Abstract
Carbohydrate−carbohydrate interactions (CCIs) are of central importance for several biological processes. However, the ultra-weak nature of CCIs generates difficulties in studying this interaction, thus only little is known about CCIs. Here we present a highly sensitive equilibrium-fluctuation-analysis of single liposome binding events to supported lipid bilayers (SLBs) based on total internal reflection fluorescence (TIRF) microscopy that allows us to determine apparent kinetic rate constants of CCIs. The liposomes and SLBs both contained natural Lex glycosphingolipids (Galβ4(Fucα3)GlcNAcβ3Galβ4Glcβ1Cer), which were employed to mimic cell−cell contacts. The kinetic parameters of the self-interaction between Lex-containing liposomes and SLBs were measured and found to be modulated by bivalent cations. Even more interestingly, upon addition of cholesterol, the strength of the CCIs increases, suggesting that this interaction is strongly influenced by a cholesterol-dependent presentation and/or spatial organization of glycosphingolipids in cell membranes.
Dynamic short lived recognition events between cells are of essential importance for several biological and pathological processes like in embryogenesis, maturation of lymphoid cells and for the metastatic spreading of cancer cells. The dynamic and transitional character of these processes implies specific adhesive forces or interactions that are repeatedly built up and destroyed. Carbohydrate-protein interactions, which are commonly weaker than most protein-protein interactions, constitute one important class of such interactions. Carbohydrate-carbohydrate interactions (CCIs) offer even lower affinity while still providing very high specificity. Consequently CCIs have been proposed as a versatile mechanism responsible for structurally and temporally dynamic processes, such as cell adhesion and recognition1,2,3,4.
The ultra-weak nature of CCIs5 is a great advantage for short lived processes as well as for modulating interactions between relatively large entities, such as cells. However, this feature simultaneously generates a great challenge as most techniques come to their limit when the weakest biological interaction known is being studied. Thus, compared to protein-protein or carbohydrate-protein interactions, there is still very little known about specific CCIs. It is just over twenty years ago since the first report about CCIs was published, in which Eggens et al. described the homotypic, glycan specific, Ca2+-mediated, self-interaction between Lex determinants (Galβ4(Fucα3)GlcNAcβ-R)) and their role in embryogenesis6, as it had been presumed earlier by Fendersson and coworkers7. Since then, several groups have identified other types of CCIs, including the interaction between GM3 (NeuAcα3Galβ4Glcβ1Cer) and lactosylceramide (Galβ4Glcβ1Cer) initiating metastasis8,9. Despite the medical relevance of the latter, the Lex-Lex interaction has been the most studied. Curiously, in the first report by Eggens et al. F9 embryonal carcinoma cell interactions were observed in buffers containing physiological concentrations of bivalent cations (0.9 mM CaCl2 + 0.5 mM MgCl2)6 whereas in the following studies, by the same group, Lex-Lex interaction was only detected in buffer containing a significantly higher cation concentration (10 mM CaCl2)10. In the studies that followed, 10 mM CaCl2 was always used (and needed) to detect the weak Lex-Lex interaction, indicating that some kind of cation bridging is required to generate sufficiently strong interactions to become measurable5,10,11,12,13,14,15,16,17. To our knowledge there is only one exception; using AFM, Penadés' group observed that the attraction force of Lex-Lex interactions (obtained by probing the interaction between Lex trisaccharides immobilized on Au via a thiol linkage) in water or in aqueous calcium solution was the same, i.e. the interaction force was observed not to depend on CaCl218,19. Studying the literature about Lex-Lex interaction we were puzzled by why it did not seem possible to reproduce, using simplified model systems, the very first experiments by Eggens et al., which showed Lex-Lex interaction at physiological concentrations of bivalent cations. In particular, it seemed strange that unphysiological high concentrations of bivalent cations were needed to observe a physiological relevant interaction. It is in this context interesting to note that the interaction seems dependent on the orientation of the Lex determinant at the membrane surface. For example, albeit dependent on Ca2+, Gourier et al. used micropipette aspiration technique to identify that Lex-Lex interactions are stronger for Lex determinants linked to lactosylceramide than linked to three alkyl chains via a long flexible spacer14,15. This result indicates that Lex-Lex interactions are favored not only by the mere presence of cations but also by ligand geometries akin to that of the natural cell membrane.
To gain biophysically relevant insights insights on the nature of Lex-Lex interactions, it thus seemed to us of great importance to choose a native-like model system, i.e. one that mimics, as much as possible, the conformation and presentation of Lex in a cell membrane. Therefore, natural glycosphingolipids (GSLs) (Fig. 1b), purified from canine or human intestines20,21 were inserted into supported lipid bilayers (SLBs) and liposomes. Both types of model membranes allow for controlled modifications facilitating studies of membrane interactions as demonstrated for the interaction with peptides and proteins22,23,24,25, virus like particles26,27, liposomes28,29, cells30,31, and also carbohydrates14,15,32.
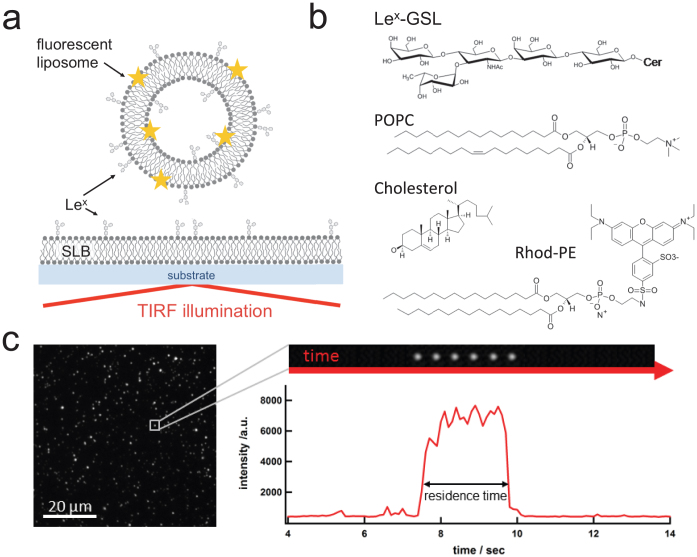
Figure 1. Scheme of the TIRF based approach.
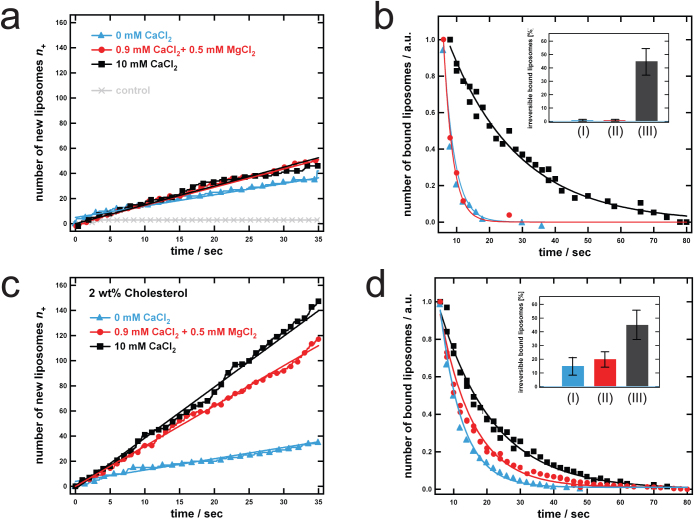
(a) Schematic illustration of the detection principle. Fluorescently labeled liposomes containing Lex-GSLs interact with an SLB containing Lex-GSL. TIRF-based illumination is used to track surface-bound liposomes. (b) Chemical structure of lipids used. (c) A typical TIRF image of surface bound liposomes together with a kymograph and the intensity profile of a small image area containing a single liposome.
Furthermore, a relevant mimicking of the cell membrane implies low concentrations of GSLs. This is likely to result in low surface densities accompanied with low signal-to-noise levels. Together with the general weak nature of CCIs it is thus a great technical challenge to enable the detection of the interaction and furthermore to facilitate the extraction of kinetic data. Traditionally used techniques that allow for the extraction of kinetic data, such as surface plasmon resonance (SPR) or quartz crystal microbalance with dissipation (QCM-D), are not sensitive enough for the detection of ultra-weak interactions under the required conditions. Other techniques like atomic force microscopy (AFM) or micropipette aspiration technique provide the required sensitivity, but can only be applied under transient conditions. We recently demonstrated an approach based on total internal reflection fluorescence (TIRF) microscopy as a versatile tool for the study of single liposome binding events allowing for the detection of association and dissociation events under equilibrium binding conditions27,33,34,35.
In the present study, we demonstrate this TIRF microscopy based equilibrium-fluctuation-analysis as a highly sensitive single liposome technique that facilitates the study of the weak interactions between SLBs and liposomes containing Lex-GSL mimicking Lex-Lex interactions at the cell-cell interface (Fig. 1a). With this technique we are able not only to monitor Lex-Lex interactions in real time with a single liposome resolution, but also to avoid diffusion limitations and determine apparent kinetic rate constants for this low affinity interaction. We measured attractive Lex-Lex interactions even in the absence of Ca2+, but also found that bivalent cations even at physiological concentrations, in particular in combination with cholesterol – a steroid that is known to modulate the orientation of lipids in membranes – in the SLB, strengthened the Lex-Lex interaction.
Results
Scheme of the TIRF experiments
In a typical experiment, GSL containing bilayers are first formed on the glass surface of a microtiter-plate which in the next step is exposed to liposomes containing 5 wt% GSL and 1 wt% fluorescently labeled lipid 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhod-PE, Fig. 1b). We use TIRF microscopy to follow the interaction of fluorescently labeled Lex-containing liposomes with the Lex-containing SLBs. Due to the evanescent field only those liposomes that are in close proximity (< 150 nm) or attached to the SLB are detected. Figure 1c shows a typical image of surface-bound liposomes (left) and a kymograph (right, top) together with the intensity profile (right, bottom) of a small image area describing the residence time of a single liposome bound to the SLB. As previously described, we are able to determine kinetic rate constants by following the attachment and detachment of labeled liposomes, i.e. analyzing equilibrium fluctuations of single liposome binding events27,33,35. In short, counting newly arrived liposomes over time gives the association rate constant, whilst analyzing the residence time of bound vesicles reveals the dissociation rate constant. (For details see Methods and Gunnarsson et al.33,34,35.) A key advantage of this approach is that we are able to study kinetics under steady-state conditions, thus avoiding diffusion limitations. Since the association events may theoretically be limited by liposome diffusion, quantification of association rate constants of the reaction (kon) is only possible if the observed association rate constant is significantly smaller than the expected diffusion rate kdiff ≈ 2.5·106 M−1s−1 of the liposomes in solution (for estimation see Methods). The apparent kon values of the interactions explored in this work were observed to be significantly smaller than kdiff (see Methods), confirming that we are indeed measuring under reaction rather than diffusion-limited conditions.
In the first part of the work we studied the interaction of liposomes and SLBs, both containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, Fig. 1b) and 5 wt% Lex. This concentration was chosen as in many animal plasma cell membranes GSLs constitute ~5% of the lipid molecules36. We compared the interaction of Lex-containing liposomes and SLBs in three different solutions: TRIS buffer without CaCl2 or MgCl2, TRIS buffer containing 0.9 mM CaCl2 and 0.5 mM MgCl2, and TRIS buffer containing 10 mM CaCl2. TRIS buffer containing physiological concentrations of bivalent cations (0.9 mM CaCl2 + 0.5 mM MgCl2) was used in accordance with the previous study by Eggens et al.6.
In the second part of this study we show the effect of cholesterol in the SLB on the interaction between the liposomes and the SLB with various concentrations of divalent cations. Here, 2 wt% cholesterol was added to the SLB and not to the liposomes to avoid side-effects of the cholesterol on the properties of the liposomes as for example size, which may affect the interaction kinetics irrespective of, for example, the influence of cholesterol on GSL presentation27.
Influence of Ca2+ on Lex-Lex trans-interaction
Figure 2a shows the association curves, i.e. the number of newly arrived liposomes versus time, for liposomes interacting with the SLB, both containing 5 wt% Lex, versus a negative control, performed with the SLB containing Lea pentaglycosylceramide instead of Lex pentaglycosylceramide. The Lea determinant (Galβ3(Fucα4)GlcNAcβ-R) is a structural isomer of Lex, the only difference being the switched positions of the terminal fucose and galactose residues. Control experiments were also performed using liposomes and SLBs without GSLs as well as liposomes and SLBs both containing Lea GSLs. No detectable interaction was observed, either in Ca2+-free solution or in solution containing 10 mM CaCl2. Remarkably, specific Lex-Lex interactions were observed for all three solution conditions, i.e. regardless of the presence of Ca2+-ions, while basically no interaction was observed for the negative controls. As described by Equation (1) (Methods) the slope of these curves is proportional to the association rate constant kon. Under the assumption that the coverage of binding sites is identical to the Lex concentration in the SLB, Equation (1) (Methods) can be used to determine the association rate constant to be kon = 0.9 × 103 M−1s−1 for TRIS buffer without cations and kon = 1.2 × 103 M−1s−1 for TRIS buffer containing additional bivalent cations (Table 1). Due to a possible influence of lateral Lex-Lex association on kon, it should be considered as an apparent rate constant which may depend not only on the strength of the Lex-Lex association, but also on GSL clustering (see Methods) as well as GSL concentration in the liposomes27. Nevertheless, the observation that this apparent association rate constant is similar at all solution conditions, was at first glance unexpected, because previously the affinity of the Lex-Lex interaction was observed to be significantly higher in buffer containing 10 mM CaCl2 than in CaCl2-free buffer5,10,11,12,13,14,15,16,17. However, it is important to distinguish between the association rate constant kon and the affinity. To provide an estimation of the affinity, i.e. the association constant Ka = kon/koff, one must also consider the dissociation rate constant, koff. Based on statistics of the residence time, i.e. the time each vesicle remains bound, dissociation plots as shown in Fig. 2b can be generated, from which koff can in turn be obtained. Here, a much slower dissociation was observed for TRIS buffer containing 10 mM CaCl2 than for TRIS buffer without or with low concentrations of bivalent cations. By exponentially fitting of the dissociation curves (see Methods) the corresponding dissociation rate constants, koff, were determined to be 0.04 s−1 for TRIS buffer containing 10 mM CaCl2 and 0.4 s−1 for the two other solutions (Table 1).
Figure 2. Bivalent cations and cholesterol modulate association and dissociation of Lex-Lex interaction.
(a) and (b) Number of newly arrived liposomes n+ as a function of time and (c) and (d) normalized number of liposomes that are still bound as a function of time for the different buffers used: TRIS buffer without additional cations (blue), TRIS buffer containing 0.9 mM CaCl2 and 0.5 mM MgCl2 (red) and TRIS buffer containing 10 mM CaCl2 (black) and the control (grey). (a) and (b) SLBs are composed of 95 wt% POPC + 5 wt% Lex GSL and liposomes of 94 wt% POPC + 5 wt% Lex GSL + 1wt% Rhod-PE. For the control Lex GSL was replaced by Lea GSL. (c) and (d) SLBs are composed of 93 wt% POPC + 5 wt% Lex GSL + 2 wt% cholesterol and liposomes of 94 wt% POPC + 5 wt% Lex GSL + 1 wt% Rhod-PE. Continuous lines represent the corresponding curve fits. Inset (b) and (d): Fraction of irreversible bound liposomes during the time of an experiment for (I) TRIS buffer without additional cations (blue), (II) TRIS buffer containing 0.9 mM CaCl2 and 0.5 mM MgCl2 (red) and (III) TRIS buffer containing 10 mM CaCl2 (black).
Table 1. Apparent kinetic constants of Lex-Lex interaction between SLBs and liposomes*.
| kon/M−1s−1 | koff/s−1 | Ka/M−1 | ||
|---|---|---|---|---|
| without cholesterol | 0 mM CaCl2 | (0.9 ± 0.2) × 103 (R2 = 0.99) | 0.40 ± 0.05 (R2 = 0.98) | 0.2 × 104 |
| 0.9 mM CaCl2 + 0.5 mM MgCl2 | (1.2 ± 0.5) × 103 (R2 = 0.98) | 0.40 ± 0.05 (R2 = 0.99) | 0.3 ×104 | |
| 10 mM CaCl2 | (1.2 ± 0.4) × 103 (R2 = 0.97) | (44 ± 5) · 10−3 (R2 = 0.98) | 2.7 × 104 | |
| 2 wt% cholesterol | 0 mM CaCl2 | (0. 9 ± 0.3) ×103 (R2 = 0.98) | 0.15 ± 0.02 (R2 = 0.95) | 0.6 × 104 |
| 0.9 mM CaCl2 + 0.5 mM MgCl2 | (3.0 ± 0.9) × 103 (R2 = 0.99) | 0.11 ± 0.01 (R2 = 0.97) | 2.7 × 104 | |
| 10 mM CaCl2 | (3.2 ± 1.0) × 103 (R2 = 0.98) | (62 ± 5) · 10−3 (R2 = 0.98) | 5.2 × 104 |
*Kinetic constants are given as the mean and its standard deviation of a triplet of experiments. R2-values for the goodness of fitting are given in parenthesis.
This analysis is based on the fraction of liposomes displaying a reversible binding behavior within the experimental time window. However, due to the distribution in size and GSL content of liposomes, interactions that rely on multiple weak interactions are typically heterogeneous27, i.e. multiple interaction patterns can be observed. Inspection of the relative number of liposomes that were irreversibly bound within the experimental time window, and thus excluded from the analysis above, thus provides an additional indicator of the binding strength (inset Fig. 2b). For TRIS buffer containing 10 mM CaCl2 about 45% of all liposomes that had been attached to the SLB at the beginning of the experiment stayed bound, i.e. did not detach from the SLB within the time of the complete experiment, while for the two other solutions no or very little irreversibly bound liposomes were detected. In the following, we focus on the fraction of vesicles displaying reversible binding behavior, since then the dissociation rate constant and thus the affinity, Ka, can be determined.
Taken together, for the fraction of lipid vesicles displaying reversibility on these time scales, the association constant Ka is one order of magnitude higher at high calcium concentration (Ka = 2.7 × 104 M−1 at 10 mM CaCl2) than at low concentrations of bivalent cations (Ka = 0.2 × 104 M−1) (Table 1).
Influence of cholesterol on Lex-Lex trans-interaction
Previous work suggests that the presentation and conformation of the GSL may play a crucial role for Lex-Lex interaction37. To investigate if this is the case also when the GSL is incorporated in its natural lipid bilayer environment, we studied the interaction between Lex-containing liposomes and SLBs containing cholesterol, which is known to have striking effects on the properties of membranes, since it regulates membrane fluidity38, domain formation39 and presentation of saccharides at the membrane surface37,40. In order to avoid possible effects of liposome size or curvature on the binding kinetics27 cholesterol was added to the SLB rather than to the liposomes.
Figures 2c and d show the association and dissociation curves for the interaction between Lex-containing liposomes and SLBs containing cholesterol. Without bivalent cations the association rate constant is similar to values observed for cholesterol-free SLBs; i.e. kon = 0.9 × 103 M−1s−1 (Fig. 2a and c). However, we observed a strong effect on the association rate constants in TRIS buffer containing 0.9 mM CaCl2 + 0.5 mM MgCl2 and in TRIS buffer containing 10 mM CaCl2, for which the corresponding association rate constants were kon = 3.0 × 103 M−1s−1 and 3.2 × 103 M−1s−1, respectively. These values are almost three times higher than those observed for the interactions without cholesterol (Fig. 2a, kon = 1.2 × 103 M−1s−1). The dissociation curves shown in Fig. 2d are also clearly different from the dissociation curves observed in Fig. 2b. Interestingly, the dissociation rate constants in TRIS buffer without bivalent cations and TRIS buffer containing 0.9 mM CaCl2 + 0.5 mM MgCl2 are found to be koff = 0.15 s−1 and koff = 0.11s−1 respectively, which is significantly smaller than for the interactions observed without cholesterol (Fig. 2b, koff = 0.4 s−1). In TRIS buffer containing 10 mM CaCl2 the dissociation rate constant is found to be koff = 62·10−3 s−1, i.e. similar to the cholesterol-free SLB (Fig. 2b, koff = 44·10−3 s−1).
We also observed the number of irreversibly bound liposomes to follow the same trend as the dissociation rate constants. For TRIS buffer containing 10 mM CaCl2 about 45% of all liposomes were irreversibly bound, i.e. did not detach from the SLB within the time of the experiment, which is comparable to the irreversible fraction observed without cholesterol. At 0.9 mM CaCl2 + 0.5 mM MgCl2 about 20% of the detected liposomes were counted as irreversibly bound, whilst in TRIS buffer 15% of all liposomes were irreversible bound within the experimental time. These numbers are significantly larger than those observed without cholesterol, and are attributed to a fraction of liposomes displaying sufficiently high number of bonds to render the interaction long lived.
Taken together, Lex-Lex interaction is significantly strengthened in the presence of cholesterol. In buffer containing 10 mM CaCl2 the association constant of the reversible fraction is Ka = 5.2 × 104 M−1, which is twice the Ka observed for cholesterol-free SLBs (Ka = 2.7 × 104 M−1). In TRIS buffer without MgCl2 or CaCl2 the association constant is Ka = 0.6 × 104 M−1, which is three times higher compared to values observed for cholesterol-free SLBs (Ka = 0.2 × 104 M−1). Most interestingly, the association constant in TRIS buffer containing physiological concentrations of MgCl2 and CaCl2 is Ka = 2.7 × 104 M−1, which almost corresponds to an order of magnitude higher affinity than the corresponding value observed for cholesterol-free SLBs: Ka = 0.3 × 104 M−1.
Discussion
Previous studies on the interaction between Lex-Lex glycoconjugates have revealed two general trends, being that the interaction is strictly dependent on carbohydrate structures and on the concentrations of Ca2+ ions. Our study confirms these trends but adds additional insights with respect to the dynamics of these interactions, which was previously difficult to quantify due to a lack of sufficient sensitivity, in particular at low Ca2+ concentrations. One important result is that with our approach, Lex-Lex interactions were easily distinguishable from unspecific interactions (Lex-Lea or Lea-Lea interactions) even in the absence of Ca2+ (Fig. 2a and b). The Lex -Lex interaction was observed to be significantly stronger in solution containing high concentrations of CaCl2 than in solution containing physiological concentrations of bivalent cations and in solution without CaCl2, while in the presence of cholesterol in the SLB, the interaction was strengthened also at physiological cation concentrations. The latter observation may help explain an inconsistency that appears from reading the literature on the subject, namely that in contrast to investigations using live cells, Lex-Lex interactions require high Ca2+ concentrations (>10 mM) to be detectable if studied using reductionist model systems. The well-established role of cholesterol as a modulator for glycolipid presentation and spatial organization in cell membranes thus motivates a separate discussion on its expected influence of Lex GSLs.
Using NMR, the binding affinities for the interaction between two Lex monomers tethered together11, as well as Lex methyl glycosides and Lex presenting liposomes12, were found to be much weaker (binding affinities 102 to 104 times lower) than the interactions observed in this work. However, in both studies the Lex saccharides were free monomers in solution, which is in contrast to our experimental system with a confined orientation of the Lex determinants in the cell membrane. Thus, the nature of the binding observed in the NMR studies is of a monovalent nature, hence much weaker than polyvalent binding probably occurring between Lex GSLs of neighboring cell membranes or cell membrane mimics as used in our study (liposomes and SLBs). Yet, Gege et al. made an interesting observation when comparing two different types of tethered Lex monomers (one was tethered through the C6-hydroxy group of GlcNAc, the other through the C1 anomeric oxygen of GlcNAc)11. It was found that the interaction was very much dependent on the conformation of the Lex moiety, i.e. no interaction could be detected in case of tethering through the C1 anomeric oxygen of GlcNAc, while an attractive interaction was clearly detectable in case of tethering through the C6-hydroxy group of GlcNAc (binding affinity Ka of 5–10 M−1). From these studies it appears tempting to suspect that a confined orientation of the Lex determinant is required in order to make an estimate of the strength of the Lex-Lex interaction, possibly relevant also for the interaction between cell membranes.
Taking a step towards membrane mimics, Hernáiz et al. studied the interaction between immobilized layers of Lex determinants where the determinants were in a confined orientation19. In particular, the group employed SPR to determine kinetic rate constants of the interaction between Lex determinants immobilized via a thiol-linkage to gold nanoparticles and planar gold surfaces19. The group observed the association rate constant to be kon = 2.3 × 103 M−1s−1 in Ca2+-containing buffer. This value is in the same range as the association rate constant observed in our study, kon = 1.2 × 103 M−1s−1. However, Hernáiz et al. determined the dissociation rate constant to be koff = 1.5 × 10−3 s−1, which is more than one magnitude lower than the dissociation rate constant observed in our study, koff = 44 × 10−3 s−1. The observed differences may be explained by the different model systems and techniques employed. The TIRF approach used in our work allows us to study the kinetic of single liposomes. Thus, only low concentrations of liposomes (pM) are required, which prevents auto-aggregation of liposomes in the bulk. In contrast, using SPR, a high concentration of glyco-nanoparticles was needed, in fact 106-times the concentration of liposomes used in our study. Consequently, Hernáiz et al. possibly also detected particle aggregates, which will have a profound influence on the residence time. Although our analysis has been focused on the weakly interacting reversible fraction of liposomes, we also observed a fraction of vesicles that were irreversibly bound over our experimental time scales. Under different imaging conditions, these vesicles will also appear reversibly bound, some of which fitting an order of magnitude lower koff value. As this fraction will display a high affinity, these are the ones that will be detected using a system with limited sensitivity, such as SPR.
Furthermore, Hernáiz et al. immobilized the Lex determinant on a gold surface via a thiol linkage whereas in our study Lex-GSLs were incorporated into a fluid lipid membrane. In other words Harnáiz et al. studied the interaction between two rigid layers of Lex whilst we studied the interaction between Lex-GSL embedded in mobile matrices (high fluidity of the SLB was verified using FRAP, see Supplementary Information). One should thus be careful when comparing the affinities of interactions between Lex-determinants in solution or immobilized on nanoparticle surfaces to those occurring on naturel biological membranes or representative mimics.
Although so far not explored for GSL-GSL interactions, it has for long been assumed that the orientation and conformation of the saccharides at the membrane surface affects their function and their interaction with proteins. Strömberg et al. showed that different saccharide orientations of globo-series GSLs (GbO3, GbO4 and GbO5) to the membrane surface play an important role in their receptor function for bacterial adhesion40. In particular, they found differential binding of three G-adhesins variants (PapGJ96, PapGAD110 and PrsGJ96) to result from differences in epitope presentation at the membrane for the isoreceptors GbO3, GbO4 and GbO5. Another study of relevance for this work, performed by Lingwood et al., showed that different orientations of the saccharides of GM1 and GbO3-GSL affected the binding of the proteins choleratoxin and verotoxin, respectively37. In particular, the receptor activity was found to be affected by cholesterol that modulates the orientation of the saccharide moiety. Through molecular dynamic (MD) simulations the group found that in the presence of cholesterol, the glycan moiety adopts a conformation that is tilted towards the membrane plane affecting the receptor activity. In the second part of our study, a striking effect of cholesterol also on a carbohydrate-carbohydrate trans-interaction was revealed, supporting that the saccharide orientation and spatial organization is crucial also in this case. In fact, the conclusion that cholesterol can strengthen Lex-Lex interactions answers the controversy raised by the very first study by Eggens et al. who observed Lex-Lex interaction in buffer containing only 0.9 mM CaCl2 + 0.5 mM MgCl2, but which for future studies by this and other groups were repeated only at concentrations of 10 mM CaCl2. Taking a closer look on previous studies, keeping the impact of cholesterol that evolves from our results in mind, we find that experiments where interactions could be observed in buffer containing 0.9 mM CaCl2 and 0.5 mM MgCl2 were indeed performed with membranes containing cholesterol. In further studies, as the systems were supposed to be simplified, cholesterol was omitted. This finding clearly indicates that, although highly specific, Lex-Lex interaction is not independent from membrane composition, in particular not from cholesterol.
With our findings the question arises about the more detailed nature of the Lex-Lex interaction. Although the determination of association rate constants was made under the assumption that a single Lex moiety is sufficient for liposome binding, it is in fact unlikely that the observed liposome binding is governed by only one single molecular bond. Under the assumption that the distance between the liposome, which is assumed to be spherical, and the underlying SLB required to establish association coincides with the dimension of the Lex moiety, is < 0.5 nm, the contact area ranges between 60 – 300 nm2. This implies a number of 5 – 25 GSLs in the contact area at the given concentration of GSLs in the SLB and in the liposomes; a number that may vary for non-spherical liposomes or in case of GSL-clustering within the contact area. However, in both cases the number of GSLs in the contact area would increase, suggesting that formation of multiple bonds is favorable. Still, it is intricate to identify if the initial binding is due to a single or a multiple bond. In the first case binding is mediated by a single GSL-GSL contact and rapidly strengthened by the formation of further bonds in the contact zone. In the second case two or more GSL-GSL contacts are simultaneously formed when a liposome binds to the SLB. Although beyond the scope of this work to make a clear conclusion about these scenarios, we stress that the observed variations in apparent rate constants may have more than one origin. Nevertheless, in the case of no cholesterol in the SLB we largely exclude the formation of clusters or domains based on very similar diffusivity as that of a GSL-free SLB. Smith and co-workers discussed the competition between enthalpic and entropic contributions on the formation of GSL clusters41,42. On the one hand, binding enthalpy favors growing clusters while dispersion is favored by gain in entropy. In essence, weak bonds are not assumed to lead to large clusters32,41,42. However in case of cholesterol-containing SLBs domain formation is likely to occur due to cholesterol's feature to induce domain or raft formation in lipid bilayers43,44. Although the number of attractive Lex-Lex contacts at the interface between a liposome and the SLB might be sufficient to overcome the entropic loss that counteract interaction-induced cluster formation, our results rather suggest that the strength of the interaction is controlled by cholesterol and/or Ca2+ induced GSL cluster formation. For instance, both the residence time and the fraction of irreversible bound liposomes increased in the presence of cholesterol, despite the fact that the lateral diffusivity in the SLB decreases. Such clustering of Lex-GSLs is expected to increase the number of attractive bounds in the contact zone, which would consequently magnify the binding avidity given that the saccharide presentation is not compromised.
Although the interaction we observe is most likely not governed by single Lex-Lex interactions, the reversible nature of the interaction and the fact that it is modulated by cholesterol serves as a base for a discussion on its molecular nature. It has been previously proposed that Lex-Lex interaction is a process that includes mainly two types of interactions: (i) hydrophobic interactions between the pyranose rings and (ii) Ca2+-bridging between Lex-molecules10. Without cholesterol, we observed that the association rate constant was essentially independent on the concentration of bivalent cations, indicating that association is mainly driven by hydrophobic interactions. However, dissociation was observed to depend on high concentration of bivalent cations, suggesting that the strength of the interaction increases due to Ca2+-induced bridging effects. In other words, Lex-Lex interaction in this system may be initiated by hydrophobic interactions and subsequently strengthened by Ca2+-ion mediated cross-linking of Lex-molecules. In contrast, in the presence of cholesterol, both association and dissociation were affected by bivalent cations. At first glance this seems to be in contradiction with the hypothesis that the first step of the interaction is due to hydrophobic interactions between pyranose rings and independent on bivalent ions. However, in addition to a positive contribution from clustering on the association rate, we have to consider that hydrophobic interactions are likely to depend on the orientation of the Lex-moiety, i.e. the presentation of the hydrophobic pyranose rings. Supported by the recent study by Lingwood et al.37 we hypothesize that cholesterol modulates saccharide orientation facilitating hydrophobic interactions. Furthermore, we assume that within this cholesterol containing system calcium, or other bivalent cations, have the ability to bind to lipid or GSL molecules involving further structural changes that increase hydrophobic interactions45,46,47,48,49,50. In other words, in the presence of both cholesterol and bivalent cations, both the local density and the orientation of individual Lex determinant are modulated such that the interaction between the pyranose rings is increased. Although the relative importance of these contributions cannot be determined, the binding seems to be further strengthened by cross-linking via bivalent cations, although hydrophobic interactions seem to prevail in the presence of cholesterol.
In summary, we introduce a highly sensitive equilibrium-fluctuation-analysis of single liposome binding events based on TIRF microscopy and cell membrane mimics to determine kinetic constants of Ca2+-modulated, carbohydrate specific Lex-Lex interactions. Using this methodology we were able not only to determine kinetic constants of this ultra-weak interaction, but more importantly, we could resolve a persisting puzzle: as cholesterol was inserted into the SLB, the Lex-Lex interaction is strengthened at physiological concentrations of bivalent cations as it had been observed in the very first study on Lex-Lex interactions by Eggens et al.6. This result is of importance for past and future studies of carbohydrate-carbohydrate interactions in particular, but also illustrates the often ignored importance of better mimicking the natural cell membrane composition in studies utilizing model membranes. Adding not only cholesterol, but an optimized lipid composition when producing model membranes will eventually become indispensable to eventually unravel the functional role of lipids in dynamic short lived recognition events between cells during embryogenesis, maturation of lymphoid cells as well as for the metastatic spreading of cancer cells.
Methods
Lipids
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rhod-PE) and cholesterol were obtained from Avanti Polar Lipids (Alabaster, AL, USA). 2-(12-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)dodecanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine (NBD-PC) was purchased from Molecular Probes (Eugene, OR, U.S.A.). Lex pentaglycosylceramide (Galβ4(Fucα3)GlcNAcβ3Galβ4Glcβ1Cer) was purified from canine intestine20 and Lea pentaglycosylceramide (Galβ3(Fucα4)GlcNAcβ3Galβ4Glcβ1Cer) from human meconium21. The Lex and Lea GSLs were >98% pure as judged from proton NMR analysis.
Liposome preparation
Unilamellar liposomes were prepared based on the extrusion protocol described before by Hope et al.51. In short, lipids were dissolved and mixed at the desired weight ratio in chloroform:methanol 2:1, and subsequently dried as a thin film in a round-bottom flask first under a gentle stream of N2, and secondly under vacuum overnight. The lipid film was then hydrated in TRIS (10 mM TRIS, 100 mM NaCl, pH 7.4). After vortexing the resulting liposome solution was extruded 13 times through 100 nm polycarbonate filter membranes (Avanti) at room temperature. Liposome solutions were either used directly, or stored for maximum 3 days at +4°C.
The intensity-weighted liposome size distribution was determined by dynamic light scattering on a Zetasizer nano (Malvern Instruments, UK). The mean liposome size was 162 nm with a corresponding full-width half maximum of about 85 nm.
TIRF microscopy assay
Cleaning
Glass-bottom microtiter wells (96 well-plate, MatTec Corporation, Ashland, MA, USA) were cleaned for at least 2 hours in 2% Hellmanex or 10 mM sodium dodecyl sulfate solution followed by thoroughly rinsing with MilliQ (at least 10 times with 300 μl). Further surface modification and assays were performed in TRIS buffer.
Supported lipid bilayer formation
Lipid bilayers were formed by incubation for at least 45 min with the liposome dispersion (total lipid concentration 0.1 mg/ml) followed by extensive rinsing (6 times with 200 μl), taking care of not drying the surface. In order to avoid defects in the lipid bilayer, POPC liposomes, which are known to rupture easily on glass, were added to fill or “heal” possible holes in the SLB. POPC liposomes were added to a final concentration of 5 μg/ml and incubated for 30 min.
Addition of fluorescent liposomes
Fluorescent GSL-containing liposomes were added to a final lipid concentration of 1 μg/ml. Imaging was performed the same day but at least 2 hours after addition of fluorescent liposomes to allow for establishing steady state conditions.
Imaging and data analysis
Time-lapse movies were acquired on a Nikon Eclipse Ti-E inverted microscope using a 60× magnification (NA = 1.49) oil immersion objective (Nikon Corporation, Tokyo, Japan). The microscope was equipped with an X-Cite 120 lamp (Lumen Dynamics Group Inc., Mississauga, Ontario, Canada), a TRITC filter cube (Nikon Corporation) and an Andor DU897E-CSBV camera (Andor Technology, Belfast, Northern Ireland). For each spot 1000 frames with an increment of 100 ms were taken.
Images were processed and analyzed with a MatLab (MathWorks, Inc., Natick, Massachusetts, USA) software package as described in detail elsewhere33,34,35. Briefly, a liposome was registered if its intensity exceeded a preset threshold, and was considered as bound if it was present for > 1.2 sec. Bleached liposomes, which did not exhibit a sudden drop in intensity, were discarded from the analysis. For accurate statistics and to avoid underrepresentation of liposomes bound for a longer time, the maximal residence time taken into consideration, was half of the total measurement time.
Calculation of rate constants
For the analysis only liposomes with a residence time > 1.2 sec were counted, in order to exclude unspecific interactions and false positives.
The association rate constant can be represented
where 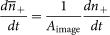 is the number of newly arrived liposomes n+ per time t and image area Aimage (determined from the experimental data, i.e. association curves), kon is the association rate constant, Clipo = 0.8 pM is the concentration of liposomes (assuming the mass of a single liposome to be 2·10−6 ng and the concentration of lipid in the well to be 1 ng/μl) and Nb the number of vesicle binding sites per μm2. Under the assumption that one Lex-Lex contact is sufficient for liposome binding Nb equals NLex, which is the number of Lex-determinants per μm2 in the SLB. Assuming the area of a lipid to be 60 Å2 and Lex-molecules to be equally distributed between the upper and lower leaflet of the SLB
is the number of newly arrived liposomes n+ per time t and image area Aimage (determined from the experimental data, i.e. association curves), kon is the association rate constant, Clipo = 0.8 pM is the concentration of liposomes (assuming the mass of a single liposome to be 2·10−6 ng and the concentration of lipid in the well to be 1 ng/μl) and Nb the number of vesicle binding sites per μm2. Under the assumption that one Lex-Lex contact is sufficient for liposome binding Nb equals NLex, which is the number of Lex-determinants per μm2 in the SLB. Assuming the area of a lipid to be 60 Å2 and Lex-molecules to be equally distributed between the upper and lower leaflet of the SLB 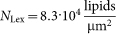 . We cannot exclude that bivalent cations and/or cholesterol may induce bridging and/or lateral association of Lex-GSLs within the SLB, with the consequence that the number of free Lex-determinants will vary. However, since it was not possible to explicitly determine such clustering, we refer to the association rate constant kon as an apparent parameter determined under the assumption that NLex does not to vary with experimental conditions. Consequently, variations in apparent kon may depend both on changes in the actual Lex-Lex interaction strength and the number of Lex-Lex contacts that are required for liposome binding, or a combination thereof. Also note that the association rate constant kon extracted by Equation (1) depends on the concentration of liposomes Clipo and not on the concentration of GSLs in the liposomes. Hence, in analogy with the observation that both the association and dissociation rates of GSL-containing liposomes to virus-like particles were shown to depend on the concentration of GSLs in the liposomes27, we expect similar trends upon changes in the concentration of Lex in the liposomes used in this work. However, with focus on how bivalent cations and cholesterol under certain conditions can modulate GSL-GSL interactions, we deliberately kept the Lex concentration fixed at a biologically relevant concentration that in addition ensured transient binding for a sufficiently large population of liposomes.
. We cannot exclude that bivalent cations and/or cholesterol may induce bridging and/or lateral association of Lex-GSLs within the SLB, with the consequence that the number of free Lex-determinants will vary. However, since it was not possible to explicitly determine such clustering, we refer to the association rate constant kon as an apparent parameter determined under the assumption that NLex does not to vary with experimental conditions. Consequently, variations in apparent kon may depend both on changes in the actual Lex-Lex interaction strength and the number of Lex-Lex contacts that are required for liposome binding, or a combination thereof. Also note that the association rate constant kon extracted by Equation (1) depends on the concentration of liposomes Clipo and not on the concentration of GSLs in the liposomes. Hence, in analogy with the observation that both the association and dissociation rates of GSL-containing liposomes to virus-like particles were shown to depend on the concentration of GSLs in the liposomes27, we expect similar trends upon changes in the concentration of Lex in the liposomes used in this work. However, with focus on how bivalent cations and cholesterol under certain conditions can modulate GSL-GSL interactions, we deliberately kept the Lex concentration fixed at a biologically relevant concentration that in addition ensured transient binding for a sufficiently large population of liposomes.
The dissociation rate constant can be determined by exponential fitting of the normalized dissociation curve:
where nbound is the number of bound liposomes, no and n1 are fitting constants and the exponent koff is the dissociation rate constant. In contrast to the association rate constant kon, where all liposomes that arrive on the SLB are counted, only liposomes that are also released within the experimental time are used for the estimation of the dissociation rate constant koff. In other words, for the calculation of kon both reversible and irreversible liposomes are counted whilst for the calculation of koff only reversible liposomes are counted.
Both kinetic constants are given as the mean and its standard deviation of a triplet of experiments.
Diffusion limited kinetics
For diffusion-limited bimolecular reactions between spherically shaped particles (here liposomes) in the solution and a spot of the radius Rb on the surface (here the radius of the binding site) the diffusion rate constant kdiff is given by52:
where Dliposome is the diffusion coefficient of liposomes in solution. The diffusion coefficient Dliposome can be calculated using:
where kB is the boltzmann constant, T is the temperature, η is the viscosity, and Rliposome it the radius of a liposome. With T = 295 K, η = 10−3 Pa·s and Rliposome = 80 nm the diffusion coefficient is Dliposome = 2.6 μm2s−1. Assuming binding to be mediated by one bond Rb is equal the radius of a Lex-GSL headgroup RLex. Given the radius RLex to be 4 Å, Equation (3) reveals the diffusion rate constant to be kdiff ≈ 2.5·106 M−1s−1. This value is significantly higher than adsorption rate constants obtained in the present study. As discussed above bivalent cations and/or cholesterol may induce bridging and/or lateral association of Lex-GSLs within the SLB, which would consequently increase the radius of the binding site Rb, and thus increase the diffusion rate constant kdiff, too. Hence, the observed binding is clearly kinetically limited.
Author Contributions
The project was formulated by G.L. and A.K. A.K. performed experiments and wrote the manuscript. Manuscript completion was performed by all authors together.
Supplementary Material
Supplementary Information
Acknowledgments
Informative discussions with Waqas Nasir and Per-Georg Nyholm are gratefully acknowledged. The work was supported by grants from Vinnova Innovations for Health (FH, GL), Swedish Research council (No 8266) and by Governmental grants to the Sahlgrenska University Hospital.
References
- Bovin N. V. Carbohydrate-carbohydrate interaction: A review. Biochemistry (Moscow) 61, 694–704 (1996). [PubMed] [Google Scholar]
- Rojo J., Morales J. C. & Penadés J. Carbohydrate-carbohydrate interactions in biological and model systems. Host-Guest Chemistry 218, 45–92 (2002). [Google Scholar]
- Bucior I. & Burger M. M. Carbohydrate-carbohydrate interactions in cell recognition. Curr. Opin. Struct. Biol. 14, 631–637 (2004). [DOI] [PubMed] [Google Scholar]
- Handa K. & Hakomori S.-I. Carbohydrate to carbohydrate interaction in development process and cancer progression. Glycoconj. J. 29, 627–637 (2012). [DOI] [PubMed] [Google Scholar]
- Pincet F. et al. Ultraweak sugar-sugar interactions for trancient cell adhesion. Biophys. J. 80, 1354–1358 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggens I. et al. Specific interaction between Lex and Lex determinants. J. Biol. Chem. 264, 9476–9484 (1989). [PubMed] [Google Scholar]
- Fenderson B. A., Zehavi U. & Hakomori S.-I. A multivalent lacto-N-fucopentaose III-lysyllysine conjugate decompacts preimplantation mouse embryos, while the free oligosaccharide is ineffective. J. Exp. Med. 160, 1591–1596 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todeschini A. G. & Hakomori S.-I. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim. Biophys. Acta 1780, 421–433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima N. et al. Cell adhesion in a dynamic flow system as compared to static system. J. Biol. Chem. 267, 17261–17270 (1992). [PubMed] [Google Scholar]
- Kojima N. et al. Further studies on cell adhesion based on Lex-Lex interaction, with new approaches: embryoglycan aggregation of F9 teratocarcinoma cells, and adhesion of various tumour cells based on Lex expression. Glycoconj. J. 11, 238–248 (1994). [DOI] [PubMed] [Google Scholar]
- Gege C., Geyer A. & Schmidt R. R. Carbohydrate-carbohydrate recognition between Lewis X blood broup antigens, bediated by calcium ions. Eur. J. Org. Chem. 2475–2485 (2002). [Google Scholar]
- Geyer A., Gege C. & Schmidt R. R. Carbohydrate-carbohydrate recognition between Lewisx glycoconjugates. Angew. Chem. Int. Ed. 38, 1466–1468 (1999). [DOI] [PubMed] [Google Scholar]
- Geyer A., Gege C. & Schmidt R. R. Calcium dependent carbohydrate-carbohydrate recognition between Lewisx blood group antigens. Angew. Chem. Int. Ed. 39, 3245–3249 (2000). [DOI] [PubMed] [Google Scholar]
- Gourier C. et al. Specific and non specific interactions involving Lex determinant quantified by lipid vesicles micromanipulation. Glycoconj. J. 21, 165–174 (2004). [DOI] [PubMed] [Google Scholar]
- Gourier C. et al. The natural Lewisx-beraing lipids promote membrane adhesion: influence of ceramide on carbohydrate-carbohydrate recognition. Angew. Chem. Int. Ed. 44, 1683–1687 (2005). [DOI] [PubMed] [Google Scholar]
- Siuzdak G. et al. Evidence of Ca2+-dependent carbohydrate associations through ion spray mass spectrometry. J. Am. Chem. Soc. 115, 2877–2881 (1993). [Google Scholar]
- Tromas C. et al. Adhesion forces between Lewisx determinant antigens as measured by atomic force microscopy. Angew. Chem. Int. Ed. 40, 3052–3055 (2001). [DOI] [PubMed] [Google Scholar]
- de la Fuente J. M. & Penadés J. Understanding carbohydrate-carbohydrate interactions by means of glyconanotechnology. Glycoconj. J. 21, 149–163 (2004). [DOI] [PubMed] [Google Scholar]
- Hernáiz M. J., de la Fuente J. M., Barrientos A. G. & Penadés J. A model system mimicking glycosphingolipid clusters to quantify carbohydrate self-interactions by surface plasmon resonance. Angew. Chem. Int. Ed. 41, 1554–1557 (2002). [DOI] [PubMed] [Google Scholar]
- McKibbin J. M. et al. Lewis blood group fucolipids and their isomers from human and canine intestine. J. Biol. Chem. 257, 755–760 (1982). [PubMed] [Google Scholar]
- Karlsson K. A. & Larson G. Molecular characterization of cell surface antigens of fetal tissue. Detailed analysis of glycosphingolipids of meconium of a human O Le(a-b+) secretor. J. Biol. Chem. 256, 3512–3524 (1981). [PubMed] [Google Scholar]
- Briand E., Zäch M., Svedhem S., Kasemo B. & Petronis S. Combined QCM-D and EIS study of supported lipid bilayer formation and interaction with pore-forming peptides. Analyst 135, 343–350 (2010). [DOI] [PubMed] [Google Scholar]
- Janshoff A. & Steinem C. Transport across artificial membranes–an analytical perspective. Anal. Bioanal. Chem. 385, 433–451 (2006). [DOI] [PubMed] [Google Scholar]
- Janshoff A., Steinem C., Sieber M. & Galla H.-J. Specific binding of peanut agglutinin to GM1-doped solid supported lipid bilayers investigated by shear wave resonator measurements. Eur. Biophys. J. 25, 105–113 (1996). [Google Scholar]
- Glasmästar K., Larsson C., Höök F. & Kasemo B. Protein adsorption on supported phospholipid bilayers. J. Colloid Interface Sci. 246, 40–47 (2002). [DOI] [PubMed] [Google Scholar]
- Rydell G. E., Dahlin A. B., Höök F. & Larson G. QCM-D studies of human norovirus VLPs binding to glycosphingolipids in supported lipid bilayers reveal strain-specific characteristics. Glycobiology 19, 1176–1184 (2009). [DOI] [PubMed] [Google Scholar]
- Bally M. et al. Interaction of single viruslike particles with vesicles containing glycosphingolipids. Phys. Rev. Letters 107, 188103 (2011). [DOI] [PubMed] [Google Scholar]
- Wikström A., Svedhem S., Sivignon M. & Kasemo B. Real-time QCM-D monitoring of electrostatically driven lipid transfer between two lipid bilayer membranes. J. Phys. Chem. B 112, 14069–14074 (2008). [DOI] [PubMed] [Google Scholar]
- Kunze A., Svedhem S. & Kasemo B. Lipid transfer between charged supported lipid bilayers and oppositely charged vesicles. Langmuir 25, 5146–5158 (2009). [DOI] [PubMed] [Google Scholar]
- Andersson A.-S., Glasmästar K., Sutherland D., Lidberg U. & Kasemo B. Cell adhesion on supported lipid bilayers. J. Biomed. Mater. Res., Part A 64, 622–629 (2003). [DOI] [PubMed] [Google Scholar]
- Svedhem S. et al. In situ peptide-modified supported lipid bilayers for controlled cell attachment. Langmuir 19, 6730–6736 (2003). [Google Scholar]
- Lorenz B. et al. Model system for cell adhesion mediated by weak carbohydrate−carbohydrate interactions. J. Am. Chem. Soc. 134, 3326–3329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnarsson A. et al. Kinetics of ligand binding to membrane receptors from equilibrium fluctuation analysis of single binding events. J. Am. Chem. Soc. 133, 14852–14855 (2011). [DOI] [PubMed] [Google Scholar]
- Gunnarsson A., Jönsson P., Marie R., Tegenfeldt J. O. & Höök F. Single-molecule detection and mismatch discrimination of unlabled DNA targets. Nano Lett. 8, 183–188 (2007). [DOI] [PubMed] [Google Scholar]
- Gunnarsson A., Jönsson P., Zhdanov V. P. & Höök F. Kinetic and thermodaynamic characterization of single-mismatch discrimination using single-molecule imaging. Nucleic Acids Res. 37, e99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. et al. In: Molecular Biology of the Cell Ch. 10, Garland Science, (2002). [Google Scholar]
- Lingwood D. et al. Cholesterol modulates glycolipid conformation and receptor activity. Nat. Chem. Biol. 7, 260–262 (2011). [DOI] [PubMed] [Google Scholar]
- Filippov A., Orädd G. & Lindblom G. Influence of cholesterol and water content on phospholipid lateral diffusion in bilayers. Langmuir 19, 6397–6400 (2003). [Google Scholar]
- Lin W.-C., Blanchette C. D. & Longo M. L. Fluid-phase chain unsaturation controlling domain microstructure and phase in ternary lipid bilayers containing GalCer and cholesterol. Biophys. J. 92, 2831–2841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strömberg N., Nyholm P. G., Pascher I. & Normark S. Saccharide orientation at the cell surface affects glycolipid receptor function. Proc. Natl. Acad. Sci. USA 88, 9340–9344 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.-S. & U S. Vesicles as a model for controlled (de-)adhesion of cells: a thermodynamic approach. Softmatter 3, 275–289 (2007). [DOI] [PubMed] [Google Scholar]
- Fenz S. F. et al. Switching from Ultraweak to Strong Adhesion. Adv. Mater. 23, 2622–2626 (2011). [DOI] [PubMed] [Google Scholar]
- Almeida P. F. F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta 1788, 72–85 (2009). [DOI] [PubMed] [Google Scholar]
- J R. S. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta 1610, 174–183 (2003). [DOI] [PubMed] [Google Scholar]
- Mashaghi A., Swann M., Popplewell J., Textor M. & Reimhult E. Optical anisotropy of supported lipid structures probed by waveguide spectroscopy and its application to study of supported lipid bilayer formation kinetics. Anal. Chem. 80, 3666–3676 (2008). [DOI] [PubMed] [Google Scholar]
- Kunze A., Zhao F., Marel A.-K., Svedhem S. & Kasemo B. Ion-mediated changes of supported lipid bilayers and their coupling to the substrate. A case of bilayer slip? Softmatter 7, 8582–8591 (2011). [Google Scholar]
- Garcia-Manyes S., Oncins G. & Sanz F. Effect of pH and ionic strength on phospholipid nanomechanics and on deposition process onto hydrophilic surfaces measured by AFM. Electrochim. Acta 51, 5029–5036 (2006). [Google Scholar]
- Binder H. & Zschörnig O. The effect of metal cations on the phase behavior and hydration characteristics of phospholipid membranes. Chem. Phys. Lipids 115, 39–61 (2002). [DOI] [PubMed] [Google Scholar]
- Nieh M. P., Harroun T. A., Raghunathan V. A., Glinka C. J. & Katsaras J. Spontaneously formed monodisperse biomimetic unilamellar vesicles: The effect of charge, dilution, and time. Biophys. J. 86, 2615–2629 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura-Lis W., Reber E. J., Cunningham B. A., Collins J. M. & Lis L. J. Ca2+ induced phase separations in phospholipid mixtures. Chem. Phys. Lipids 39, 119–124 (1986). [DOI] [PubMed] [Google Scholar]
- Hope M. J., Bally M. B., Webb G. & Cullis P. R. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta 812, 55–65 (1985). [DOI] [PubMed] [Google Scholar]
- Potanin A. A., Verkhusha V. V., Belokoneva O. S. & Wiegel F. W. Kinetics of ligand binding to a cluster of membrane-associated receptors. Eur. Biophys. J. 23, 197–205 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information