Abstract
The identification of potent spliceosome modulators that demonstrate antitumor activity indicates that this complex may be a target for drug development. Several natural products have been demonstrated to bind to the SF3b1 subunit of this macromolecule and these agents modulate alternative RNA splicing. In this article we describe their biological properties, discuss the validity of the spliceosome as a therapeutic target, and propose that alteration of alternative splicing represents a viable approach for inducing tumor-selective cytotoxicity.
Keywords: Spliceosome, alternative splicing, sudemycin, SF3b
Splicing of pre-mRNA into mature mRNAs by the removal of intronic sequences is an essential process for the generation of high-fidelity transcripts and enables for the production of appropriately encoded proteins [1–4]. The spliceosome, a complex macromolecular structure containing numerous proteins and RNA molecules, coordinates the multiple cleavage and rejoining events required for completion of the splicing cycle. An absolute requirement for this process is a series of domains present within the pre-mRNA that direct the catalytic machinery towards the correct ribonucleotides. In mammalian cells, this is achieved by a series of signal sequences, including a 5' splice site, branch point sequence, polypyrimidine tract, and 3' splice site (Figure 1) [2]. It is well documented that point mutations within these sequences can result in the loss of splicing fidelity and/or the usage of novel ‘cryptic’ splice sites [5]. Recent work has also shown that pre-mRNA splicing patterns can be linked to histone modifications, the transcription machinery, and non-coding RNAs [6]. Because of this complexity, and the relatively focused scope of this review, the reader is directed to recent review articles that describe this process in much greater detail and with excellent clarity [2, 6].
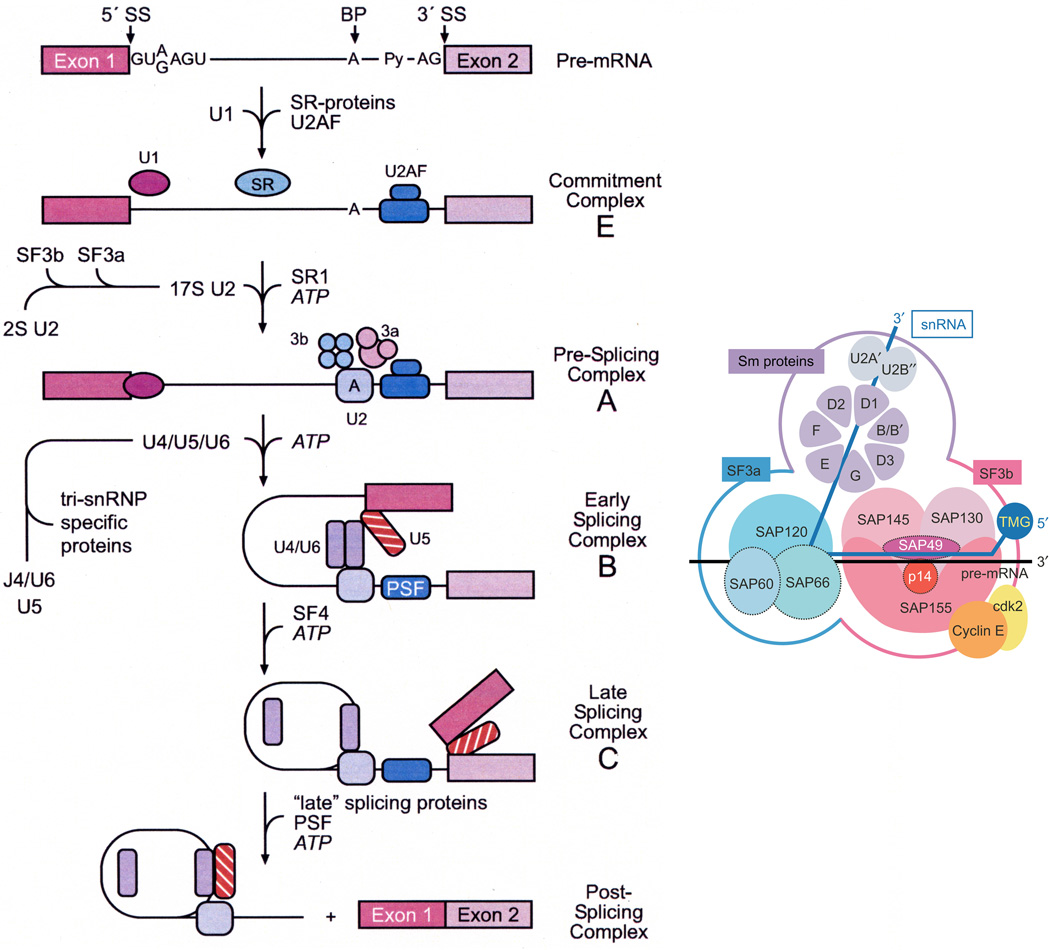
Figure 1.
An overview of the pre-mRNA splicing process (left panel, adapted from Kramer [60]). The structure and function of proteins involved in mammalian pre-mRNA splicing [60] and a representation of the SF3b subunit (right panel, adapted from Kotake et al [20]). The target for FR901464, pladienolide B, E7107, spliceostatin and sudemycin is the SF3b subunit.
Abbreviations: BP: xxx; PSF: xxx; SR: xxx.[RE3]
Alternative splicing occurs as a result of modulation of the above processes. Recent evidence indicates that up to 95% of all mammalian exonic genes are alternatively spliced [7]. This process is beneficial because it results in the generation of many unique proteins with differing function and therefore increases the diversity that can be generated from each transcript. However, if this process is compromised, the resulting changes in splicing can lead to various disease states. Point mutations within splice sites that result in loss of the wild-type protein, and potentially the generation of proteins with alternative new functions, contribute to a spectrum of human diseases (e.g. the NF1 gene for neurofibromatosis [8] and the ATM gene for ataxia-telangiectasia) [9]. In addition, it is not only the presence of the different transcripts, but also the levels and/or ratios of the different mRNA isoforms that are important. One such example has recently been documented with the MAPT gene. This gene encodes a microtubule-associated protein Tau that can be alternatively spliced to yield six variants [10]. Mutations in this gene alter mRNA splicing and consequently the cellular levels of the different Tau isoforms, which results in neurodegeneration and fronto-temporal dementia. It has been demonstrated that the ratio of the Tau proteins is important for cell survival, implying that this process is essential for neuronal cell homeostasis [10].
The above examples clearly demonstrate that the maintenance of high-fidelity mRNA splicing is important, because the translation of potentially thousands of mis-spliced mRNAs into proteins with aberrant function could be disastrous. Due to the complexity of the genomic structure of mammalian genes, it could be anticipated that splicing errors might occur with sufficient frequency to be problematic. It is therefore not surprising that eukaryotes have evolved a protective mechanism, nonsense-mediated decay (NMD) to deal with such abnormal events, which can target and eliminate many inappropriately spliced mRNAs. Briefly, NMD can recognize premature termination codons in a positional context in relation to the exon and/or exon[RE1] junctions within the transcript and if necessary, initiate a program that results in uncapping of the mRNA and subsequent degradation by an exonuclease. NMD has also been the subject of several recent reviews, so the details of this process will not be covered here [11].
Natural product spliceosome modulators
Recently, three bacterial natural products, pladienolides [12–14], FR901464 [15–17], and herboxidiene [18, 19] (Figure 2), have all been shown to modulate the function of the spliceosome (Figure 1) [20–22]. Since pladienolide B and FR901464 are structurally distinct, and derived from distantly related bacterial genera (Streptomyces and Pseudomonas, respectively), convergent evolution has led to divergent prokaryotes producing spliceosome modulators that share a common pharmacophore. The recent discovery that herboxidiene (which shows structural similarity to the Streptomyces-derived pladienolide) also targets SF3b [22], suggests that splicing modulation may be a relatively common mechanism by which bacteria compete with eukaryotes. Biochemical studies have identified the spliceosomal SF3b subunit [which includes the SF3b1 protein (Figure 1: right panel)] as the target of these modulators.
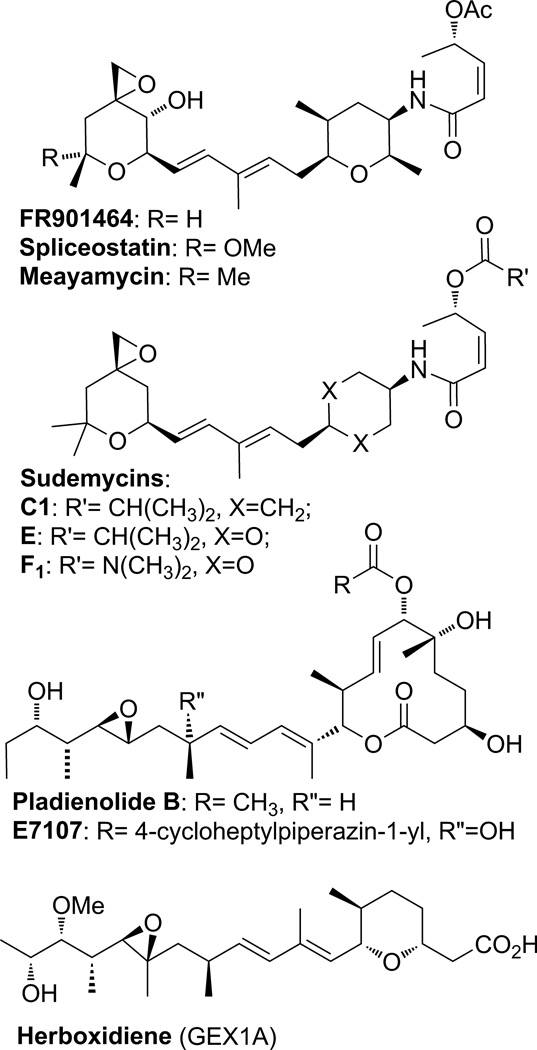
Figure 2.
The molecular structures of FR901464, spliceostatin A, meamycin, the sudemycins, pladienolide B, E7107 and herboxidiene (GEX1A).
A derivative of pladienolide b (E7107), demonstrated dramatic, selective antitumor activity in human tumor xenograft models, without significant reported toxicity. E7107 subsequently rapidly progressed to the point where it entered Phase I clinical trials in 2007 [20]. However these studies are currently suspended, (Clinicaltrials.gov: http://www.clinicaltrials.gov/ct2/show/record/NCT00459823?term=E+7107&rank=1) although the reasons for this have not been reported. While pladienolide derivatives appear to be promising candidates for further drug development owing to their excellent in vivo efficacy in animal models and low toxicity, their highly complex structures (e.g. ten chiral centers and a macrolactone ring) do not lend themselves well to standard medicinal chemistry manipulations, or to practical total analog synthesis on a production scale. For these reasons, it will be extremely challenging to develop facile synthetic approaches that could enable for cost-effective production of sufficient quantities of these compounds for widespread distribution to patients with cancer.
Synthetic spliceosome modulators
Following the discovery that SF3b was the target for pladienolide and FR901464 [20, 21], new opportunities in the design of totally synthetic specific modulators of splicing arose [20, 21]. The first synthetic compounds of this class were active analogs of FR901464 and were reported before the identification of the mode of action of these drugs. The groundbreaking work of the Jacobsen group described the first total synthesis of FR901464 [23] and subsequently, important structure–activity relationship (SAR) data [24]. This was followed by several novel approaches to the total synthesis that also reported the generation of several novel analogs [25–29]. Just before the identification of the target of FR901464, workers reported the synthesis of an analog of this compound (meayamycin) that was more stable and more cytotoxic than the parent molecule [29]. This group has also provided a substantial amount of information on the overall SAR of FR901464, the mechanism of action, and its in vivo activity [30, 31].
Because the natural products and their analogs are highly complex (containing 7–10 stereocenters!, and require elaborate synthetic schemes, there has been a significant need for more elegant scaffolds that would give much more drug-like chemical matter exhibiting the same pharmacophore. The first such set of ‘concise’ active molecules was designed and prepared by the Webb group [32, 33] and named the sudemycins [34]. These molecules were designed using a hypothetical consensus pharmacophore model that was derived from known SAR [24, 29] and molecular overlays of FR901464 and pladienolide B [32, 33]. The sudemycins are analogs of FR901464 in which six of the nine stereocenters have been symmetrized, and include modifications that dramatically improved their chemical stability [33]. This class of compounds can be readily generated in multi-gram quantities using relatively facile medicinal chemistry approaches. Importantly, the elegant nature of the sudemycin scaffold has enabled SAR development for numerous new modifications, including some that were beyond what was readily approachable with FR901464 [32, 33].
Biology of spliceosome modulators
The spliceosome modulators (pladienolide B, E7107, FR901464, meayamycin, spliceostatin A, and the sudemycins) are all potent cytotoxic agents, exhibiting IC50 values in the nM range against a panel of human tumor cell lines [14, 15, 20, 21, 29, 34]. Interestingly, the latter drug is considerably less toxic to normal cells in culture. For example, sudemycin F1 is 5- to 15-fold less cytotoxic towards normal human myoblasts as compared to a rhabdomyosarcoma cell line [34]. In addition, normal human foreskin fibroblasts are one of the most resistant cell types that have been evaluated to date [33].
Preclinical studies using the spliceosome modulators indicate that these compounds demonstrate potent antitumor activity, with remarkably little toxicity. Indeed, complete regressions were observed in a human lung tumor xenograft model following five daily doses of E7107. Additionally, the sudemycins have demonstrated modest activity in lymphoma models, with no detectable systemic damage to normal tissues [33]. One hypothetical mechanism that could account for the selective antitumor activity these compounds is based on could be that these compounds potently modulate alternate splicing in tumor cells [34, 35]. Furthermore, one can speculate that the selective toxicity induced by these small molecules might result from differences in the capacity of tumor, versus normal, cells to respond appropriately to changes in mRNA splicing. This would result in an irreversible disruption in the balance of the ‘splicing program’ of susceptible cells [36]. If this hypothesis (Figure 3) is correct, this would enable the development of novel anticancer agents by targeting an, as yet, relatively unexplored area. Due to the limited efficacy and therapeutic scope of current chemotherapeutic drugs it would be important to validate this new class of tumor vulnerability (mRNA splicing and/or NMD) for small molecule modulator design.
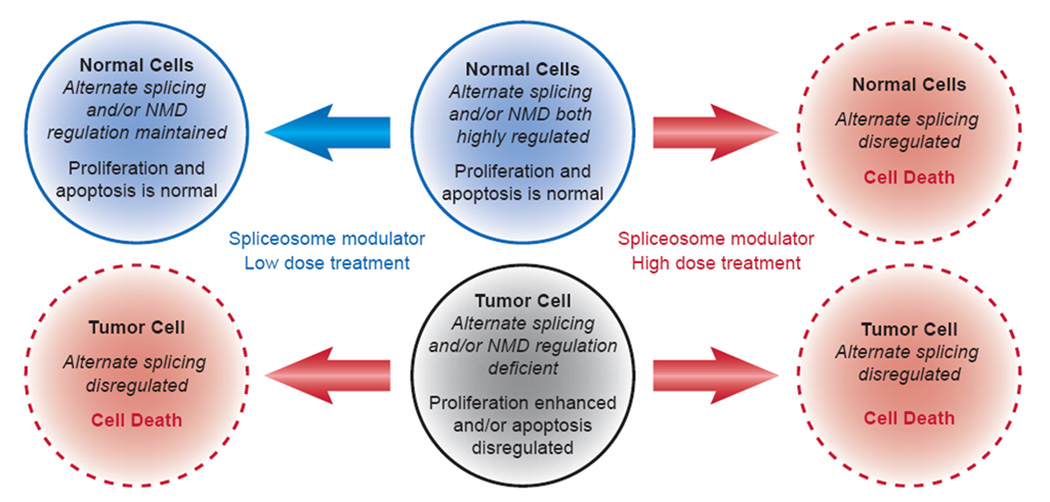
Figure 3.
Hypothetical schematic that may account for the tumor-selective toxicity seen with modulators of alternative splicing.
Abbreviation: NMD: nonsense-mediated decay.
Modulation of pre-mRNA splicing
Both biochemical and molecular analyses suggest the involvement of the SF3b spliceosomal subunit in RNA splice site selection. This has been further strengthened by recent reports that indicate that spliceostatin A (a derivative of FR901464) [35], E7107 [37], and the sudemycins [34] can modulate alternative splicing in mammalian cells. This has been observed both in vitro and in vivo, and can result in almost complete loss of the full-length mRNA for certain genes (e.g. MDM2[RE2] in rhabdomyosarcoma cells). Because these changes can be identified in drug-treated human tumor xenografts, this suggests that alteration of gene splicing may represent a suitable marker for spliceosome modulator exposure. The advantage of using the latter approach is that because it is a PCR-based assay, the sensitivity is such that even very minor changes in the pattern of mRNA expression can be observed. Hence, this may be used in combination with traditional pharmacokinetic methods to evaluate exposure of tumor cells to drug. Furthermore, in clinical trials with such agents, it should be possible to determine the effects of the drugs on splicing in readily available normal cells (e.g. peripheral blood lymphocytes), and potentially, biopsied tumor tissue. Again, similar to that seen in the preclinical experiments, this may provide not only a measure of drug exposure, but may also enable for dose adjustment to ensure tumor penetration.
Mechanism of selective action of spliceosome modulators
Recently, two articles have undertaken a detailed examination of the mechanism of modulation of alternative gene splicing afforded by spliceostatin A [35] and E7107 [37]. It has been shown that these drugs act by preventing binding of the U2 snRNA to the pre-mRNA, thereby altering the conformation of the branch point sequence. This leads to non-productive base-paring and the inappropriate selection of 3' splice acceptor sites within the unspliced transcript, resulting in the generation of alternatively spliced mRNAs [35]. Due to the differential base pairing of ribonucleotides within the drug and/or spliceosome complex, apparent selectivity for aberrant alternative splicing occurs. Hence, as detailed above, treatment of tumor cells with sudemycins results in a modulation in the splicing pattern for MDM2 due to alternative splice site selection afforded by the SF3b1/U2 snRNA subunit. This would indicate that the exon and/or intron boundaries for this gene likely contain nucleotide sequences at the branch points that are more susceptible to alternative splicing. A comparison of these sequences with other splice junctions may enable for a more detailed description of the panels of genes that might be affected by spliceosome modulators. Because genome-wide bioinformatics approaches have recently identified parameters for branch point use in transcripts, the validity of these hypotheses can be readily tested [38].
As indicated above, the details of the mechanism of selective tumor cytotoxicity remain elusive. It would be presumed that modulation of RNA splicing would be a highly deleterious event in all cells, potentially resulting in the loss of critical proteins and the expression of variants with aberrant function. Indeed, in studies using the sudemycins, aberrantly spliced mRNAs encoding proteins involved in the apoptotic cascade, in addition to the regulation of p53, have been demonstrated [34]. Furthermore, in spliceostatin A-treated cells, changes in the splicing of a series of cell cycle control genes have been observed [21]. In normal cells, these events apparently do not significantly perturb the processes involved in damage response (and/or are modulated by events in other intersecting pathways) [34]. However, in tumor cells, such a change in the levels of the proteins appears to have catastrophic consequences, resulting in the initiation of apoptosis. Potentially, the cytotoxicity of these compounds in tumor cells may result from an alteration in the pattern of expressed transcripts. Since with sudemycin, this includes changes in the splice variant mRNAs that encode proteins involved in apoptosis (e.g. caspases 2 and 9), it is conceivable that the panel of alternatively expressed proteins in these cells contributes to cytotoxicity.
Recent studies have indicated that spliceostatin A can modulate angiogenesis by inhibiting a wide variety of genes, including VEGF, as well as a those involved in cell proliferation, angiogenesis, and antiapoptosis [39]. Although the authors did not conclude that these changes were directly responsible for biological activity following drug exposure, one can speculate that modulation of mRNA expression may account for the effects observed for spliceostatin A. However, in these studies, the authors did not describe the presence of alternative transcripts, and it is unclear whether their assays were designed to enable for identification of such mRNAs [39]. Hence, it is possible that the dramatic changes in gene expression that they observed were due to inappropriately spliced RNAs produced by spliceostatin A treatment. Because recent articles have demonstrated that the effects of spliceosome modulators on alternative splicing tends to occur in a subset of mRNAs [35], a detailed examination of the change in ratios of the different transcripts present in these cells should be highly informative.
A detailed genome-wide analysis of alternative mRNA splicing in medulloblastoma has demonstrated that many of the genes that are inappropriately spliced in these tumors were also alternatively spliced and expressed in normal cerebellar tissue [40]. However, the ratio of these transcripts was significantly different in tumors, suggesting that it is not the actual presence of the mRNA molecule itself that is important, but rather the levels of the message and presumably the encoded protein. If indeed this represents a global phenomenon, then the spliceosome modulators would alter the ratios of the various transcripts in tumor cells, potentially perturbing proteome expression in a detrimental fashion (Figure 3). Whether this is sufficient to account for the antitumor activity of these compounds is unclear, but these results suggest that both transcriptome sequencing and proteomic analyses of both normal and tumor cells exposed to spliceosome modulators may elucidate key patterns and/or profiles of transcript expression that may correlate with drug response. These studies provide additional weight that the spliceosome may represent a valid target that could be exploited for anticancer drug development.
The mechanism behind the tumor-selective toxicity of the agents that act at SF3b remains an important question; several testable hypothesis can be proposed to guide future work in this area. One possibility is that the drugs are more efficiently transported into the sensitive tumor lines, or possibly the molecules are more efficiently subjected to efflux by resistant tumors and normal cells. Another possibility is that some very sensitive lines may possess tumor suppressors that have been modified by aberrant splicing [41] and that drug treatment may functionally reverse this defect, which then can lead to selective cytotoxicity. Because normal cells maintain appropriate levels of tumor suppressor proteins this would explain the relative resistance of normal cell lines. This hypothesis is consistent with recent published observations including results related to the modulation of splicing of p27 splicing and MDM-2 [21, 34, 42, 43], but hypothetically many other tumor suppressor genes could also be involved, with their importance dependent on the type of tumor line under evaluation.
Alternatively, changes in the cellular response to stress elicited by the presence of aberrant proteins encoded by the alternative spliced RNAs may be more prominent in tumor versus normal cells. For example, the unfolded protein response (UPR) may be initiated as a consequence of these events. Because UPR is known to downregulate mTOR signaling [44], in normal cells, this would likely result in transient inhibition of protein translation and cell growth, and potentially, the initiation of specific repair processes. However, as many tumors are known to have constitutive activation of the mTOR pathway (by mutation of key enzymes within the signaling cascade [45, 46]), catastrophic failure may occur in response to spliceosome modulators due to the loss of crucial cellular components thereby resulting in cytotoxicity. Hence, apparent tumor selectivity would be observed. Finally, the simple loss of proteins crucial for cell growth (e.g. DNA polymerases, key metabolic enzymes, among others) by the disruption of RNA splicing, may also result in the inhibition of mTOR signaling, the effects of which would be more pronounced in tumor cells. Clearly, numerous potential mechanisms could be envisaged for apparent tumor selectivity and elucidating these events will be highly informative with regard to future drug design.
Consequently, we advocate that any future studies detailing the cellular response to spliceosome modulators should include extensive molecular and proteomic analyses, in an attempt to correlate results from these experiments with cytotoxicity. Such studies will enable for a more complete understanding of the role of alternative splicing in cancer biology, and potentially elucidate novel spliceosomal protein targets and processes for therapeutic intervention.
Oncogenic mutations in SF3b1
Beginning in the year 2011, a remarkable convergence of independent lines of genomic research led to a series of publications reporting the association of defined ‘hotspot’ mutations in SF3b1 with several tumor types [47–57]. In one study, transcriptome sequencing identified somatically acquired point mutations in nine patients diagnosed with myelodysplasia [51]. Targeted re-sequencing of this gene was also explored by this group in a larger sample of 2087 patients. Surprisingly, these SF3b1 mutations were identified in 20% of all patients with myelodysplastic syndromes (MDS) [51]. These workers noted that these mutations were associated with the downregulation of significant parts of key gene networks that include some core mitochondrial pathways. These authors also reported mutations in SF3b1 in numerous other cancers, including acute myeloid leukemia, primary myelofibrosis, chronic myelomonocytic leukemia, breast cancer, chronic lymphocytic leukemia (CLL) and multiple myeloma [51]. Figure 4 demonstrates the spectrum of mutations identified in SF3b1 in both MDS and tumor samples. Similarly, the evaluation of the transcriptomes of samples obtained from patients with fludarabine-refractory CLL, identified recurrent mutations at some of the same ‘hotspots’ found in MDS and noted that these were mutually exclusive with p53 mutations [53]. Furthermore, recent reports indicate that recurrent mutations in the U2AF1 splicing factor are associated with another MDS patient population [48, 58], suggesting that alterations in spliceosome-associated proteins likely contribute to the pathogenesis of this disease.
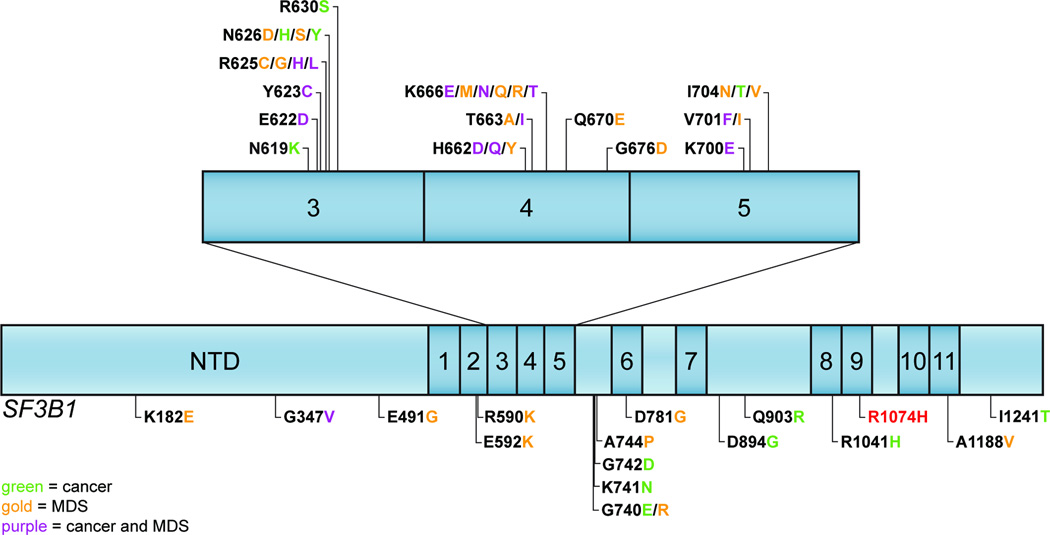
Figure 4.
The spectrum of mutations that have been identified in SF3B1, either in clinical samples, or in cell lines exposed to high concentrations of pladienolide [48, 50–55, 59, 61]. The N-terminal domain (NTD) is indicated, and the numbered boxes represent the HEAT repeats within the protein. Mutations identified in MDS and cancer are indicated in gold and green text (purple for common to both), respectively. The R1074H mutation (marked in red) has been identified in pladienolide resistant cells and expression in a sensitive line confers resistance to this drug. Since this likely encompasses the binding site for the spliceosome modulators, this suggests that tumors expressing the mutant SF3B1 isoforms would be susceptible to treatment with such agents.
Abbreviation: MDS: myelodysplastic syndromes.
Overall, these results suggest that SF3b1 mutations could affect a large total number of patients, across a broad array of tumor types. It is clear that such changes provide a selective advantage for tumor cells, although it is likely that additional cooperative mutations are required for the conversion to the malignant phenotype. Therefore, understanding the significance of these mutations in SF3b1, with regard to both tumor cell biology and the response to small molecules that target these proteins, will be highly informative. In addition, because the potentially oncogenic SF3b1 mutations identified in MDS and tumor samples occur in regions distinct from the apparent drug binding site [59] (Figure 4), it is likely that small molecule inhibitors that target this protein would have activity against cells that harbor these alterations.
Concluding remarks
The spliceosome represents an under-explored and unusual ‘network’ target; however, the recent identification of small molecules that interact with SF3b1, and the identification of mutant SF3b1 in tumor samples, all tend to validate this complex as a viable target for chemotherapeutic intervention. While the exact mechanism of cytotoxicity induced by these agents is unclear, evidence indicates that interaction with SF3b is crucial for their antitumor activity. Because this subunit regulates the fidelity of splice-site selection, these results imply that alternative splicing represents a valid target for drug development. Potentially, the interaction of spliceosome modulators with SF3b leads to an imbalance in the splicing program in susceptible cells, which may induce apoptosis by changing the levels and/or ratios of essential (and aberrant) proteins in tumor cells. The observed selectivity by spliceosome modulators for some tumor types, versus normal cells, warrants the development of additional new molecules that target this process. Additionally, a better understanding of this process may enable the development of prognostic biomarkers useful for the selection of patients that could benefit from the next generation of drugs that target the splicing machinery.
Finally, such compounds are likely to be highly valuable research reagents that can be used to probe the function of the spliceosome. Ultimately however, the development of new efficacious splicing modulators that are suitable for clinical trials, and the success of these agents, will determine whether the spliceosome can be considered a truly valid therapeutic target. There is growing, and well-justified, optimism that this research area may enable new opportunities in cancer chemotherapy.
Highlights.
Details the potential of developing the spliceosome as an anticancer target
Reviews currently available natural product and synthetic spliceosome modulators
Discusses the role of alternative splicing in cancer therapy
Reviews the selective antitumor activity of spliceosomal targeting agents
Elucidates potential mechanisms of cytotoxicity induced by spliceosome modulators
Acknowledgements
Work in the Webb and Potter laboratories is supported in part by the NIH, by the American Lebanese Syrian Associated Charities and St Jude Children’s Research Hospital (SJCRH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Egecioglu DE, Chanfreau G. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA. 2011;17:383–389. doi: 10.1261/rna.2454711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204. doi: 10.4161/rna.7.2.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 6.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr. Opin. Genet. Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 8.Baser ME, et al. The location of constitutional neurofibromatosis 2 (NF2) splice site mutations is associated with the severity of NF2. J. Med. Genet. 2005;42:540–546. doi: 10.1136/jmg.2004.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 10.Kar A, et al. Tau alternative splicing and frontotemporal dementia. Alzheimer Dis. Assoc. Disord. 2005;19(Suppl 1):S29–S36. doi: 10.1097/01.wad.0000183082.76820.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol. Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizui Y, et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J. Antibiot. (Tokyo) 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- 13.Sakai T, et al. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. I. Taxonomy, fermentation, isolation and screening. J. Antibiot. 2004;57:173–179. doi: 10.7164/antibiotics.57.173. [DOI] [PubMed] [Google Scholar]
- 14.Kanada RM, et al. Total synthesis of the potent antitumor macrolides pladienolide B and D. Angew. Chem. Int. Ed. Engl. 2007;46:4350–4355. doi: 10.1002/anie.200604997. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima H, et al. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J. Antibiot. (Tokyo) 1996;49:1204–1211. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima H, et al. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. (Tokyo) 1996;49:1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima H, et al. New antitumor substances, FR901463, FR901464 and FR901465. III. Structures of FR901463, FR901464 and FR901465. J. Antibiot. (Tokyo) 1997;50:96–99. doi: 10.7164/antibiotics.50.96. [DOI] [PubMed] [Google Scholar]
- 18.Miller-Wideman M, et al. Herboxidiene, a new herbicidal substance from Streptomyces chromofuscus A7847. Taxonomy, fermentation, isolation, physico-chemical and biological properties. J. Antibiot. (Tokyo) 1992;45:914–921. doi: 10.7164/antibiotics.45.914. [DOI] [PubMed] [Google Scholar]
- 19.Sakai Y, et al. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J. Antibiot. (Tokyo) 2002;55:855–862. doi: 10.7164/antibiotics.55.855. [DOI] [PubMed] [Google Scholar]
- 20.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 21.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa M, et al. Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS Chem. Biol. 2011;6:229–233. doi: 10.1021/cb100248e. [DOI] [PubMed] [Google Scholar]
- 23.Thompson CF, et al. Total synthesis of FR901464. Convergent assembly of chiral components prepared by asymmetric catalysis. J. Am. Chem. Soc. 2000;122:10482–10483. [Google Scholar]
- 24.Thompson CF, et al. FR901464: total synthesis, proof of structure, and evaluation of synthetic analogues. J. Am. Chem. Soc. 2001;123:9974–9983. doi: 10.1021/ja016615t. [DOI] [PubMed] [Google Scholar]
- 25.Horigome M, et al. A synthesis of FR901464. Tetrahedron Lett. 2001;42:8207–8210. [Google Scholar]
- 26.Motoyoshi H, et al. Structure-activity relationship for FR901464: a versatile method for the conversion and preparation of biologically active biotinylated probes. Biosci. Biotechnol. Biochem. 2004;68:2178–2182. doi: 10.1271/bbb.68.2178. [DOI] [PubMed] [Google Scholar]
- 27.Albert BJ, et al. Total synthesis of FR901464, an antitumor agent that regulates the transcription of oncogenes and tumor suppressor genes. J. Am. Chem. Soc. 2006;128:2792–2793. doi: 10.1021/ja058216u. [DOI] [PubMed] [Google Scholar]
- 28.Motoyoshi H, et al. Total synthesis of FR901464: second generation. Tetrahedron. 2006;62:1378–1389. [Google Scholar]
- 29.Albert BJ, et al. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J. Am. Chem. Soc. 2007;129:2648–2659. doi: 10.1021/ja067870m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albert BJ, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol. Cancer Ther. 2009;8:2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osman S, et al. Structural requirements for the antiproliferative activity of pre-mRNA splicing inhibitor FR901464. Chemistry. 2011;17:895–904. doi: 10.1002/chem.201002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagisetti C, et al. Antitumor compounds based on a natural product consensus pharmacophore. J. Med. Chem. 2008;51:6220–6224. doi: 10.1021/jm8006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagisetti C, et al. Synthetic mRNA splicing modulator compounds with in vivo antitumor activity. J. Med. Chem. 2009;52:6979–6990. doi: 10.1021/jm901215m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan L, et al. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 2011;6:582–589. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrionero A, et al. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germann S, et al. Splicing programs and cancer. J. Nucleic Acids. 2012;269570 doi: 10.1155/2012/269570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folco EG, et al. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corvelo A, et al. Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 2010;6:E1001016. doi: 10.1371/journal.pcbi.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furumai R, et al. Spliceostatin A blocks angiogenesis by inhibiting global gene expression including VEGF. Cancer Sci. 2010;101:2483–2489. doi: 10.1111/j.1349-7006.2010.01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menghi F, et al. Genome-wide analysis of alternative splicing in medulloblastoma identifies splicing patterns characteristic of normal cerebellar development. Cancer Res. 2011;71:2045–2055. doi: 10.1158/0008-5472.CAN-10-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghigna C, et al. Alternative splicing and tumor progression. Curr. Genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allende-Vega N, et al. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene. 2012 doi: 10.1038/onc.2012.38. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 43.Gundluru MK, et al. Design, synthesis and initial biological evaluation of a novel pladienolide analog scaffold. Medchemcomm. 2011;2:904–908. doi: 10.1039/C1MD00040C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin L, et al. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 45.Emerling BM, Akcakanat A. Targeting PI3K/mTOR signaling in cancer. Cancer Res. 2011;71:7351–7359. doi: 10.1158/0008-5472.CAN-11-1699. [DOI] [PubMed] [Google Scholar]
- 46.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr. Opin. Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damm F, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2011;26:1137–1140. doi: 10.1038/leu.2011.321. [DOI] [PubMed] [Google Scholar]
- 48.Hahn CN, Scott HS. Spliceosome mutations in hematopoietic malignancies. Nat. Genet. 2011;44:9–10. doi: 10.1038/ng.1045. [DOI] [PubMed] [Google Scholar]
- 49.Lasho TL, et al. SF3B1 mutations in primary myelofibrosis: clinical, histopathology and genetic correlates among 155 patients. Leukemia. 2011;26:1135–1137. doi: 10.1038/leu.2011.320. [DOI] [PubMed] [Google Scholar]
- 50.Malcovati L, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 53.Rossi D, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 56.Hirabayashi S, et al. Spliceosomal gene aberrations are rare, coexist with oncogenic mutations, and are unlikely to exert a driver effect in childhood MDS and JMML. Blood. 2012;119:E96–E99. doi: 10.1182/blood-2011-12-395087. [DOI] [PubMed] [Google Scholar]
- 57.Patnaik MM, et al. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119:569–572. doi: 10.1182/blood-2011-09-377994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokoi A, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 60.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 61.Visconte V, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26:542–545. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]