9:145–161. [Google Scholar]
- 40.Chen J, Wabuyele M, Chen H, Patterson D, Hupert M, Shadpour H, Nikitopoulos D, Soper SA. Anal. Chem. 2005;77:658–666. doi: 10.1021/ac048758e. [DOI] [PubMed] [Google Scholar]
- 41.Johnson TJ, Ross D, Locascio LE. Anal. Chem. 2002;74:45–51. doi: 10.1021/ac010895d. [DOI] [PubMed] [Google Scholar]
- 42.Liu YJ, Ganser D, Schneider A, Liu R, Grodzinski P, Kroutchinina N. Anal. Chem. 2001;73:4196–4201. doi: 10.1021/ac010343v. [DOI] [PubMed] [Google Scholar]
- 43.Ogonczyk D, Wegrzyn J, Jankowski P, Dabrowski B, Garstecki P. Lab Chip. 2010;10:1324–1327. doi: 10.1039/b924439e. [DOI] [PubMed] [Google Scholar]
- 44.Qiu X, Mauk MG, Chen D, Liu C, Bau HH. Lab Chip. 2010;10:3170–3177. doi: 10.1039/c0lc00038h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker H, Gartner C. Electrophoresis. 2000;21:12–26. doi: 10.1002/(SICI)1522-2683(20000101)21:1<12::AID-ELPS12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 46.Becker H, Gartner C. Anal. Bioanal. Chem. 2008;390:89–111. doi: 10.1007/s00216-007-1692-2. [DOI] [PubMed] [Google Scholar]
- 47.Soper SA, Ford SM, Qi S, McCarley RL, Kelly K, Murphy MC. Anal. Chem. 2000;72:642A–651A. doi: 10.1021/ac0029511. [DOI] [PubMed] [Google Scholar]
- 48.Fredrickson CK, Xia Z, Das C, Ferguson R, Tavares FT, Fan ZH. J. Microelectromech. Syst. 2006;15:1060–1068. [Google Scholar]
- 49.Madou MJ. Fundamentals of Microfabrication: The Science of Miniaturization. Boca Raton: CRC Press; 2002. [Google Scholar]
- 50.Hupert ML, Guy WJ, Llopis SD, Shadpour H, Rani S, Nikitopoulos DE, Soper SA. Microfluid. Nanofluid. 2007;3:1–11. [Google Scholar]
- 51.Klank H, Kutter JP, Geschke O. Lab Chip. 2002;2:242–246. doi: 10.1039/b206409j. [DOI] [PubMed] [Google Scholar]
- 52.Sauer-Budge AF, Mirer P, Chatterjee A, Klapperich CM, Chargin D, Sharon A. Lab Chip. 2009;9:2803–2810. doi: 10.1039/b904854e. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, Yang S, Lee CS, DeVoe DL. Electrophoresis. 2008;29:2241–2250. doi: 10.1002/elps.200700608. [DOI] [PubMed] [Google Scholar]
- 54.Yang J, Liu Y, Rauch CB, Stevens RL, Liu RH, Lenigk R, Grodzinski P. Lab Chip. 2002;2:179–187. doi: 10.1039/b208405h. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Sun X, Lee ML. Anal Chem. 2007;79:1926–1931. doi: 10.1021/ac0617621. [DOI] [PubMed] [Google Scholar]
- 56.Paul D, Pallandre A, Miserere S, Weber J, Viovy JL. Electrophoresis. 2007;28:1115–1122. doi: 10.1002/elps.200600503. [DOI] [PubMed] [Google Scholar]
- 57.Shah JJ, Geist J, Locascio LE, Gaitan M, Rao MV, Vreeland WN. Anal Chem. 2006;78:3348–3353. doi: 10.1021/ac051883l. [DOI] [PubMed] [Google Scholar]
- 58.Koesdjojo MT, Koch CR, Remcho VT. Anal. Chem. 2009;81:1652–1659. doi: 10.1021/ac802450u. [DOI] [PubMed] [Google Scholar]
- 59.Brown L, Koerner T, Horton JH, Oleschuk RD. Lab Chip. 2006;6:66–73. doi: 10.1039/b512179e. [DOI] [PubMed] [Google Scholar]
- 60.Mair DA, Rolandi M, Snauko M, Noroski R, Svec F, Frechet JM. Anal. Chem. 2007;79:5097–5102. doi: 10.1021/ac070220w. [DOI] [PubMed] [Google Scholar]
- 61.Chen HY, McClelland AA, Chen Z, Lahann J. Anal. Chem. 2008;80:4119–4124. doi: 10.1021/ac800341m. [DOI] [PubMed] [Google Scholar]
- 62.Tsao CW, DeVoe DL. Microfluid. Nanofluid. 2009;6:1–16. [Google Scholar]
- 63.Zhang J, Das C, Fan ZH. Microfluid. Nanofluid. 2008;5:327–335. [Google Scholar]
- 64.Llopis SL, Osiri J, Soper SA. Electrophoresis. 2007;28:984–993. doi: 10.1002/elps.200600435. [DOI] [PubMed] [Google Scholar]
- 65.Zangmeister RA, Tarlov MJ. Langmuir. 2003;19:6901–6904. [Google Scholar]
- 66.Dudek MM, Gandhiraman RP, Volcke C, Cafolla AA, Daniels S, Killard AJ. Langmuir. 2009;25:11155–11161. doi: 10.1021/la901455g. [DOI] [PubMed] [Google Scholar]
- 67.Raj J, Herzog G, Manning M, Volcke C, MacCraith BD, Ballantyne S, Thompson M, Arrigan DW. Biosens. Bioelectron. 2009;24:2654–2658. doi: 10.1016/j.bios.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 68.Sun X, Li D, Lee ML. Anal. Chem. 2009;81:6278–6284. doi: 10.1021/ac9001832. [DOI] [PubMed] [Google Scholar]
- 69.Sun X, Liu J, Lee ML. Anal. Chem. 2008;80:856–863. doi: 10.1021/ac701948n. [DOI] [PubMed] [Google Scholar]
- 70.Liu J, Sun X, Lee ML. Anal. Chem. 2005;77:6280–6287. doi: 10.1021/ac0580060. [DOI] [PubMed] [Google Scholar]
- 71.Zhao Z, Peytavi R, Diaz-Quijada GA, Picard FJ, Huletsky A, Leblanc E, Frenette J, Boivin G, Veres T, Dumoulin MM, Bergeron MG. J. Clin. Microbiol. 2008;46:3752–3758. doi: 10.1128/JCM.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y, Wang H, Huang J, Yang J, Liu B, Yang P. Anal. Chim. Acta. 2009;650:77–82. doi: 10.1016/j.aca.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 73.Wittmann-Liebold B, Graack HR, Pohl T. Proteomics. 2006;6:4688–4703. doi: 10.1002/pmic.200500874. [DOI] [PubMed] [Google Scholar]
- 74.Meldrum DR. Science. 2001;292:515–517. doi: 10.1126/science.292.5516.515. [DOI] [PubMed] [Google Scholar]
- 75.Paegel BM, Emrich CA, Weyemayer GJ, Scherer JR, Mathies RA. Proc. Natl. Acad. Sci. U. S. A. 2002;99:574–579. doi: 10.1073/pnas.012608699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen H, Fan ZH. Electrophoresis. 2009;30:758–765. doi: 10.1002/elps.200800566. [DOI] [PubMed] [Google Scholar]
- 77.Tia S, Herr AE. Lab Chip. 2009;9:2524–2536. doi: 10.1039/b900683b. [DOI] [PubMed] [Google Scholar]
- 78.Li Y, Buch JS, Rosenberger F, DeVoe DL, Lee CS. Anal. Chem. 2004;76:742–748. doi: 10.1021/ac034765b. [DOI] [PubMed] [Google Scholar]
- 79.Griebel A, Rund S, Schonfeld F, Dorner W, Konrad R, Hardt S. Lab Chip. 2004;4:18–23. doi: 10.1039/b311032j. [DOI] [PubMed] [Google Scholar]
- 80.Usui K, Hiratsuka A, Shiseki K, Maruo Y, Matsushima T, Takahashi K, Unuma Y, Sakairi K, Namatame I, Ogawa Y, Yokoyama K. Electrophoresis. 2006;27:3635–3642. doi: 10.1002/elps.200600221. [DOI] [PubMed] [Google Scholar]
- 81.Das C, Zhang J, Denslow ND, Fan ZH. Lab Chip. 2007;7:1806–1812. doi: 10.1039/b712794d. [DOI] [PubMed] [Google Scholar]
- 82.Osiri JK, Shadpour H, Park S, Snowden BC, Chen ZY, Soper SA. Electrophoresis. 2008;29:4984–4992. doi: 10.1002/elps.200800496. [DOI] [PubMed] [Google Scholar]
- 83.Liu J, Chen CF, Yang S, Chang CC, Devoe DL. Lab Chip. 2010;10:2122–2129. doi: 10.1039/c003505j. [DOI] [PubMed] [Google Scholar]
- 84.Pang H, Pavski V, Yeung ES. J. Biochem. Biophys. Methods. 1999;41:121–132. doi: 10.1016/s0165-022x(99)00042-1. [DOI] [PubMed] [Google Scholar]
- 85.Emrich CA, Medintz IL, Chu WK, Mathies RA. Anal. Chem. 2007;79:7360–7366. doi: 10.1021/ac0711485. [DOI] [PubMed] [Google Scholar]
- 86.Moorthy J, Mensing GA, Kim D, Mohanty S, Eddington DT, Tepp WH, Johnson EA, Beebe DJ. Electrophoresis. 2004;25:1705–1713. doi: 10.1002/elps.200405888. [DOI] [PubMed] [Google Scholar]
- 87.Ohashi T, Mawatari K, Sato K, Tokeshi M, Kitamori T. Lab Chip. 2009;9:991–995. doi: 10.1039/b815475a. [DOI] [PubMed] [Google Scholar]
- 88.Yu L, Li CM, Liu Y, Gao J, Wang W, Gan Y. Lab Chip. 2009;9:1243–1247. doi: 10.1039/b816018j. [DOI] [PubMed] [Google Scholar]
- 89.Kong J, Jiang L, Su X, Qin J, Du Y, Lin B. Lab Chip. 2009;9:1541–1547. doi: 10.1039/b818430e. [DOI] [PubMed] [Google Scholar]
- 90.Herrmann M, Veres T, Tabrizian M. Lab Chip. 2006;6:555–560. doi: 10.1039/b516031f. [DOI] [PubMed] [Google Scholar]
- 91.Wang J, Ibanez A, Chatrathi MP, Escarpa A. Anal. Chem. 2001;73:5323–5327. doi: 10.1021/ac010808h. [DOI] [PubMed] [Google Scholar]
- 92.Sun S, Yang M, Kostov Y, Rasooly A. Lab Chip. 2010;10:2093–2100. doi: 10.1039/c003994b. [DOI] [PubMed] [Google Scholar]
- 93.Lee BS, Lee JN, Park JM, Lee JG, Kim S, Cho YK, Ko C. Lab Chip. 2009;9:1548–1555. doi: 10.1039/b820321k. [DOI] [PubMed] [Google Scholar]
- 94.Morier P, Vollet C, Michel PE, Reymond F, Rossier JS. Electrophoresis. 2004;25:3761–3768. doi: 10.1002/elps.200406093. [DOI] [PubMed] [Google Scholar]
- 95.Yang W, Sun X, Wang HY, Woolley AT. Anal. Chem. 2009;81:8230–8235. doi: 10.1021/ac901566s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Honda N, Lindberg U, Andersson P, Hoffmann S, Takei H. Clin. Chem. 2005;51:1955–1961. doi: 10.1373/clinchem.2005.053348. [DOI] [PubMed] [Google Scholar]
- 97.Andersson P, Jesson G, Kylberg G, Ekstrand G, Thorsen G. Anal. Chem. 2007;79:4022–4030. doi: 10.1021/ac061692y. [DOI] [PubMed] [Google Scholar]
- 98.Li Y, Ou LM, Yu HZ. Anal. Chem. 2008;80:8216–8223. doi: 10.1021/ac8012434. [DOI] [PubMed] [Google Scholar]
- 99.Madou M, Zoval J, Jia GY, Kido H, Kim J, Kim N. Annu. Rev. Biomed. Eng. 2006;8:601–628. doi: 10.1146/annurev.bioeng.8.061505.095758. [DOI] [PubMed] [Google Scholar]
- 100.Garcia-Cordero JL, Barrett LM, O’Kennedy R, Ricco AJ. Biomed. Microdevices. 2010;12:1051–1059. doi: 10.1007/s10544-010-9459-5. [DOI] [PubMed] [Google Scholar]
- 101.Steigert J, Grumann M, Brenner T, Riegger L, Harter J, Zengerle R, Ducree J. Lab Chip. 2006;6:1040–1044. doi: 10.1039/b607051p. [DOI] [PubMed] [Google Scholar]
- 102.Lange SA, Roth G, Wittemann S, Lacoste T, Vetter A, Grassle J, Kopta S, Kolleck M, Breitinger B, Wick M, Horber JK, Dubel S, Bernard A. Angew. Chem., Int. Ed. 2005;45:270–273. doi: 10.1002/anie.200501243. [DOI] [PubMed] [Google Scholar]
- 103.Roper MG, Easley CJ, Landers JP. Anal. Chem. 2005;77:3887–3893. doi: 10.1021/ac050756m. [DOI] [PubMed] [Google Scholar]
- 104.Sundberg SO, Wittwer CT, Gao C, Gale BK. Anal. Chem. 2010;82:1546–1550. doi: 10.1021/ac902398c. [DOI] [PubMed] [Google Scholar]
- 105.Chen D, Mauk M, Qiu X, Liu C, Kim J, Ramprasad S, Ongagna S, Abrams WR, Malamud D, Corstjens PL, Bau HH. Biomed. Microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu RH, Yang J, Lenigk R, Bonanno J, Grodzinski P. Anal. Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 107.Li Y, Wang Z, Ou LM, Yu HZ. Anal. Chem. 2007;79:426–433. doi: 10.1021/ac061134j. [DOI] [PubMed] [Google Scholar]
- 108.Myers FB, Lee LP. Lab Chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 109.Grumann M, Steigert J, Riegger L, Moser I, Enderle B, Riebeseel K, Urban G, Zengerle R, Ducree J. Biomed. Microdevices. 2006;8:209–214. doi: 10.1007/s10544-006-8172-x. [DOI] [PubMed] [Google Scholar]
- 110.Okagbare PI, Emory JM, Datta P, Goettert J, Soper SA. Lab Chip. 2010;10:66–73. doi: 10.1039/b908759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yuk JS, Trnavsky M, McDonagh C, MacCraith BD. Biosens. Bioelectron. 2010;25:1344–1349. doi: 10.1016/j.bios.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 112.Becker H. Lab Chip. 2010;10:271–273. doi: 10.1039/b925993g. [DOI] [PubMed] [Google Scholar]