Abstract
A common synonymous single nucleotide polymorphism in exon 12 of the low-density lipoprotein receptor (LDLR) gene, rs688, has been associated with increased plasma total and LDL cholesterol in several populations. Using immortalized lymphoblastoid cell lines from a healthy study population, we confirmed an earlier report that the minor allele of rs688 is associated with increased exon 12 alternative splicing (P < 0.05) and showed that this triggered nonsense-mediated decay (NMD) of the alternatively spliced LDLR mRNA. However, since synonymous single nucleotide polymorphisms may influence structure and function of the encoded proteins by co-translational effects, we sought to test whether rs688 was also functional in the full-length mRNA. In HepG2 cells expressing LDLR cDNA constructs engineered to contain the major or minor allele of rs688, the latter was associated with a smaller amount of LDLR protein at the cell surface (−21.8 ± 0.6%, P = 0.012), a higher amount in the lysosome fraction (+25.7 ± 0.3%, P = 0.037) and reduced uptake of fluorescently labeled LDL (−24.3 ± 0.7%, P < 0.01). Moreover, in the presence of exogenous proprotein convertase subtilisin/kexin type 9 (PCSK9), a protein that reduces cellular LDL uptake by promoting lysosomal degradation of LDLR, the minor allele resulted in reduced capacity of a PCSK9 monoclonal antibody to increase LDL uptake. These findings are consistent with the hypothesis that rs688, which is located in the β-propeller region of LDLR, has effects on LDLR activity beyond its role in alternative splicing due to impairment of LDLR endosomal recycling and/or PCSK9 binding, processes in which the β-propeller is critically involved.
INTRODUCTION
The low-density lipoprotein receptor (LDLR), a cell surface glycoprotein, is responsible for the binding and uptake of plasma LDL particles and plays a critical role in maintaining cellular cholesterol homeostasis (1). Mutations in the LDLR gene can lead to elevated plasma LDL levels, resulting in an increased risk for atherosclerosis and coronary heart disease (2). Recent genome-wide association studies have identified several common single nucleotide polymorphisms (SNPs) at the LDLR locus that contribute to inter-individual variation in serum lipid concentrations (3). Among these, the minor variant of rs688 (Asn591 ACC→ACT), a synonymous SNP located within exon 12, has been reported to be associated with a 4–10% increase in plasma cholesterol levels in several independent populations (3–7). Although a number of SNPs within LDLR (e.g. rs12983082, rs2738446, rs1799898, rs9789302, rs5925) are in linkage disequilibrium (LD) with rs688, r2 > 0.8 (Supplementary Material, Fig. S1), none are as strongly associated with plasma lipids, suggesting that rs688 is the causative underlying polymorphism. Indeed, the minor ‘T’ allele of rs688 has been found to disrupt a SRp40 exonic splicing enhancer in exon 12, causing a modest (<10%) reduction in splicing efficiency that results in the generation of an LDLR transcript lacking exon 12, designated LDLR12(−) (7). It was hypothesized that LDLR12(−) encodes a soluble LDL ‘receptor’ that acts in a dominant negative fashion by binding plasma LDL, thus inhibiting its uptake by the full length or classical form of the receptor (7). However, since no LDLR isoform consistent with the translation of the LDLR12(−) transcript has been identified, the functional consequences of LDLR exon 12 skipping remain unknown.
Exon 12 skipping generates a premature termination codon downstream of the splicing event (7,8). Premature termination codons are known to trigger nonsense-mediated decay (NMD), a widespread post-transcriptional regulatory mechanism whereby alternative splicing has been shown to down-regulate gene expression (9,10). Thus, it is possible that the LDLR12(−) transcript is subject to NMD and is not translated into a protein; however, it has not yet been shown if LDLR12(−) is an NMD target.
Although rs688 directly affects exon 12 alternative splicing, this exon would be expected to be retained in nearly 90% of LDLR transcripts of T/T homozygotes, in whom exon 12 skipping is greatest (7). Whereas rs688 is a synonymous SNP, recent studies have shown that synonymous SNPs can alter protein conformation by transforming frequent codons into rare codons, thus reducing translational efficiency and disrupting the process of co-translational protein folding (11–16). For example, a synonymous SNP in the multidrug resistance 1 (MDR1) gene was reported to change substrate specificity of the resulting protein by impacting its conformation, presumably through changes in translational efficiency (15). Since the majority of LDLR transcripts containing the rs688 minor allele are predicted to be protein coding, we were interested in determining whether rs688 had functional effects beyond its impact on exon 12 alternative splicing. rs688 is located in the epidermal growth factor-like repeat (EGF)-β-propeller region of LDLR, a region that is crucial for displacing bound LDL particles and regulating LDLR recycling between endosomes and the cell surface (17,18). Proprotein convertase subtilisin/kexin type 9 (PCSK9), a protein that directs LDLR to lysosomes for degradation, binds specifically to the EGF-A domain, adjacent to the β-propeller region (18). Hence, we hypothesized that rs688 might impact PCSK9 regulation of LDLR.
In the present study, we systematically tested the functional impact of rs688 on several aspects of LDLR regulation. First we determined whether LDLR12(−) undergoes NMD. Next, to assess the effect of rs688 on LDLR regulation independently of its effects on alternative splicing, we generated the expression of constructs containing LDLR12(+) cDNA with the rs688 ‘C’ or ‘T’ allele to test the impact of the SNP on LDLR localization, activity and regulation by PCSK9.
RESULTS
rs688 promotes LDLR exon 12 alternative splicing
To validate the relationship between rs688 and LDLR exon 12 alternative splicing, we quantified LDLR12(+) and LDLR12(−) transcript levels in 173 immortalized lymphoblastoid cell lines derived from participants of the Cholesterol and Pharmacogenetics (CAP) clinical trial. Consistent with previous reports, T/T homozygotes had a 6% reduction in exon 12 splicing efficiency compared with either the C/C or C/T cell lines (P < 0.05, Supplementary Material, Fig. S2A). Although there were no statistically significant differences in total LDLR mRNA levels between C and T, there was a trend for T/T homozygotes to have lower LDLR total mRNA levels compared with either the C/C or C/T cell lines (Supplementary Material, Fig. S2B).
LDLR12(−) is subject to nonsense-mediated decay
Exon 12 skipping disrupts the open reading frame and introduces a premature stop codon, an effect that usually triggers the NMD response (19). To determine whether the LDLR12(−) transcript is subject to NMD, CAP LCLs (n = 12) were incubated with cycloheximide, a protein synthesis inhibitor that represses NMD. Compared with LDLR12(+), LDLR12(−) transcripts were significantly increased after 1 h of cycloheximide treatment, and remained elevated for an additional 2 h (Fig. 1A). There were no statistically significant differences in change of LDLR 12(+) levels between C/C and T/T homozygotes after cycloheximide treatment (data not shown). To verify the splice variant-specific differences in mRNA decay rates, HepG2 cells were incubated with 1 µg/ml actinomycin D to inhibit mRNA synthesis. LDLR12(−) had a dramatically shorter half-life than LDLR 12(+), 1.64 ± 0.03 versus 3.66 ± 0.39 h, respectively, P = 0.0007, n = 12 (Fig. 1B). These results strongly suggest that LDLR12(−) is subject to NMD.
Figure 1.

LDLR12(−) is subject to nonsense-mediated mRNA decay (NMD). (A) LCLs were treated with 1 µg/ml cycloheximide for 4 h (n = 12), and harvested over 240 min, after which LDLR12(−) and LDLR12(+) were quantified by qPCR. (B) HepG2 cells (n = 12) were incubated with 1 µg/ml actinomycin D and collected over 6 h. LDLR12(−) and LDLR12(+) were quantified by qPCR. Time points represent means of mRNA levels relative to the 0 time point, which was made equal to 1. The inset at the top-right corner shows the half-life of LDLR12(−) and LDLR12(+). P-values were calculated with paired two-tailed t-tests. Values plotted are mean ± SEM.
The rs688 ‘T’ allele increases accumulation of LDLR in lysosomes and reduces LDL uptake
Given the possibility that a synonymous SNP can affect structure of the translated LDLR protein, and the critical role of the EGF-β-propeller domain in determining whether LDLR recycles between endosomes and the cell surface or is shunted to the lysosome for degradation, we next tested whether rs688 alters cellular LDLR distribution. Using western blot analysis, we quantified LDLR protein in total cell lysates, at the cell surface, and in the lysosome fractions of HepG2 cells transiently transfected with a plasmid containing LDLR12(+) with either the rs688 ‘C’ or ‘T’ alleles. Endogenous LDLR was inhibited by incubation with 25-hydroxycholesterol (1 µg/ml) and was not detectable (data not shown). As shown in Figure 2A and B, while there was no significant allelic difference in the amount of LDLR protein in the total cell lysates, there was greater LDLR protein in the lysosome fraction (25.7 ± 0.3%, P = 0.037) and less at the cell surface (21.8 ± 0.6%, P = 0.0012) in HepG2 cells transfected with the ‘T’ allele expression construct compared with those expressing the ‘C’ allele construct. Changes in intracellular localization were verified by immunofluoresence imaging, which demonstrated that LDLR was more densely concentrated in the lysosomes of HepG2 cells transfected with the rs688 ‘T’ allele than in those expressing the ‘C’ allele (Fig. 2C and D). Co-localization coefficients, used to assess the degree of co-localization between LDLR and lysosome signals, were significantly different for the two alleles (r2 = 0.01 for the ‘C’ containing construct versus r2 = 0.61 for the ‘T’ containing construct) (Supplementary Material, Fig. S3A and B). Lastly, we found that DiI-LDL uptake was reduced 24.3 ± 0.7% in ‘T’ versus ‘C’ allele carriers (n = 8, P < 0.05, Fig. 2E), whereas there was no statistically significant difference in DiI-acetyl-LDL uptake between the genotypes (Fig. 2F), indicating that the reduced LDL uptake is LDLR mediated. Our findings of reduced cell surface LDLR protein and LDL uptake between the ‘T’ and ‘C’ alleles are consistent with in vivo observations that the ‘T’ allele is associated with a higher plasma LDL cholesterol level and suggest that the rs688-induced change in LDLR distribution has a functional impact on LDLR activity.
Figure 2.
Effects of rs688 on LDLR localization and activity. (A) HepG2 cells were transfected with the LDLR constructs (pCMV-LDLR) containing rs688 ‘C’ or ‘T’ for 48 h, and total cell lysates, cell surface protein and lysosomal protein were isolated and subject to western blot analysis using an anti-LDLR and anti-neomycin phosphotransferase (NPTII) antibody. NPTII is a selection marker included in the pCMV-LDLR plasmid. One representative western blot is shown. (B) Quantification of band intensity of LDLR normalized to NPTII. (C and D) Colocalization of LDLR protein and lysosomes in transiently transfected cells expressing human LDLR containing either rs688 ‘C’ or ‘T’, respectively. Shown for each image set of the immunofluoresence staining are: nuclei (blue), LDLR (green) and lysosome (red). Yellow indicates colocalization between LDLR and lysosome in the merged image. The arrowheads point to examples of LDLR protein that are not colocalized with lysosomes. The arrows indicate examples of colocalization of LDLR protein and lysosomes. The scale bar in the top-right corner represents 10 µm. Immunohistochemistry imaging at ×63 magnification of cells was observed by confocal microscopy on a Zeiss LSM 710 confocal inverted microscope. (E and F) Transiently transfected HepG2 cells (n = 8) were incubated with varying concentrations of DiI-LDL or DiI-Ac LDL and quantified as previously described (29). P-values were calculated with the two-tailed unpaired t-test and values plotted are average ± SEM. *MFI, mean fluorescence intensity.
rs688 impacts PCSK9 regulation of LDLR protein
PCSK9 binds the EGF-A domain of LDLR at the cell surface and directs it to the lysosome for degradation (20). Given the impact of rs688 on LDLR intracellular localization, we next tested for an effect of rs688 on PCSK9 regulation of LDLR. We measured DiI-LDL uptake in HepG2 cells transiently transfected with the LDLR overexpression plasmid containing either the rs688 ‘C’ or ‘T’ allele, after incubation with varying concentrations of exogenous PCSK9 protein (0, 2, 20 µg/ml). The lower concentration of PCSK9 attenuated the difference in DiI-LDL uptake observed between the ‘C’ versus ‘T’ allele, whereas the higher concentration completely abolished this difference (Fig. 3A), indicative of an allelic difference in LDL uptake in response to exogenous PCSK9. We then assessed the effects of further incubation with 1B20, a PCSK9 monoclonal antibody directed against the catalytic domain of PCSK9 that disrupts PCSK9 binding to LDLR. For these experiments, cells were incubated with 2 µg/ml of PCSK9 protein in order to achieve a reduction in DiI-LDL uptake while maintaining the differential effects of the rs688 alleles. There was a striking resistance of the rs688 ‘T’ allele to the PCSK9 antibody mediated increase in LDL uptake, with an IC50 of ∼5.25 versus ∼0.026 pm for the ‘C’ allele (P < 0.001, n = 8, Fig. 3B). Taken together, these results strongly suggest that the rs688 ‘T’ allele reduces LDL uptake in response to the PCSK9 monoclonal antibody 1B20.
Figure 3.
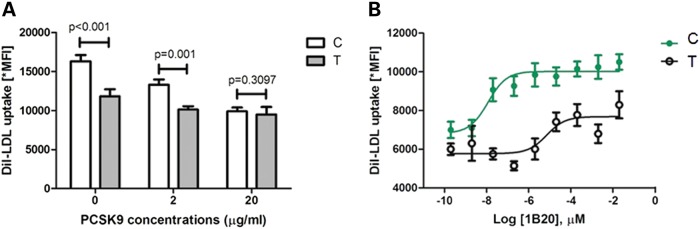
Effects of rs688 on PCSK9 regulation of LDLR protein. (A) Transiently transfected HepG2 cells (n = 8) expressing LDLR containing either rs688 ‘C’ or ‘T’ were incubated with DiI-LDL (10 µg/ml) mixed with purified PCSK9 protein (0, 2, 20 µg/ml) for 5 h. DiI-LDL uptake was determined as previously described (29). (B) Exogenous PCSK9 protein (2 µg/ml) was pre-incubated with varying concentrations of IB20, a PCSK9-antibody and subsequently mixed with DiI-LDL (10 µg/ml). The rate of DiI uptake was quantified in transiently transfected HepG2 cells (n = 8) as described in (A). *MFI, mean fluorescence intensity.
For comparison, we carried out similar incubations in HepG2 cells that transiently expressed allelic variants of a synonymous coding SNP in exon 1 of the LDLR gene (rs2228671 Cys27 TGC→TGT), for which the minor ‘T’ allele has been associated with decreased LDL-C (21,22). This SNP, which is located in the ligand-binding domain of the LDLR, is outside of the LD block containing rs688 (Supplementary Material, Fig. S1) and is not predicted to interact with PCSK9. Consistent with in vivo observations, the ‘T’ allele of rs2228671 was significantly associated with an increase in DiI-LDL uptake (Supplementary Material, Fig. S4A). However, in contrast to rs688, the magnitude of difference between the two alleles was similar at all PCSK9 protein concentrations. Moreover, there was no significant allelic difference in the effect of incubation with the PCSK9 antibody on DiI-LDL uptake (IC50 of ∼0.37 pm for ‘T’ versus ∼0.21 pm for ‘C’, P > 0.5, n = 8) (Supplementary Material, Fig. S4B).
DISCUSSION
Although several common LDLR SNPs have been found to be strongly associated with plasma LDL cholesterol (3), the effects of these SNPs on LDLR function and/or regulation are not well understood. The minor allele of rs688, a coding synonymous SNP in exon 12 of LDLR, has been associated with increased plasma LDL cholesterol levels in several independent populations and has been reported to modestly decrease the splicing efficiency of LDLR exon 12 (7). However, the present results indicate that rs688 has functional effects on LDLR activity beyond regulation of alternative splicing, namely alteration of both LDLR intracellular localization and PCSK9 regulation.
LDLR exon 12 skipping introduces a premature termination codon +400 bp downstream of the site of alternative splicing, which led us to hypothesize that LDLR12(−) is a subject of NMD. Here, we report LDLR12(−) undergoes NMD in LCLs and HepG2 cells, which is consistent with the trend of reduced total LDLR transcript observed in rs688 homozygotes carriers. Coupling alternative splicing and NMD could account at least in part for the rs688 association with plasma total and LDL cholesterol levels. The non-sense mutation S78X in LDLR exon 3, which was found in Norwegian familial hypercholesterolemia patients, was recently found to induce NMD (23), further supporting the functional significance of NMD in the regulation of LDLR.
Although rs688 clearly impacts LDLR through alternative splicing, the majority of LDLR transcripts retain exon 12 even in minor allele homozygotes. There has been increasing evidence that synonymous SNPs are involved in mechanisms of human diseases by interfering with mRNA structure, codon usage and/or conformation of the encoded protein, thereby possibly altering protein function (24). Bartoszewski et al. recently reported that a synonymous SNP in the coding region of the human cystic fibrosis transmembrane conductance regulator (CFTR) gene alters mRNA structure and reduces translation rate and expression of CFTR protein compared with wild-type (25). Co-translational folding can be hindered by synonymous SNPs that transform frequent codons into rare codons (26). Moreover, it has been shown that the amount of cognate tRNA is directly proportional to the frequency of codon usage (27). Using the Codon Usage Database (http://kazusa.or.jp/codon/), rs688 was found to convert a common codon ACC (36% usage) to a less commonly used codon, ACT (25% usage).
The potential effects of synonymous coding SNPs raise the possibility that by altering the dynamics of protein folding, rs688 may affect LDLR protein structure and function. Here, we demonstrate that the ‘T’ allele of rs688 reduces LDLR at the cell surface, increases localization to the lysosome and decreases PCSK9 effects on LDLR activity. Since the β-propeller region of LDLR is located downstream of the EGF-A, the PCSK9-binding domain, and has been shown to be critical to both LDLR recycling from the endosome to the cell surface and to PCSK9-mediated lysosomal LDLR degradation (28), these findings raise the possibility that rs688 alters protein conformation in a manner that affects the EGF region. Our observation that overexpression of LDLR protein containing the rs688 ‘T’ allele results in reduced DiI-LDL uptake compared with overexpression with the ‘C’ allele supports a functional effect of this mechanism on LDL metabolism. It has been reported that the minimal level of secreted PCSK9 in HepG2 cells has little impact on cell surface LDLR protein (29), suggesting that rs688 may alter cellular LDLR recycling independently of PCSK9 in this cell line. However, the addition of exogenous PCSK9 to HepG2 cells revealed that the rs688 ‘T’ allele hinders PCSK9 regulation of LDLR protein. Therefore, these results suggest that rs688 regulates LDLR protein in both PCSK9-dependent and -independent manners. Further investigation is required to directly determine whether rs688 alters LDLR β-propeller conformation and/or PCSK9 binding.
The ‘T’ allele of the LDLR SNP rs2228671 has previously been reported to be associated with significant lower plasma LDL cholesterol levels in European populations (21,22). To date, no functional effect of this SNP has been identified. Interestingly, like rs688, rs2228671 is a synonymous SNP. There are no significant differences in predicted mRNA secondary structure and minimum free energy between major and minor alleles of either rs688 and rs2228671 using several software programs (Genebee, mfold and RNAfold, data not shown). However, both polymorphisms result in changes from frequent to rare codons that could affect co-translational folding and hence LDLR functionality. The present findings indicate that the putative effect of rs2228671 on LDLR function is distinct from that of rs688 in that it does not alter the effects of the inhibitory PCSK9 antibody on LDL uptake in HepG2 cells.
It should be noted that overexpression of LDLR may alter its intracellular distribution and cause cholesterol and cholesteryl ester aggregate in cells. Heeren et al. (30) demonstrated that overexpression of LDLR under the Rous sarcoma virus long-terminal repeat promoter causes pathological intracellular lipid accumulation and the formation of cholesterol and cholesteryl ester crystals in vitro. In our study system, no such crystals were found. We used 25-hydroxycholesterol to inhibit endogenous LDLR and tested much lower DiI-LDL concentrations (5∼20 µg/ml) than that used by Heeren et al. (50 µg/ml). Moreover, we observed relatively modest (∼30%) increases in DiI-LDL uptake in our transiently transfected cells compared with those without 25-hydroxycholesterol treatment (data not shown).
In summary, we have found that rs688 has two distinct mechanisms for regulating LDLR post-transcriptionally. First, the minor allele stimulates exon 12 alternative splicing to generate an LDLR transcript that is targeted to NMD. Secondly, in the majority of transcripts containing exon 12, the minor allele reduces LDLR cell surface protein and alters LDLR intracellular distribution, presumably by altering LDLR protein conformation. This effect also appears to inhibit PCSK9-mediated reduction in LDL uptake as manifest by a reduced response to a PCSK9 monoclonal antibody. There has been recent evidence for the efficacy of PCSK9 monoclonal antibodies in reducing plasma LDL levels (31,32), and our findings raise the possibility that rs688 may influence the magnitude of the therapeutic response.
MATERIALS AND METHODS
LCL culture conditions
Lymphocytes were isolated from each CAP subject using IsoPrep TM (Robbins Scientific Corporation) and transformed by Epstein-Barr Virus (33). Cell lines were grown at 37°C, 5% CO2 in RPMI Medium 1640 (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 500 units/ml Penicillin/Streptomycin and 2 nm GlutaMax (Invitrogen).
LDLR mRNA quantitation
Total RNA was isolated using the RNAeasy mini kit (Qiagen). cDNA was synthesized from 1 mg of total RNA with the High-Capacity cDNA Archive Kit (Life Technologies). LDLR12(+) and LDLR12(−) transcripts were quantified with two SYBR-Green qPCR assays spanning exon 10 to 12 and exon 10 to 13, respectively, via pre-quantitated serially diluted standards using the following primers: LDLR12(+): ATCACCCTAGATCTCCTCAGTG and GCACTGAAAATGGCTTCGTT; LDLR12(−):GGCATCACCCTAGGACAAAGT and GGGTGAGGTTGTGGAAGAGAA. Total LDLR mRNA was calculated by adding LDLR12(+) and LDLR12(−) transcripts. LDLR exon 12 splicing efficiency was calculated as follows: LDLR12(+)/total LDLR. Each qPCR reaction was performed in triplicate on the ABI PRISM 7900 Sequence Detection System with standard reagents and protocols (Applied Biosystems). CLPTM1 was quantified and used for data normalization as previously described (34).
Studies of nonsense-mediated decay
LCLs (n = 12) were treated with 1 µg/ml cycloheximide and harvested after 0, 40, 60, 90, 120 and 240 min. HepG2 cells (n = 12) were incubated with 1 µg/ml actinomycin D and collected after 0, 0.5, 1, 2, 4, 6, 24 and 48 h. LDLR12(+) and LDLR12(−) transcripts were quantified as described earlier. mRNA quantity values were log transformed and plotted versus time. Linear regression was used to calculate the slope of the resulting line, and mRNA half-life was calculated as: t1/2 (h) = ln2/(−2.303 × slope). Transcript half-life was calculated using only time points consistent with first-order decay kinetics (35). P-values were calculated with paired two-tailed t-tests.
LDLR overexpression studies
pCMV6-LDLR-FLAG plasmid (Origene), a mammalian expression vector that contains the entire LDLR cDNA with a FLAG tag (also known as MYC-DDK tag) fused to the 3′ end was used for overexpressing LDLR protein. Site-directed mutagenesis was performed using the GeneTailor™ Site-Directed Mutagenesis System kit (Life Technologies) to introduce the rs688 ‘T’ allele or rs2228671 ‘T’ allele. Plasmid sequences were confirmed by Sanger sequencing. Primers used for rs688 were: CTATGACACCGTCATCAGCAGAGACATCCAGGC and CTGCTGATGACGGTGTCACTTAGGAAGAGACGC.
Primers used for rs2228671 were: ACTGCAGTGGGCGACAGATGCGAAAGAAACGA and CATCT GTCGCCCACTGCAGTCCCCGCCGCG.
HepG2 cells were plated at a density of 3–5 × 104 cells/cm2 in a six-well dish and transiently transfected with 3 μg of each of the LDLR expression plasmids described earlier (pCMV-LDLR-FLAG, pCMV-LDLR-FLAG-rs688) using the GenJetTM In Vitro DNA Transfection Reagent (Signagen). 25-hydroxycholesterol (1 µg/ml) was added to the cell culture medium to suppress the endogenous LDLR synthesis. Transfection efficiency was evaluated by western blot analysis using both anti-LDLR (Fitzgerald) and anti-FLAG (Origene) monoclonal antibodies for LDLR and anti- neomycin phosphotransferase (NPTII) (Millipore).
Western blot analyses
The Cell Surface Protein Isolation Kit (Pierce) and the Lysosome Isolation Kit (Sigma-Aldrich) were used to purify proteins from the cell surface and lysosome fractions of the HepG2 transfected cells following manufacturer's instructions. Briefly, HepG2 cells were transfected with the LDLR constructs (pCMV-LDLR) containing rs688 ‘C’ or ‘T’ for 48 h, then half of cells were processed for total cell lysates and cell surface protein isolation and the other half of cells were processed for lysosome protein isolation. The protein concentrations were determined by the Bradford assay (Bio-Rad) and used to normalize each sample. Transfection efficiency was monitored by analysis using an anti-neomycin phosphotransferase (NPTII) antibody (Millipore), which is included in the pCMV-LDLR as a selection marker for transfection. Total cell lysates, isolated cell surface proteins, and lysosome proteins were analyzed by western blotting using anti-LDLR polyclonal antibody (Fitzgerald). Bands were quantified using an Alpha Imager and normalized to the transfection efficiency control NPTII. Two-tailed unpaired t-tests were used to identify statistically significant differences in LDLR signal intensity between rs688 ‘C’ and ‘T’. All experiments were performed in triplicate.
Immunofluoresence
Transiently expressing LDLR HepG2 cells were grown on cover slips and stained with 50 nm of LysoTracker® Red DND-99 (Invitrogen) under growth conditions for 30 min with the LysoTracker®, fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. To detect the LDLR-FLAG fusion proteins, fixed cells were incubated with an anti-FLAG monoclonal primary antibody, and an Alexa-488 (green) conjugated secondary antibody (Invitrogen). Nuclei (blue) were counterstained with DAPI (Invitrogen). Stained cells were mounted in Slow Fade™ Light antifade solution (Molecular Probes) and the fluorescence was observed under DIC optics 63 × oil by confocal microscopy on a Zeiss LSM 710 confocal inverted microscope. Potential cross-reaction of the various staining procedures was assessed by incubating cells with each stain alone. The results of fluorescence colocalization were represented graphically in a scatter plot where the intensity of green (LDLR) was plotted against the intensity of red (lysosome) for each pixel and colocalization analysis and the coefficients of determination were determined by the Zeiss ZEN software.
DiI-LDL or DiI-Acetyl-LDL uptake
HepG2 cells transfected with either LDLR expression constructs (rs688 ‘C’ or ‘T’) were incubated with varying concentrations (0–20 µg/ml) of DiI-LDL (Biomedical Technologies) or DiI-acetyl-LDL (0–20 µg/ml Kalen Biomedical, LLC) in the MEM medium supplemented with 10% lipoprotein-deficient serum (LPDS) and 25-hydroxycholesterol (1 µg/ml) for 5 h. Levels of DiI-LDL or DiI-acetyl-LDL uptake were quantified by Victor Fluorescence Plate Reader at excitation/emission at 530/580 nm and normalized by cell lysates protein concentrations measured by the Bradford Assay (Bio-Rad) using the Molecular Devices Spectramax 340 VIS Plate Reader. Background fluorescence values were measured from transfected cells incubated with unlabeled LDL or acetyl-LDL. Two-tailed unpaired t-tests were used to identify statistically significant differences in DiI-LDL or DiI-acetyl-LDL uptake. All experiments were performed eight times.
Exogenous PCSK9 and anti-PCSK9 antibody 1B20 treatments in HepG2 cells
Transiently transfected HepG2 cells were incubated with purified PCSK9 proteins (provided by Merck) at varying concentrations (0, 2, 20 µg/ml) mixed with 10 µg/ml DiI-LDL for 5 h in the MEM medium containing 1 µg/ml of 25-hydroxycholesterol and 10% LPDS. DiI-LDL uptake was measured as previously described (36). 1B20, a PCSK9 antibody also obtained as a gift from Merck, was serially diluted (0–20 nm), combined with media-containing PCSK9 protein (2 µg/ml) for 20 min at room temperature and added to the transfected cells. The IC50 values were determined using the sigmoidal dose–response curve-fitting program in GraphPad 5.0. Two-tailed t-tests were used to identify statistically significant differences in DiI-LDL uptake in response to PCSK9 and IC50 values for the1B20 treatment. All experiments were performed eight times.
Statistical analysis
For gene expression quantification by qPCR, the Grubb's test for outliers was calculated for each three triplicate measurement. All statistical analyses were performed using either JMP 7.0.1 (SAS Institute) or GraphPad Prism 5.
SUPPLEMENTARY MATERIAL
FUNDING
This project was supported by an investigator initiated grant from Merck, and NIH grants HL069757 and HL104133–01.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr David Neff (Merck) for helpful discussions, Dr Xi Ai (Merck) for her excellent technical support and Merck for providing the purified PCSK9 protein and monoclonal PCSK9 antibody 1B20. Dr Elizabeth Theusch contributed to data analysis and Jeremy Lee (University of California, Berkeley) assisted in cell culture.
Conflict of Interest statement. Dr Krauss is a member of the Merck Global Atherosclerosis Advisory Board.
REFERENCES
- 1.Goldstein J.L., Anderson R.G., Brown M.S. Receptor-mediated endocytosis and the cellular uptake of low density lipoprotein. Ciba Foundation symposium. 1982:77–95. doi: 10.1002/9780470720745.ch5. in press. [DOI] [PubMed] [Google Scholar]
- 2.Brown M.S., Goldstein J.L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 3.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boright A.P., Connelly P.W., Brunt J.H., Morgan K., Hegele R.A. Association and linkage of LDLR gene variation with variation in plasma low density lipoprotein cholesterol. Journal of human genetics. 1998;43:153–159. doi: 10.1007/s100380050060. [DOI] [PubMed] [Google Scholar]
- 5.Fu Y., Katsuya T., Higaki J., Asai T., Fukuda M., Takiuchi S., Hatanaka Y., Rakugi H., Ogihara T. A common mutation of low-density lipoprotein receptor gene is associated with essential hypertension among Japanese. Journal of human hypertension. 2001;15:125–130. doi: 10.1038/sj.jhh.1001132. [DOI] [PubMed] [Google Scholar]
- 6.Schaefer E.J., Lamon-Fava S., Johnson S., Ordovas J.M., Schaefer M.M., Castelli W.P., Wilson P.W. Effects of gender and menopausal status on the association of apolipoprotein E phenotype with plasma lipoprotein levels. Results from the Framingham Offspring Study. Arterioscler Thromb. 1994;14:1105–1113. doi: 10.1161/01.atv.14.7.1105. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H., Tucker H.M., Grear K.E., Simpson J.F., Manning A.K., Cupples L.A., Estus S. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum Mol Genet. 2007;16:1765–1772. doi: 10.1093/hmg/ddm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou F., Gopalraj R.K., Lok J., Zhu H., Ling I.F., Simpson J.F., Tucker H.M., Kelly J.F., Younkin S.G., Dickson D.W., et al. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer's disease. Human molecular genetics. 2008;17:929–935. doi: 10.1093/hmg/ddm365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Sanchez F., Mittnacht S. Nonsense-mediated decay: paving the road for genome diversification. Bioessays. 2008;30:926–928. doi: 10.1002/bies.20825. [DOI] [PubMed] [Google Scholar]
- 10.Wen J., Brogna S. Nonsense-mediated mRNA decay. Biochemical Society transactions. 2008;36:514–516. doi: 10.1042/BST0360514. [DOI] [PubMed] [Google Scholar]
- 11.Waldman Y.Y., Tuller T., Keinan A., Ruppin E. Selection for translation efficiency on synonymous polymorphisms in recent human evolution. Genome biology and evolution. 2011;3:749–761. doi: 10.1093/gbe/evr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H., Wang P., You J., Zheng Y., Fu Y., Tang Q., Zhou L., Wei Z., Lin B., Shu Y., et al. Screening of Kozak-motif-located SNPs and analysis of their association with human diseases. Biochemical and biophysical research communications. 2010;392:89–94. doi: 10.1016/j.bbrc.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Nackley A.G., Shabalina S.A., Lambert J.E., Conrad M.S., Gibson D.G., Spiridonov A.N., Satterfield S.K., Diatchenko L. Low enzymatic activity haplotypes of the human catechol-O-methyltransferase gene: enrichment for marker SNPs. PloS one. 2009;4 doi: 10.1371/journal.pone.0005237. e5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee P.H., Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic acids research. 2008;36:D820–824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 16.Wang D., Johnson A.D., Papp A.C., Kroetz D.L., Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenetics and genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 17.Zhang D.W., Garuti R., Tang W.J., Cohen J.C., Hobbs H.H. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13045–13050. doi: 10.1073/pnas.0806312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beglova N., Jeon H., Fisher C., Blacklow S.C. Structural features of the low-density lipoprotein receptor facilitating ligand binding and release. Biochemical Society transactions. 2004;32:721–723. doi: 10.1042/BST0320721. [DOI] [PubMed] [Google Scholar]
- 19.Schell T., Kulozik A.E., Hentze M.W. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-3-reviews1006. REVIEWS1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tveten K., Holla O.L., Cameron J., Strom T.B., Berge K.E., Laerdahl J.K., Leren T.P. Interaction between the ligand-binding domain of the LDL receptor and the C-terminal domain of PCSK9 is required for PCSK9 to remain bound to the LDL receptor during endosomal acidification. Human molecular genetics. 2012;21:1402–1409. doi: 10.1093/hmg/ddr578. [DOI] [PubMed] [Google Scholar]
- 21.Martinelli N., Girelli D., Lunghi B., Pinotti M., Marchetti G., Malerba G., Pignatti P.F., Corrocher R., Olivieri O., Bernardi F. Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid profile. Blood. 2010;116:5688–5697. doi: 10.1182/blood-2010-03-277079. [DOI] [PubMed] [Google Scholar]
- 22.Linsel-Nitschke P., Gotz A., Erdmann J., Braenne I., Braund P., Hengstenberg C., Stark K., Fischer M., Schreiber S., El Mokhtari N.E., et al. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease–a Mendelian Randomisation study. PloS one. 2008;3 doi: 10.1371/journal.pone.0002986. e2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holla O.L., Kulseth M.A., Berge K.E., Leren T.P., Ranheim T. Nonsense-mediated decay of human LDL receptor mRNA. Scandinavian journal of clinical and laboratory investigation. 2009;69:409–417. doi: 10.1080/00365510802707163. [DOI] [PubMed] [Google Scholar]
- 24.Angov E. Codon usage: nature's roadmap to expression and folding of proteins. Biotechnology journal. 2011;6:650–659. doi: 10.1002/biot.201000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartoszewski R.A., Jablonsky M., Bartoszewska S., Stevenson L., Dai Q., Kappes J., Collawn J.F., Bebok Z. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. The Journal of biological chemistry. 2010;285:28741–28748. doi: 10.1074/jbc.M110.154575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai C.J., Sauna Z.E., Kimchi-Sarfaty C., Ambudkar S.V., Gottesman M.M., Nussinov R. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. Journal of molecular biology. 2008;383:281–291. doi: 10.1016/j.jmb.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 28.Surdo P.L., Bottomley M.J., Calzetta A., Settembre E.C., Cirillo A., Pandit S., Ni Y.G., Hubbard B., Sitlani A., Carfi A. Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO reports. 2011;12:1300–1305. doi: 10.1038/embor.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L., McCabe T., Condra J.H., Ni Y.G., Peterson L.B., Wang W., Strack A.M., Wang F., Pandit S., Hammond H., et al. An anti-PCSK9 antibody reduces LDL-cholesterol on top of a statin and suppresses hepatocyte SREBP-regulated genes. International journal of biological sciences. 2012;8:310–327. doi: 10.7150/ijbs.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heeren J., Steinwaerder D.S., Schnieders F., Cichon G., Strauss M., Beisiegel U. Nonphysiological overexpression of low-density lipoprotein receptors causes pathological intracellular lipid accumulation and the formation of cholesterol and cholesteryl ester crystals in vitro. J Mol Med (Berl) 1999;77:735–743. doi: 10.1007/s001099900045. [DOI] [PubMed] [Google Scholar]
- 31.Roth E.M., McKenney J.M., Hanotin C., Asset G., Stein E.A. Atorvastatin with or without an Antibody to PCSK9 in Primary Hypercholesterolemia. The New England journal of medicine. 2012 doi: 10.1056/NEJMoa1201832. in press. [DOI] [PubMed] [Google Scholar]
- 32.Stein E.A., Mellis S., Yancopoulos G.D., Stahl N., Logan D., Smith W.B., Lisbon E., Gutierrez M., Webb C., Wu R., et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. The New England journal of medicine. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 33.Pressman S., Rotter J.I. Epstein-Barr virus transformation of cryopreserved lymphocytes: prolonged experience with technique. American journal of human genetics. 1991;49:467. [PMC free article] [PubMed] [Google Scholar]
- 34.Medina M.W., Gao F., Ruan W., Rotter J.I., Krauss R.M. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina M.W., Gao F., Naidoo D., Rudel L.L., Temel R.E., McDaniel A.L., Marshall S.M., Krauss R.M. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PloS one. 2011;6 doi: 10.1371/journal.pone.0019420. e19420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan J.C., Piper D.E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., et al. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9820–9825. doi: 10.1073/pnas.0903849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



