Abstract
The corneal endothelium is composed of a monolayer of corneal endothelial cells (CECs), which is essential for maintaining corneal transparency. To better characterize CECs in different developmental stages, we profiled mRNA transcriptomes in human fetal and adult corneal endothelium with the goal to identify novel molecular markers in these cells. By comparing CECs with 12 other tissue types, we identified 245 and 284 signature genes that are highly expressed in fetal and adult CECs, respectively. Functionally, these genes are enriched in pathways characteristic of CECs, including inorganic anion transmembrane transporter, extracellular matrix structural constituent and cyclin-dependent protein kinase inhibitor activity. Importantly, several of these genes are disease target genes in hereditary corneal dystrophies, consistent with their functional significance in CEC physiology. We also identified stage-specific markers associated with CEC development, such as specific members in the transforming growth factor beta and Wnt signaling pathways only expressed in fetal, but not in adult CECs. Lastly, by the immunohistochemistry of ocular tissues, we demonstrated the unique protein localization for Wnt5a, S100A4, S100A6 and IER3, the four novel markers for fetal and adult CECs. The identification of a new panel of stage-specific markers for CECs would be very useful for characterizing CECs derived from stem cells or ex vivo expansion for cell replacement therapy.
GEO accession number: GSE41616.
INTRODUCTION
Corneal endothelial cells (CECs) are a monolayer of endothelial cells lining the Descemet's membrane of the cornea. CECs exhibit a typical hexagonal shape and form tight junctions with the neighboring cells. The main function of CECs is to serve as a water pump via Na+–K+-ATPase, which actively transports bicarbonate ion from the stroma into the anterior chamber. The active bicarbonate ion flux provides the driving force for pumping the corneal fluid out of the stroma, thus keeping the stroma in a constant state of hydration to achieve corneal transparency. Adult CECs have limited capacity for cell proliferation in vivo as the cell cycle is arrested in the G1 phase (1), and CEC deficiency due to genetic diseases, trauma and aging would lead to irreversible corneal edema, opacity and eventual blindness (2).
CEC genetic disorders include Fuchs endothelial corneal dystrophy (FECD), posterior polymorphous corneal dystrophy (PPCD), congenital hereditary endothelial dystrophy type 1 and 2 (CHED1 and 2) and X-linked endothelial corneal dystrophy (3). The genetic defects underlying some of these hereditary diseases have been recently reported. For example, mutations in Collagen type VIII A2 (COL8A2), a major component of the Descemet's membrane secreted by CECs, are associated with either early-onset FECD or PPCD, whereas mutations in the solute carrier family 4 member A11 (SLC4A11) gene are related to late-onset FECD and CHED2 (4). Nevertheless, many disease target genes remain to be discovered.
Clinically, besides entire corneal transplantation, surgical procedures such as endokeratoplasty or Descemet's stripping endothelial keratoplasty is developed to replace dysfunctional CECs with healthy corneal endothelium (5). However, due to the global shortage of tissue donors and limited transplant-grade cornea tissues, new methods to generate functional CECs need to be established. Recent advancements in stem cell biology have opened new avenues to expand adult or fetal CECs ex vivo (6) or derive CECs from neural crest cells (7), or even with human somatic stem cells such as corneal stroma stem cells, mesenchymal stem cells and bone marrow-derived endothelial progenitor cells (8–10). Theoretically, in vitro differentiation of CECs from pluripotent stem cells such as human embryonic stem cells (hESCs) or induced pluripotent stem cells would be a very feasible approach to provide unlimited source of CECs for cell replacement therapy.
Several protein markers have been used to identify adult CECs, most related to cell adhesion, gap junction, tight junctions and pump function such as N-cadherin, connexin-43 (11), ZO-1 (12), Na+–K+-ATPase (13) and Occludin (14). However, molecular markers unique to fetal and adult CECs and molecular pathways associated with maturation of CECs are not well studied. We therefore took a comprehensive approach to profile mRNA transcriptomes in both fetal and adult human CECs (HCECs) by RNA-sequencing (RNA-seq) and examine unique molecular markers in fetal and adult CECs. Bioinformatics analysis identified major gene expression changes between fetal and adult CECs in cell metabolism, cell adhesion and the transforming growth factor beta (TGF-β) signaling pathway. By further comparing CEC gene expression profiles with 12 other cell types, we identified 245 and 284 tissue-specific genes in fetal and adult CECs, respectively. In fact, several endothelial dystrophy genes are highly expressed in adult CECs, consistent with their implicated role in CEC physiology. Significantly, we validated a portion of the fetal and adult-specific markers using immunohistochemistry, demonstrating that our data analysis is a valuable resource for identifying protein biomarkers unique to fetal and adult CECs.
RESULTS
Tissue-specific gene expression in fetal and adult CECs
Because markers for either fetal or adult CEC are not well studied, we performed RNA-Seq in three adult and two fetal CEC tissues and produced ∼179 million uniquely mapped reads. To identify genes that were specific to either fetal or adult CEC, we procured 97 samples from the public domain representing 12 different types of cells and tissues (Supplementary Material, Table S2). For analytical reasons, we only selected RNA-Seq datasets generated by the Illumina system to prevent potential platform biases.
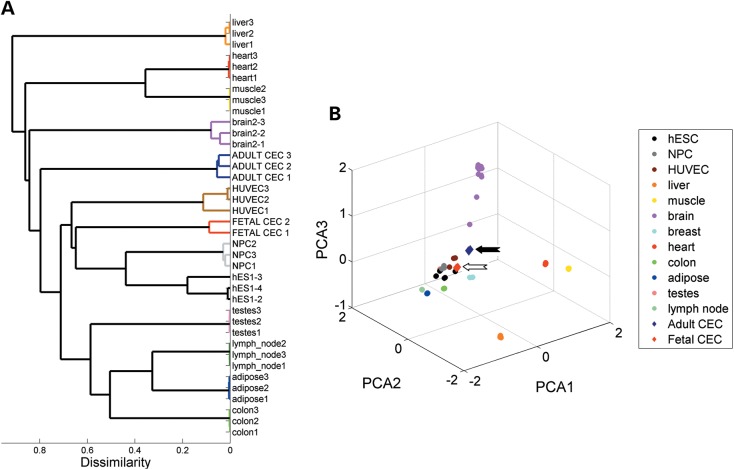
Hierarchal clustering revealed that fetal CECs cluster closer to relatively immature cell types, such as hESCs, neural progenitor cells (NPCs) and human umbilical vein endothelial cells (HUVECs; Fig. 1A). In contrast, adult CECs clustered away from these early stage cells and grouped together with other terminally differentiated somatic cell types. By principal component analysis (Fig. 1B), we found that adult CECs are positioned closer to adult brain samples, consistent with the fact that CECs are derived from neural crest cells that share the same cell origin of the neural tube. Overall, identical cell types clustered tightly together, suggesting that each cell type exhibits well-defined transcriptional patterns. In particular, adult and fetal CEC clustered closely together compared with other cell types, indicating that CECs also have a unique transcriptional profile.
Figure 1.
Adult and fetal CECs exhibit distinct global mRNA expression patterns. (A) Unbiased hierarchical clustering shows fetal and adult CEC relationship with representative samples from other tissues and cell types. (B) Three-dimensional scatterplot of the first three principal components (PCA1, PCA2 and PCA3) demonstrate that fetal samples cluster to hESCs, NPCs and HUVECs, and adult CEC cluster closer to brain samples. Ninety-seven samples from 12 different tissue and cell types were downloaded from the GEO database. Solid and open arrows point out adult and fetal CECs.
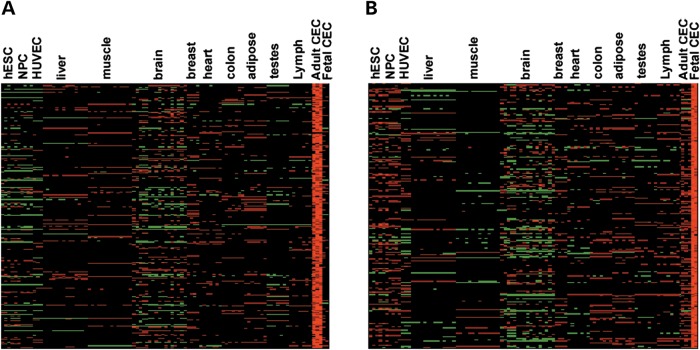
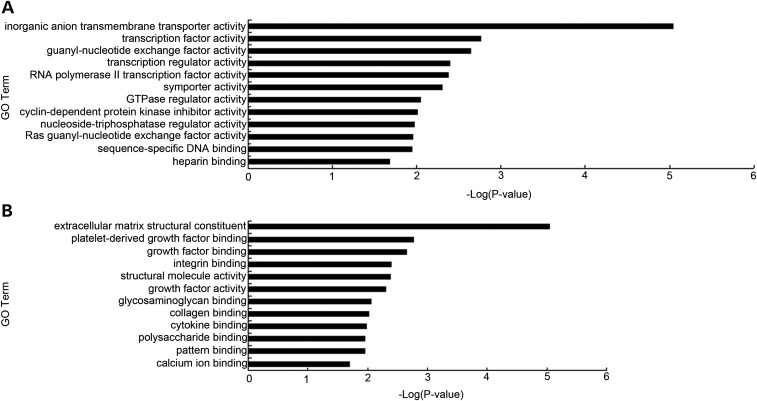
To gain insight into the exact molecular differences between adult and fetal CECs from other cell types, we performed statistical analysis (one-way analysis of variance, or ANOVA) and identified genes that were specifically overexpressed in either adult or fetal CECs. Using the criteria of ANOVA Tukey–Kramer multiple comparison corrected P-values <0.01 and an additional cutoff of at least 2-fold greater expression compared with other 12 tissues, we identified a total of 284 (Fig. 2A) and 245 genes (Fig. 2B) that are specifically enriched in adult and fetal CECs, respectively. Gene ontology (GO) analysis revealed that the 284 up-regulated genes in adult CECs mainly participate in inorganic anion transmembrane transporter activity, transcription factor activity and cyclin-dependent protein kinase inhibitor activity (Fig. 3A), reflecting the unique features of mature CECs. Genes unique to fetal CECs are involved in extracellular matrix structural constituent, platelet-derived growth factor binding and growth factor activity (Fig. 3B). These results probably reflect the specific functional traits of fetal CECs for growth factor contribution during fetal CEC development.
Figure 2.
Heatmap analysis of the up-regulated gene in adult and fetal CECs. Heatmap of relative expression levels for genes that are highly expressed in (A) adult or (B) fetal CECs.
Figure 3.
GO analysis of CEC tissue-specific signature genes. Bar graphs showing the significance of enrichment terms for a set of uniquely expressed genes in (A) adult CECs and (B) fetal CECs compared with 12 tissue and cell types. P-values <0.05.
We next took a systems approach and asked whether adult or fetal CECs express a unique transcriptional network (or module) of genes. By weighted gene co-expression network analysis (WGCNA), we identified single modules that represent adult (n = 300) and fetal (n = 343) CECs transcriptional networks (Supplementary Material, Figs S1 and S2). When we overlapped this result with our single-gene statistical analysis, we found fairly good concordance with 70% overlap with adult (n = 209) and 44% in fetal (n = 160) CECs (Supplementary Material, Table S1). Taken together, our multi-approach analysis suggests that the unique gene expression profiles in fetal and adult CECs are highly robust.
WGCNA can also lead to a natural measure of gene similarity (or membership) to a particular module. Genes with high membership in a module are generally regarded as hub genes, and tend to play central roles in the network. Interestingly, in adult CECs, we identified SLC4A11 as one of the top-ranking hub genes. SLC4A11 has been linked with hereditary corneal dystrophies (15,16) and knockdown of SLC4A11 in cultured CECs has been shown to induce apoptosis (17). Thus, our network analysis helped to refine and rank the set of key players in the adult or fetal CEC networks. We provided a ranked list of adult and fetal CEC stage-specific hub genes (Supplementary Material, Table S1). Together, our analysis has uncovered a clear transcriptional signature in adult and fetal CECs that are not present in other cell and tissue types examined, including other types of endothelial cells.
Differential expression pattern between adult and fetal CECs
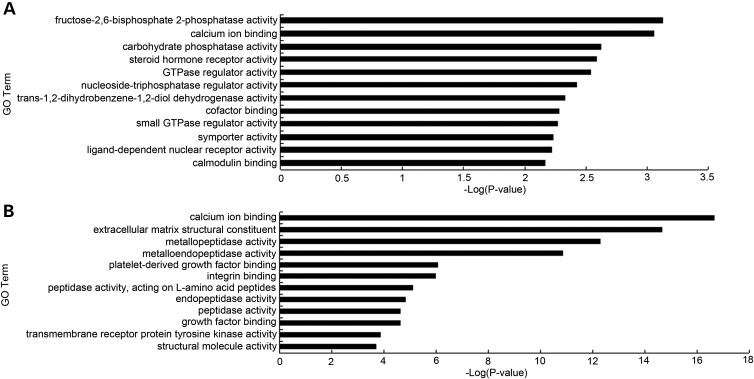
We next directly compared adult and fetal CECs and identified 688 and 1763 genes that are expressed by more than 2-fold [false discovery rate (FDR) < 5%] in adult and fetal CECs, respectively. To increase the stringency and identify the most differentially expressed genes between adult and fetal CECs, we used a 5-fold change cutoff, and by this criterion, we identified 518 and 668 genes that are highly expressed in adult and fetal CECs, respectively. Remarkably, functional annotation of the differentially expressed genes suggests distinct functional differences between adult and fetal CECs. For example, GO and kyoto encyclopedia of genes and genomes (KEGG) pathway analysis indicated that many genes involved in metabolic processes are up-regulated in adult CECs (Fig. 4A and Table 1), whereas fetal CECs showed GO terms enriched in metallopeptidase activity, extracellular structure and TGF-β signaling (Fig. 4B and Table 2). Interestingly, calcium ion binding proteins such as S100 family members (S100A4, S100A5 and S100A6) are expressed higher in adult CECs. Collectively, our analysis revealed clear differences between adult and fetal CECs, indicating that CECs acquire transcriptional changes in select pathways during developmental maturation.
Figure 4.
GO analysis of CEC stage-specific signature genes. Bar graphs showing the significance of enrichment terms for a set of higher expressed genes in (A) adult over fetal CECs and (B) fetal over adult CECs. P-values <0.05.
Table 1.
Analysis of KEGG pathway via DAVID software for 518 adult stage-specific CECs compared with fetal CECs
| Term | Count | Genes | P-value |
|---|---|---|---|
| Glycolysis/Gluconeogenesis | 10 | ALDOA, GPI, LDHA, ALDOC, PKM2, ENO2, FBP1, PGK1, ALDH3B1, ENO1 | 1.96E−04 |
| Arginine and proline metabolism | 8 | GLS2, SAT1, GLUL, GOT1, ACY1, GATM, NAGS, NOS3 | 0.002188 |
| Fructose and mannose metabolism | 6 | ALDOA, PFKFB4, AKR1B10, PFKFB2, ALDOC, FBP1 | 0.005918 |
| Pentose phosphate pathway | 5 | ALDOA, GPI, ALDOC, FBP1, RBKS | 0.010124 |
| p53 signaling pathway | 7 | STEAP3,TNFRSF10B, CCND3,ZMAT3,CYCS, GADD45G, RRM2B | 0.030056 |
| Nitrogen metabolism | 4 | GLS2, CTH, GLUL, CA3 | 0.044035 |
P-value <0.05.
Table 2.
Analysis of KEGG pathway via DAVID software for fetal CEC stage-specific gene expression
| Term | Count | Genes | P-value |
|---|---|---|---|
| ECM-receptor interaction | 22 | COL4A2, COL4A1, COL3A1, COL2A1, COL5A, COL5A1, SDC3, LAMA2, GP5, LAMA4, ITGA5, TGA8, ITGA7, COL6A3, COL1A2, COL6A2, COL6A1, TNN, COL1A1, THBS2, COL11A1, FN1 | 1.63E−12 |
| Axon guidance | 22 | PLXNC1, PLXNA1, GNAI1, EFNB1, EFNB2, EPHB3, CXCL12, EPHA3, SLIT3, EPHB2, SEMA5B, EPHB6, SEMA6B, EPHA6, CXCR4, ROBO1, FYN, SEMA3F, SEMA3D, ROBO2, SEMA3A, SRGAP1 | 8.34E − 09 |
| Focal adhesion | 26 | COL3A1, COL2A1, BCL2, COL6A3, COL6A2, COL6A1, TNN, PIK3R3, THBS2, COL11A1, FN, COL4A2, COL4A1, IGF1, FLNC, COL5A2, COL5A1, LAMA2, LAMA4, ITGA5, FYN, CCND2 ITGA8, ITGA7, COL1A2, COL1A1 | 8.22E−08 |
| Arrhythmogenic right ventricular cardiomyopathy | 10 | LAMA2, SLC8A1, CACNA2D1, PKP2, ITGA5, ITGA8, ITGA7, LEF1, CACNG4, SGCD | 0.001988 |
| Hypertrophic cardiomyopathy | 10 | LAMA2, SLC8A1, CACNA2D1, ITGA5, ITGA8, ITGA7, IGF1, CACNG4, SGCD, TGFB2 | 0.004299 |
| Pathways in cancer | 23 | FGF18, RET, AR, COL4A2, BMP2, COL4A1, LEF1, KITLG, IGF1, CDK6, FGF12, GLI2, MMP2, FZD7, TGFB2, LAMA2, SMO, FZD10, CBLB, LAMA4, BCL2, PIK3R3, FN1 | 0.005296 |
| Dilated cardiomyopathy | 10 | LAMA2, SLC8A1, CACNA2D1, ITGA5, ITGA8, ITGA7, IGF1, CACNG4, SGCD, TGFB2 | 0.007243 |
| Cell adhesion molecules | 12 | NCAM2, CADM3, CLDN19, CD34, ITGA8, CD276, NLGN1, NLGN3, CDH3, JAM2, SDC3, CLDN15 | 0.010309 |
| Small cell lung cancer | 8 | LAMA2, LAMA4, COL4A2, COL4A1, BCL2, CDK6, PIK3R3, FN1 | 0.037463 |
| TGF-beta signaling pathway | 8 | BMP2, LTBP1, ACVRL1, LEFTY2, DCN, THBS2, CHRD, TGFB2 | 0.044046 |
| Gap junction | 8 | TUBB2B, GNAI1, TUBB6, LPAR1, PLCB1, ITPR3, TUBA1A, TUBB4 | 0.048826 |
P-value <0.05.
Stage-specific marker genes in adult and fetal CECs are confirmed at the protein level
Although we have identified many putative molecular markers to distinguish adult and fetal CECs on the RNA level, it is still unclear whether these markers can be confirmed at the protein level. We therefore collected fetal and adult corneal tissues for immunohistochemistry analysis. As expected, immunostaining analysis showed expression of tight junction protein ZO-1 in both fetal (17 weeks in gestation) and adult CECs (>37 years old; Supplementary Material, Fig. S3A and B). Na+–K+-ATPase, which is one of the most important markers for the pump function in CECs, is highly expressed in adult CECs but not in fetal CECs (Supplementary Material, Fig. S3C and D), consistent with the observations in our RNA-Seq analysis (Supplementary Material, Table S3). This result suggests that pump function is not yet fully established in 17-week-old fetal CECs.
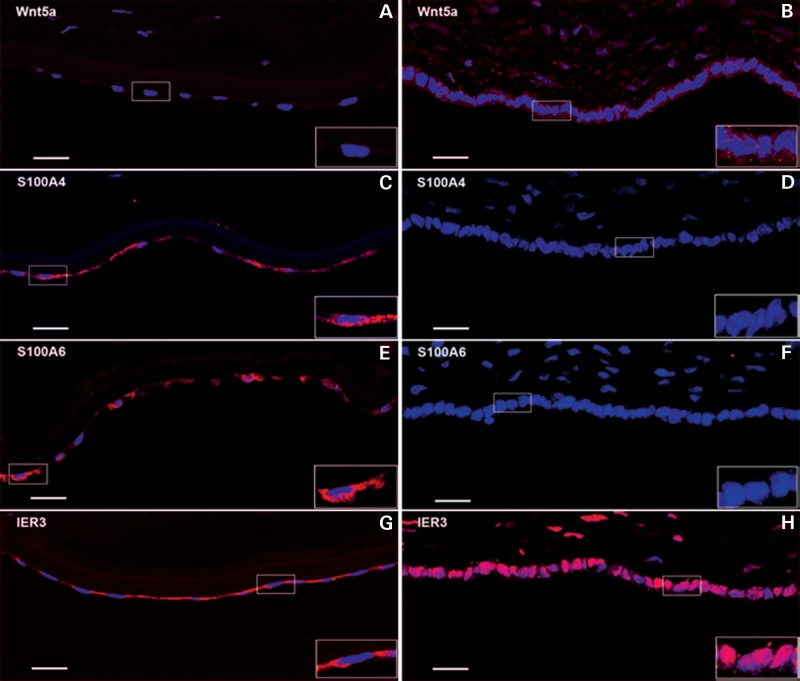
We next examined components of the Wnt signaling pathways since it is known to be an important regulator of ocular tissue development (18,19). Indeed, genes involved in Wnt signaling are highly enriched in fetal CECs (Supplementary Material, Table S1). Our immunocytochemistry experiment showed that Wnt intracellular signaling molecules such as axin2 (Supplementary Material, Fig. S3E and F) and beta-catenin (Supplementary Material, Fig. S3G and H) are readily detected in both fetal and adult CECs. However, we found that Wnt5a is only expressed in fetal, but not adult CECs (Fig. 5A and B), suggesting that, at the protein level, the Wnt5a pathway is only functional in fetal CECs.
Figure 5.
Immunostaining of novel marker proteins expressed in adult and fetal CECs. The bottom monolayer of the samples depicts corneal endothelium. A red signal shows the expression of each antigen as detected by a specific antibody. The blue signal is Hoesch dye. Left panels show adult CEC samples and right panels display fetal CEC samples. (A) and (B) are immunofluorescent labeling of Wnt5a in adult and fetal CECs; (C) and (D) detect S100A4 expression in adult and fetal CECs; (E) and (F) for S100A6; (G) and (H) showing IER3 protein localization in adult and fetal CECs. Scale bar, 20 μm.
The S100 protein family consists of several calcium-binding proteins and have been shown to regulate a variety of cellular processes (20). We found that mRNAs in sub-members of the S100A protein family (S100A4, S100A5 and S100A6) are highly expressed in adult, but not fetal CECs (Supplementary Material, Table S1). Our immunohistochemistry confirmed that S100A4 and S100A6 proteins were readily detected in adult CECs (Fig. 5C and E) or fetal cornea epithelium (Supplementary Material, Fig. S4), but not in fetal cornea endothelium (Fig. 5D and F). Therefore, S100A4 and S100A6 calcium-binding proteins are two additional novel markers for distinguishing mature adult versus immature fetal CECs.
We also examined cellular localization for proteins that are highly expressed in both adult and fetal CECs. Significantly, we found novel localizations of intracellular proteins such as a stress-induced protein Immediate Early Response 3 (IER3) during CEC maturation. Our immunostaining experiment demonstrated that IER3 can be readily detected in both fetal and adult CECs, but with a stark contrast in subcellular localization. IER3 localizes in the cytoplasm of adult CECs, whereas it is found in the nucleus of fetal CECs. Thus, IER3 is another useful protein marker to characterize the maturation of CECs in development (Fig. 5G and H).
DISCUSSION
CECs are critical for maintaining corneal transparency through regulating stromal hydration. Although previous studies have examined select CEC gene expression via RT–PCR, no information is available concerning the entire transcriptome of either fetal or adult CECs. In this study, we conducted a comprehensive mRNA-Seq analysis of fetal and adult CECs, uncovering both tissue- and stage-specific gene transcripts. These molecular signatures of fetal and adult CECs would be very useful to develop biomarkers to characterize different stages of CECs both in vivo and in vitro.
Here, we have found that genes highly expressed in adult CECs are tightly associated with special CEC functions. For example, the barrier function of CECs is set up by tight junction, which is well correlated with a high level of ZO-1 and N-cadherin mRNAs in adult CECs. Similarly, AQP1, Na+–K+-ATPase, Na+/HCO3− and other genes relevant to pump function of mature CECs are detected at a higher level in adult CECs than in fetal CECs. Adult CECs are in a quiescent state with very limited proliferation ability. Consistently, in our analysis, we found that Ki67 and cyclinD2 are dramatically down-regulated in adult CECs compared with fetal CECs.
It has been postulated that both Wnt and TGF-β pathways play a role in CEC development. The Wnt pathways can be separated into two different pathways: canonical frizzle receptor/beta-catenin-dependent T cell factor pathway and non-canonical Ryk receptor/Jun NH2-terminal kinase and calcium/calmodulin-dependent kinase pathways. These two pathways are targeted by different Wnt molecules such as Wnt1, Wnt3a and Wnt8 for the canonical pathway, whereas Wnt5a, Wnt4 and Wnt11 for the non-canonical pathway (21). Of note, we found that 27 genes in the Wnt pathway are highly expressed in fetal CECs (Supplementary Material, Tables S1 and S4), particularly for the non-canonical Wnt pathway genes such as Wnt5a and Wnt4, raising the possibility that the non-canonical Wnt pathway may be important for CEC development. Similarly, eight TGF-β signaling pathway members (TUBB2B, GNAI1, TUBB6, LPAR1, PLCB1, ITPR3, TUBA1A and TUBB4) are detected at a much higher level in fetal CECs than in adult CECs. TGF-β family genes are involved in regulating cell differentiation, proliferation and extracellular matrix production in ocular tissue (22). In fact, TGF-β2 null mice exhibit an absence of endothelium and fusion of the lens with the cornea (23). In coupling with high levels of TGF-β signaling molecules, we also observed enrichment of genes in the extracellular matrix structural constituent in fetal CECs. Overall, our stage-specific gene expression analysis supports the notion that both Wnt and TGF-β pathways are important for the differentiation and maturation of fetal CECs into adult CECs.
Recently, derivation of CEC-like cells in vitro from progenitor cells or somatic stem cells has been investigated for transplantation treatment of CEC deficiency. Hatou et al. (8) isolated cornea-derived precursors from the adult corneal stroma to make tissue-engineered corneal endothelium. Joyce et al. (9) investigate the potential of umbilical cord blood mesenchymal stem cells to become HCECs. Recently, Ju et al. (7) tested the feasibility of differentiating neural crest cells into functional corneal endothelial cells. In all these studies, the markers used for characterizing CEC-like cells were limited to ZO-1, Na+–K+-ATPase and N-cadherin. Our demonstration of four novel protein markers including S100A4, S100A6, Wnt5a and IER3 in this study will certainly aid the characterization of CEC-like cells from the above studies. S00A4 is one of S100 family genes encoding calcium-binding proteins involved in cell morphogenesis in multiple cell lineages (24). S100A6 was previously shown in the plasma membrane of corneal and limbal epithelial cells (25). It has been debated whether S100 expression is specific in human corneal endothelium (26,27). In the present study, we found that S100A4 and S100A6 are detectable in adult CECs as well as in adult and fetal corneal epithelium, but not in fetal CECs. Thus, S100A4 or S100A6 should be considered as a mature marker for adult CECs. IER3 is a stress-induced protein (28) encoded by radiation-inducible immediate-early gene IEX-1, previously detected as a potential target gene in human cancer, inflammatory diseases or hypertension (29,30). In keratinocytes, it is observed that IER3 can translocate from nucleus to cytoplasm in response to 1alpha,25-dihydroxyvitamin D3 for reduced cell growth (31). Consistently, our observed IER3 translocation from the nucleus in fetal CECs to the cytoplasm in adult CECs may also be associated with reduced cell proliferation and growth in adult CECs. Taken together, our newly identified CEC protein markers as well as stage-specific CEC mRNAs would be very useful to characterize and quality-control a variety of CEC-like cells derived from either ex vivo expansion of corneal progenitors or in vitro differentiation of somatic and pluripotent stem cells.
Lastly, corneal endothelial dystrophies are a set of hereditary corneal diseases characterized by slowly progressive edema of the cornea in different modes, each associated with different target genes. We noted that several of our newly identified CEC-tissue-specific genes are also corneal disease target genes. For example, COL8A2 and SLC4A11, two major PPCD-related genes (32), are expressed at an extremely high level in both fetal and adult CECs. Similarly, CDKN1A exhibits a much higher expression level in adult CECs than fetal CECs. CDKN1A is an important marker of cellular senescence (33) and mediates part of the cellular stress response in the pathogenesis of both early and late-onset FECD (34). Our current study identifies over two hundred genes that are highly expressed in adult and fetal CECs when compared with 12 other tissue types (Supplementary Material, Table S2). Because these CEC signature genes are enriched for the genes critical for CEC functions, we speculate that some of these highly expressed signature genes in adult and fetal CECs could be good candidates of disease target genes underlying many other hereditary CEC dystrophies.
MATERIALS AND METHODS
Adult and fetal CEC samples
Adult cornea samples (age 31, 56 and 64 years old) were obtained from normal subjects who donated to the eye bank in UCLA Jules Stein Eye Institute. Fetal CEC samples were dissected from fetal ocular samples (16–18 weeks of gestation) provided by UCLA Department of Pathology & Laboratory as approved by UCLA IRB. The whole corneal endothelial layers were carefully peeled off under a dissecting microscope and were immediately lysed in reagents for RNA purification.
RNA isolation
RNA were extracted from those adult and fetal tissues using the RNeasy Micro Kit (Qiagen, USA). RNA concentration was detected by nanodrop spectrophotometer.
Library construction and high-throughput sequencing
RNA-Seq library construction followed the protocol described in Illumina TruSeq™ RNA Sample Preparation Guide. Library construction was started with 100 ng of total RNA, via poly-T oligo-attached magnetic beads to purify the poly-A containing mRNA molecules. RNA fragments were reverse-transcribed into first cDNA strand, followed by synthesis second cDNA strand. After end repaired with a single ‘A’ base, cNDAs were ligated with different adapters for PCR amplification. After being amplified for 15 cycles, the concentration of the product was tested by the Qubit Fluorometer (Invitrogen, USA). We subjected 10 nmol of each sample to sequencing according to the manufacturer's instruction with Illumin Hi-seq 2000. Reads were mapped to the hg19 genome using burrows-wheeler alignment (35).
Statistical and bioinformatics analysis
For all datasets analyzed, we first transformed mapped transcript read counts to the reads per kilobase per million mapped reads (RPKM) metric, and genes with an average RPKM < 1 were filtered out, followed by quantile normalization. We performed Student's t-test with Storey's multiple testing correction to produce the FDR. Genes that had greater than a mean fold-change of 2 and FDR <5% were considered differentially expressed. All statistical analysis was performed using built-in Matlab functions. GO and KEGG pathway analyses were performed via David bioinformatics on-line software.
Tissue-specific expression
We curated multiple samples from the public domain, restricting ourselves to datasets generated by the Illumina Solexa platform to reduce platform biases. We carried out one-way ANOVA, and then selected for genes that were specifically overexpressed in either adult or fetal CEC samples. Two cutoffs were used: (1) Tukey–Kramer multiple comparison corrected P-values <0.01, which is a relatively conservative method for one-way ANOVA with different sample sizes and (2) mean 2-fold expression over all tissue types. These analyses were performed in Matlab using built-in functions (anova1, and multcomp).
Weighted gene co-expression network analysis
We constructed a signed weighted correlation network as previously described (36). Briefly, a matrix of pairwise correlations was generated between all pairs of genes across all samples. This matrix was raised to the power 12 to produce the adjacency matrix, which is used to calculate the Topological Overlap (TO), a robust and biologically meaningful measure of network interconnectedness. Genes with highly similar co-expression relationships were grouped together by performing average linkage hierarchical clustering on the TO. Module membership was calculated for each gene i in module q, as MMiq = cor(xi, Eq), where xi is the expression profile of gene i and Eq is the eigengene of module q. Genes with high module memberships are generally regarded as hub genes. WGCNA is a publically available R package that has been described in detail in the reference (36).
Immunostaining
Adult and fetal cornea tissues were fixed with 4% PFA for 30 min at room temperature, and cryo-protected in 30% sucrose/PBS overnight. After embedded in OCT, frozen section were cut using a microtome-cryostat (Leica, USA) in 10 μm. The sections were washed three times in phosphate-buffered saline (PBS) and then permeabilized in 0.4% Triton-X 100 for 1 h. Blocking was carried out using 4.5% normal donkey serum, then sections were incubated overnight at 4°C in primary antibodies diluted in blocking solution. The primary antibodies used in the experiment were anti-Wnt5a (Santa Cruz), S100A4 (Abnova), S100A6 (Abnova), IER3 (Abgent), ZO-1 (Invitrogen), Na+–K+-ATPase (Millipore), Axin2 (abcam) and Beta-catenin (BD). After being washed three times in PBS, sections were incubated in fluorochrome-conjugated secondary antibodies for 1 h at room temperature, cell nuclei staining was done by Hoechst, and the slides were mounted with 5% n-propylgallate (Sigma) in 50% glycerol. Immunostaining results were taken using a 40× objective under fluorescent light with a confocal microscope (Leica TCS-SP; Leica Microsystems).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH grant GM081621, International Science & Technology Cooperation Program of China #2011DFB30010, Ministry of Science and Technology 973 Basic Research program in China (2011CB965102, 2011CB966204 and 2012CB966303), National Natural Science Foundation of China (81271258). Y.C. is partly supported by Chinese Scholarship Council (CSC) funding (No. 2011626117). K.H. is supported by the California Institute of Regenerative Medicine Training Grant (TG2-01169).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the assistance of UCLA Broad Stem Cell Research Center Sequencing Core for high-throughput sequencing.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Koizumi N., Okumura N., Kinoshita S. Development of new therapeutic modalities for corneal endothelial disease focused on the proliferation of corneal endothelial cells using animal models. Exp. Eye Res. 2012;95:60–67. doi: 10.1016/j.exer.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Peh G.S., Beuerman R.W., Colman A., Tan D.T., Mehta J.S. Human corneal endothelial cell expansion for corneal endothelium transplantation: an overview. Transplantation. 2011;91:811–819. doi: 10.1097/TP.0b013e3182111f01. [DOI] [PubMed] [Google Scholar]
- 3.Weiss J.S., Moller H.U., Lisch W., Kinoshita S., Aldave A.J., Belin M.W., Kivela T., Busin M., Munier F.L., Seitz B., et al. [The IC3D classification of the corneal dystrophies] Klin. Monbl. Augenheilkd. 2011;228(Suppl. 1):S1–S39. doi: 10.1055/s-0029-1245895. [DOI] [PubMed] [Google Scholar]
- 4.Schmedt T., Silva M.M., Ziaei A., Jurkunas U. Molecular bases of corneal endothelial dystrophies. Exp. Eye Res. 2012;95:24–34. doi: 10.1016/j.exer.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosheh F.R., Cremona F.A., Rapuano C.J., Cohen E.J., Ayres B.D., Hammersmith K.M., Raber I.M., Laibson P.R. Trends in penetrating keratoplasty in the United States 1980–2005. Int. Ophthalmol. 2008;28:147–153. doi: 10.1007/s10792-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y., Zhou Q., Qu M., Yang L., Wang Y., Shi W. In vitro culture of human fetal corneal endothelial cells. Graefes Arch. Clin. Exp. Ophthalmol. 2011;249:663–669. doi: 10.1007/s00417-010-1547-y. [DOI] [PubMed] [Google Scholar]
- 7.Ju C., Zhang K., Wu X. Derivation of corneal endothelial cell-like cells from rat neural crest cells in vitro. PLoS One. 2012;7:e42378. doi: 10.1371/journal.pone.0042378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatou S., Yoshida S., Higa K., Miyashita H., Inagaki E., Okano H., Tsubota K., Shimmura S. Functional corneal endothelium derived from corneal stroma stem cells of neural crest origin by retinoic acid and Wnt/beta-catenin signaling. Stem Cells Dev. 2012 doi: 10.1089/scd.2012.0286. doi:10.1089/scd.2012.0286. [DOI] [PubMed] [Google Scholar]
- 9.Joyce N.C., Harris D.L., Markov V., Zhang Z., Saitta B. Potential of human umbilical cord blood mesenchymal stem cells to heal damaged corneal endothelium. Mol. Vis. 2012;18:547–564. [PMC free article] [PubMed] [Google Scholar]
- 10.Shao C., Fu Y., Lu W., Fan X. Bone marrow-derived endothelial progenitor cells: a promising therapeutic alternative for corneal endothelial dysfunction. Cells Tissues Organs. 2011;193:253–263. doi: 10.1159/000319797. [DOI] [PubMed] [Google Scholar]
- 11.Jongen W.M., Fitzgerald D.J., Asamoto M., Piccoli C., Slaga T.J., Gros D., Takeichi M., Yamasaki H. Regulation of connexin 43-mediated gap junctional intercellular communication by Ca2+ in mouse epidermal cells is controlled by E-cadherin. J. Cell Biol. 1991;114:545–555. doi: 10.1083/jcb.114.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petroll W.M., Barry-Lane P.A., Cavanagh H.D., Jester J.V. ZO-1 reorganization and myofibroblast transformation of corneal endothelial cells after freeze injury in the cat. Exp. Eye Res. 1997;64:257–267. doi: 10.1006/exer.1996.0211. [DOI] [PubMed] [Google Scholar]
- 13.Crawford K.M., Ernst S.A., Meyer R.F., MacCallum D.K. NaK-ATPase pump sites in cultured bovine corneal endothelium of varying cell density at confluence. Invest. Ophthalmol. Vis. Sci. 1995;36:1317–1326. [PubMed] [Google Scholar]
- 14.McCarthy K.M., Skare I.B., Stankewich M.C., Furuse M., Tsukita S., Rogers R.A., Lynch R.D., Schneeberger E.E. Occludin is a functional component of the tight junction. J. Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- 15.Vilas G.L., Loganathan S.K., Quon A., Sundaresan P., Vithana E.N., Casey J. Oligomerization of SLC4A11 protein and the severity of FECD and CHED2 corneal dystrophies caused by SLC4A11 mutations. Hum. Mutat. 2012;33:419–428. doi: 10.1002/humu.21655. [DOI] [PubMed] [Google Scholar]
- 16.Vilas G.L., Morgan P.E., Loganathan S.K., Quon A., Casey J.R. A biochemical framework for SLC4A11, the plasma membrane protein defective in corneal dystrophies. Biochemistry. 2011;50:2157–2169. doi: 10.1021/bi101887z. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Seet L.F., Koh L.W., Venkatraman A., Venkataraman D., Mohan R.R., Praetorius J., Bonanno J.A., Aung T., Vithana E.N. Depletion of SLC4A11 causes cell death by apoptosis in an immortalized human corneal endothelial cell line. Invest. Ophthalmol. Vis. Sci. 2012;53:3270–3279. doi: 10.1167/iovs.11-8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuhrmann S. Wnt signaling in eye organogenesis. Organogenesis. 2008;4:60–67. doi: 10.4161/org.4.2.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakatsu M.N., Ding Z., Ng M.Y., Truong T.T., Yu F., Deng S.X. Wnt/beta-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. Invest. Ophthalmol. Vis. Sci. 2011;52:4734–4741. doi: 10.1167/iovs.10-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schafer B.W., Heizmann C.W. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem. Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi A., Yamamoto H., Kishida S. Multiplicity of the interactions of Wnt proteins and their receptors. Cell Signal. 2007;19:659–671. doi: 10.1016/j.cellsig.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Motegi Y., Usui T., Ishida K., Kato S., Yamashita H. Regulation of bovine corneal endothelial cell cycle by transforming growth factor-beta. Acta Ophthalmol. Scand. 2003;81:517–525. doi: 10.1034/j.1600-0420.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- 23.Saika S., Saika S., Liu C.Y., Azhar M., Sanford L.P., Doetschman T., Gendron R.L., Kao C.W., Kao W.W. TGFbeta2 in corneal morphogenesis during mouse embryonic development. Dev. Biol. 2001;240:419–432. doi: 10.1006/dbio.2001.0480. [DOI] [PubMed] [Google Scholar]
- 24.Barraclough R., Dawson K.J., Rudland P.S. Elongated cells derived from rat mammary cuboidal epithelial cell lines resemble cultured mesenchymal cells in their pattern of protein synthesis. Biochem. Biophys. Res. Commun. 1984;120:351–358. doi: 10.1016/0006-291x(84)91261-0. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Riau A.K., Setiawan M., Mehta J.S., Ti S.E., Tong L., Tan D.T., Beuerman R.W. S100A expression in normal corneal-limbal epithelial cells and ocular surface squamous cell carcinoma tissue. Mol. Vis. 2011;17:2263–2271. [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer T.R., Grossniklaus H.E., Vigneswaran N., Waring G.O., Kozarsky A. Cytokeratin expression in corneal endothelium in the iridocorneal endothelial syndrome. Invest. Ophthalmol. Vis. Sci. 1992;33:3581–3585. [PubMed] [Google Scholar]
- 27.Hayashi K., Sueishi K., Tanaka K., Inomata H. Immunohistochemical evidence of the origin of human corneal endothelial cells and keratocytes. Graefes Arch. Clin. Exp. Ophthalmol. 1986;224:452–456. doi: 10.1007/BF02173362. [DOI] [PubMed] [Google Scholar]
- 28.Akilov O.E., Wu M.X., Ustyugova I.V., Falo L.D., Jr, Geskin L.J. Resistance of Sezary cells to TNF-alpha-induced apoptosis is mediated in part by a loss of TNFR1 and a high level of the IER3 expression. Exp. Dermatol. 2012;21:287–292. doi: 10.1111/j.1600-0625.2012.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arlt A., Schafer H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur. J. Cell Biol. 2011;90:545–552. doi: 10.1016/j.ejcb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 30.De Keulenaer G.W., Wang Y., Feng Y., Muangman S., Yamamoto K., Thompson J.F., Turi T.G., Landschutz K., Lee R.T. Identification of IEX-1 as a biomechanically controlled nuclear factor-kappaB target gene that inhibits cardiomyocyte hypertrophy. Circ. Res. 2002;90:690–696. doi: 10.1161/01.res.0000012922.40318.05. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi T., Pittelkow M.R., Warner G.M., Squillace K.A., Kumar R. Regulation of a novel immediate early response gene, IEX-1, in keratinocytes by 1alpha,25-dihydroxyvitamin D3. Biochem. Biophys. Res. Commun. 1998;251:868–873. doi: 10.1006/bbrc.1998.9556. [DOI] [PubMed] [Google Scholar]
- 32.Kannabiran C. Genetics of corneal endothelial dystrophies. J. Genet. 2009;88:487–494. doi: 10.1007/s12041-009-0067-1. [DOI] [PubMed] [Google Scholar]
- 33.Muller M. Cellular senescence: molecular mechanisms, in vivo significance, and redox considerations. Antioxid. Redox Signal. 2009;11:59–98. doi: 10.1089/ars.2008.2104. [DOI] [PubMed] [Google Scholar]
- 34.Jun A.S., Meng H., Ramanan N., Matthaei M., Chakravarti S., Bonshek R., Black G.C., Grebe R., Kimos M. An alpha 2 collagen VIII transgenic knock-in mouse model of Fuchs endothelial corneal dystrophy shows early endothelial cell unfolded protein response and apoptosis. Hum. Mol. Genet. 2012;21:384–393. doi: 10.1093/hmg/ddr473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.