Abstract
The formation of alternative DNA secondary structures can result in DNA breakage leading to cancer and other diseases. Chromosomal fragile sites, which are regions of the genome that exhibit chromosomal breakage under conditions of mild replication stress, are predicted to form stable DNA secondary structures. DNA breakage at fragile sites is associated with regions that are deleted, amplified or rearranged in cancer. Despite the correlation, unbiased examination of the ability to form secondary structures has not been evaluated in fragile sites. Here, using the Mfold program, we predict potential DNA secondary structure formation on the human chromosome 10 sequence, and utilize this analysis to compare fragile and non-fragile DNA. We found that aphidicolin (APH)-induced common fragile sites contain more sequence segments with potential high secondary structure-forming ability, and these segments clustered more densely than those in non-fragile DNA. Additionally, using a threshold of secondary structure-forming ability, we refined legitimate fragile sites within the cytogenetically defined boundaries, and identified potential fragile regions within non-fragile DNA. In vitro detection of alternative DNA structure formation and a DNA breakage cell assay were used to validate the computational predictions. Many of the regions identified by our analysis coincide with genes mutated in various diseases and regions of copy number alteration in cancer. This study supports the role of DNA secondary structures in common fragile site instability, provides a systematic method for their identification and suggests a mechanism by which DNA secondary structures can lead to human disease.
INTRODUCTION
Alternative DNA secondary structures, which vary in conformation from the customary right-handed B form, are suggested to have a role both in biological processes such as transcription and telomere maintenance and in genomic mutational events including deletions, amplifications and chromosomal rearrangements (1). At least 10 alternative conformations have been identified to date, including hairpins/cruciforms, Z-DNA, triplexes, tetraplexes, slipped DNA and sticky DNA (2). Formation of these structures can occur when the DNA duplex is unwound during metabolic DNA processes such as DNA replication and transcription, and cause abnormalities in these processes. DNA secondary structures are strongly associated with ∼20 hereditary neurological diseases (due to simple sequence amplifications), ∼50 human diseases (caused by genomic rearrangements and deletions) and several psychiatric diseases (resulting from polymorphisms in simple repeat sequences) (3).
Triplet repeats, which form hairpin loops or slipped conformations, can give rise to expansions resulting in diseases such as myotonic dystrophy, fragile X syndrome, Friedreich's ataxia and Huntington's disease (4). Z-DNA, triplex and tetraplex formation potentials at the oncogene c-MYC correspond to the major breakpoint hotspots found in lymphomas and leukemias. Similarly, the major breakpoint cluster region in BCL-2 follicular lymphomas can form triplex DNA structures (5). Multiple stem-loop structures have been predicted or identified in several human fragile sites examined so far (6). Genome-wide analysis of palindrome formation, due to large inverted repeats, revealed these sequences to cluster in cancer cells at regions which undergo gene amplification, implicating these alternative structures in tumor progression (7). Purine–pyrimidine tracts and other repetitive elements capable of forming alternative DNA structures are overrepresented in DNA sequences surrounding breakpoints involved in chromosomal rearrangements (8–10). Detailed analysis of 11 gross deletions resulting in various diseases revealed that alternative DNA conformations could explain the formation of DNA breaks at known breakpoints in patients (10). While these studies provide further evidence supporting a role of various alternative DNA secondary structures in disease development and progression, no unbiased study has been performed analyzing the formation of multiple stem loop DNA secondary structures.
Chromosomal fragile sites exhibit gaps or breaks on metaphase chromosomes under conditions that partially inhibit DNA synthesis (11). Many genes deleted, amplified or rearranged in cancer are located within the fragile sites (12). In a comprehensive survey of simple recurrent cancer-specific translocations, we found that over half of gene pairs involved in these translocations have breakpoints of at least one gene mapped to fragile sites (13). Bignell et al. also found that, by deriving mutation signatures of unexplained homozygous deletions in cancer cell lines, some matched those of recessive cancer genes while others had signatures similar to fragile sites (14). In addition to cancer, fragile site instability is associated with neuropsychiatric diseases, including schizophrenia and autism (15). More importantly, fragile site-induced DNA breakage can produce deletions within the FHIT gene and the formation of RET/PTC1 rearrangements, resembling those found in human tumors (16,17).
Fragile sites are divided into two classes, common or rare, based on their frequency in the population, and are further divided according to their mode of induction in cultured cells. While rare fragile sites are present in <5% of the population and inherited in a Mendelian manner, common fragile sites are present in all individuals. Most common fragile sites are induced by low doses of aphidicolin (APH) (18), an inhibitor of DNA polymerases α, δ and ɛ (19,20). The precise mechanism of instability at fragile sites remains elusive, but analysis of several common fragile sites has revealed AT-rich sequences displaying the potential to form highly stable secondary structures (21,22), which may stall DNA replication fork progression. The CGG repeats, which are present in all rare, folate-sensitive fragile sites, can form quadraplex (23) and hairpin (24) structures in vitro, and display significant blocks to DNA replication both in vitro (25) and in vivo (26). Our study of the AT-rich rare fragile site FRA16B demonstrated the formation of secondary structure and DNA polymerase stalling within this sequence in vitro, as well as reduced replication efficiency and increased instability in human cells (27). The examination of replication intermediates from cells containing AT-rich sequences within common fragile site FRA16D in yeast showed site-specific replication fork stalling depending on the length of the AT repeat (28). DNA synthesis of the same fragile site by human replicative polymerases δ and α using an in vitro primer extension assay confirmed polymerase stalling at sites predicted to form inhibitory DNA structures (29). Similar findings were observed for eukaryotic replicative polymerases at hairpin and tetraplex structures formed within CGG repeat expansions (30). Replication fork stalling also occurs at endogenous AT-rich sequences within the common fragile site FRA16C in human lymphoblastoid cells, which is enhanced by APH treatment (31). These data suggest that the ability of fragile sites to form stable secondary structures during DNA replication likely contributes to their breakage by stalling replication fork progression. Despite the connection between DNA secondary structure formation and DNA fragility, no comprehensive study has evaluated the extent of DNA secondary structure-forming potential at fragile versus non-fragile regions.
In this study, we use computational predictions of the chromosome 10 sequence to form multiple stem loop DNA secondary structures, and compare the stability and clustering of these structures at fragile sites versus non-fragile sites. On chromosome 10, three APH-induced common fragile sites contained more sequence segments with potential high secondary structure-forming ability compared with non-fragile regions, and in the APH-induced sites these segments occurred in more dense clusters than non-fragile regions. Traditionally, fragile sites are defined cytogenetically as unstained gaps of an average 3 Mb in size on metaphase chromosomes. However, fragility is not present in the entire region. Using secondary structure-forming ability in combination with the information derived from molecularly defined rare and common fragile sites, we developed a threshold to predict DNA fragility; using this threshold, we can narrow down sites of true fragility within the current cytogenetically defined fragile sites. These computational analyses were further validated using an in vitro alternative DNA structure formation assay and a DNA breakage cell assay. Similarly applying this threshold, we uncovered potential new fragile sites previously unidentified or too small to be observed cytogenetically. Furthermore, the regions identified by this DNA fragility prediction threshold were correlated with regions known to be mutated in human diseases and cancer.
RESULTS
Analysis of chromosome 10 for DNA secondary structure-forming potential
Chromosome 10 was chosen to investigate the differences in DNA secondary structure-forming ability between fragile and non-fragile DNA, due to its moderate length and relatively even distribution of non-fragile (∼60%) versus fragile (∼40%) sequences (NCBI GeneBank, http://www.ncbi.nlm.nih. gov/sites). In addition, the fragile sites present on chromosome 10 include both common and rare sites, as well as different modes of induction [APH, bromodeoxyuridine (BrdU) and folic acid] (Supplementary Material, Table S1). In contrast to the rare fragile sites on chromosome 10, the common fragile sites have yet to be defined on the molecular level, and a computational approach would be beneficial for predicting their underlying fragility.
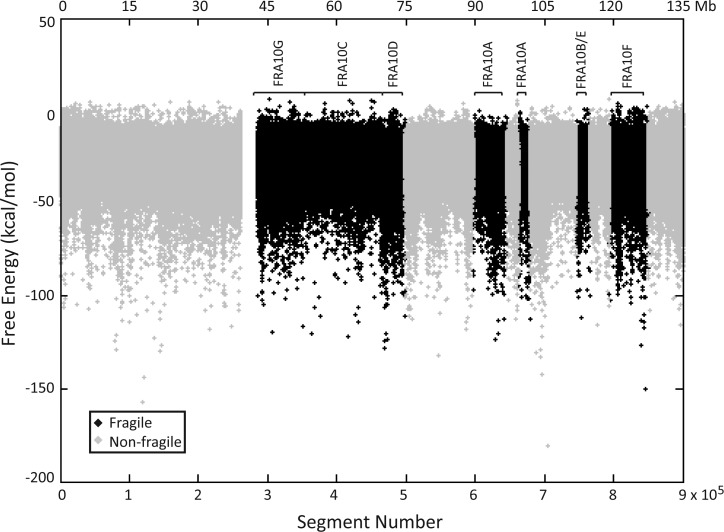
The approximately 136 Mb sequence of chromosome 10 was used, and un-sequenced gap, centromeric (32) and subtelomeric (33) sequences were then removed. The secondary structure-forming potential of the remaining chromosome 10 sequence (131 Mb) was analyzed using the Mfold program (34) by inputting sequentially 300 nt segments with a 150 nt shift window (Fig. 1). We chose the 300 nt length because it equals the Okazaki initiation zone of the DNA replication fork in mammalian cells, which possesses a single-stranded property during DNA replication (35,36). For each DNA segment, the program predicts various potential DNA secondary structures along with a corresponding free-energy value. A more negative free-energy value indicates a more stable DNA secondary structure. Therefore, the most favorable structure and free-energy value were selected for each segment. The average free-energy value for chromosome 10 is −27.2 ± 9.3 kcal/mol, with a range of −180.5 to +6.1 kcal/mol.
Figure 1.
Free-energy values for predicted DNA secondary structures on chromosome 10. The free-energy value for the most favorable Mfold-predicted DNA secondary structure for each 300 nt segment, with 150 nt increments, of the chromosome 10 sequence is presented. The lower x-axis depicts the segment number in the p- to q-arm direction on chromosome 10, the upper x-axis depicts the corresponding nucleotide number, and the y-axis displays the free-energy value of the predicted structure. Un-sequenced gap, centromeric and subtelomeric sequences were removed. Non-fragile and fragile DNAs are depicted in gray and black, respectively. The locations of fragile sites are marked by brackets.
To examine whether there is clustering of sequences capable of forming stable DNA secondary structures, 50 consecutive 300 nt segments were grouped into sections, with each section consisting of 7650 nt. The level of clustering was calculated based on the proportion of segments within each section with a free-energy value less than −40 kcal/mol. This value was chosen because it is approximately one standard deviation (SD) below the mean free-energy value of chromosome 10 and theoretically predicts a more energetically favorable structure. A higher proportion indicates that more segments are capable of forming highly stable secondary structures. The mean proportion of segments per section below −40 kcal/mol for chromosome 10 is 0.087 ± 0.116, with a range of 0–0.96.
Predicted DNA secondary structure-forming ability at fragile sites compared with non-fragile region
Delayed replication within fragile sites due to the formation of stable DNA secondary structures is suggested to contribute to fragile site breakage (22). To investigate if fragile DNA can form significantly more stable secondary structures than non-fragile DNA, the 131 Mb chromosome 10 sequence was divided into non-fragile or fragile regions, with the fragile sequence being further defined into individual fragile sites based on class and mode of induction (Supplementary Material, Fig. S1). A total of 373 537 segments were analyzed for fragile sites and 517 301 segments for non-fragile regions (Supplementary Material, Table S1). The mean free energies of the predicted secondary structures in the non-fragile and fragile regions are shown in Supplementary Material, Table S2. While the non-fragile DNA exhibited a comparable mean free-energy value to the entire chromosome 10, no significant difference was seen between non-fragile and fragile sites. This result is not unexpected, since the mean free-energy values are averaged over very large regions and differences between non-fragile and fragile sites may be masked.
Next, we examined the number of segments with free-energy values less than −40 kcal/mol for each fragile site and non-fragile DNA; such segments are energetically more favorable to fold into secondary structures compared with the overall chromosome 10 sequence. Three APH-induced common fragile sites FRA10G, FRA10D and FRA10F all contain significantly more segments with free-energy values less than −40 kcal/mol compared with non-fragile DNA (P< 1E−100), indicating more stable DNA secondary structures forming within these regions (Table 1). In contrast, the BrdU-induced common fragile site FRA10C, folate-sensitive rare fragile site FRA10A and the co-mapped BrdU-induced rare fragile site FRA10B and APH-induced common fragile site FRA10E had fewer segments with free-energy values less than −40 kcal/mol compared with non-fragile DNA. Similar results were obtained when analyzing the percentage of segments with free-energy values below −50 kcal/mol (Supplementary Material, Table S3), which is approximately 2 SD below the mean free-energy value of chromosome 10, suggesting increasing stability in the predicted secondary structures.
Table 1.
Percentage of segments with free energy below −40 kcal/mol
| Region | Non-fragile | FRA10G | FRA10C | FRA10D | FRA10A | FRA10B/E | FRA10F |
|---|---|---|---|---|---|---|---|
| Segments ΔG > −40 kcal/mola | 471 257 (91.1%) | 56 047 (87.8%) | 114 086 (96.7%) | 22 991 (80.2%) | 63 094 (93.7%) | 18 724 (93.6%) | 48 948 (87.9%) |
| Segments ΔG < −40 kcal/molb | 46 044 (8.9%) | 7820 (12.2%) | 3914 (3.3%) | 5677 (19.8%) | 4241 (6.3%) | 1277 (6.4%) | 6717 (12.1%) |
| P-value* | – | 2.34E−166 | 0 | 0 | 1.21E−133 | 7.04E−35 | 4.67E−133 |
aTotal number of segments with a free-energy value higher or lower than −40 kcal/mol, with the percentage of the total in parentheses.
bFragile sites highlighted in gray have a significantly greater proportion of segments with a free-energy value less than −40 kcal/mol compared with non-fragile DNA.
*P-value calculated by comparing each fragile site with non-fragile DNA using a chi-square test. A P-value of 0 represents a P < 1E−200.
To examine any differences in the clustering of DNAs capable of forming stable DNA secondary structures between non-fragile and fragile regions, 50 consecutive 300 nt segments were grouped into sections and analyzed. A total of 7069 sections were analyzed for fragile sites and 10 334 sections for non-fragile regions (Supplementary Material, Table S1). The proportion of segments within each section having a free-energy value less than −40 kcal/mol was then calculated (Supplementary Material, Fig. S2) and averaged for chromosome 10, non-fragile DNA and each fragile site (Table 2). Analysis of variance revealed a significant difference in the proportion of segments below −40 kcal/mol among all fragile and non-fragile sites (P = 7.7E−292). A subsequent pairwise comparison of each fragile site with a non-fragile site showed that APH-induced common fragile sites FRA10G, FRA10D and FRA10F exhibited a significantly greater proportion of low free-energy segments per section when compared with non-fragile DNA (P < 1E−13). Inversely, BrdU-induced common fragile site FRA10C, rare fragile site FRA10A and co-mapped rare fragile site FRA10B and APH-induced common fragile site FRA10E each had a lesser proportion of segments below −40 kcal/mol compared with non-fragile DNA.
Table 2.
Distribution of proportion of segments per section with free energy below −40 kcal/mol
| Region | Chr 10 | Non-fragile | FRA10G | FRA10C | FRA10D | FRA10A | FRA10B/E | FRA10F |
|---|---|---|---|---|---|---|---|---|
| Number of sections | 17 403 | 10 334 | 1272 | 2360 | 574 | 1349 | 401 | 1113 |
| Mean | 0.087 | 0.089 | 0.122 | 0.033 | 0.198 | 0.063 | 0.064 | 0.121 |
| SD | 0.116 | 0.119 | 0.144 | 0.047 | 0.153 | 0.081 | 0.073 | 0.122 |
| Mean difference from non-fragile | – | – | 0.033 | −0.056 | 0.109 | −0.026 | −0.025 | 0.032 |
| Standard error | – | – | 0.0042 | 0.0015 | 0.0065 | 0.0025 | 0.0038 | 0.0038 |
| P-value* | – | – | 2.27E−14 | 8.19E−279 | 1.17E−51 | 1.11E−23 | 1.39E−4 | 2.77E−15 |
*Overall, the mean difference among fragile and non-fragile sites analyzed by analysis of variance is highly significant (P = 7.7E−292). Subsequently, post-hoc Student's t-tests (two-tailed) were performed between each fragile site and non-fragile DNA. The P-values were adjusted for multiple comparisons by Bonferroni's correction.
**Fragile sites highlighted in gray have a significantly greater proportion of segments per section with a free-energy value less than −40 kcal/mol.
Interestingly, in both analyses only APH-induced common fragile sites were statistically significant in favor of forming a stable secondary structure compared with non-fragile regions, except for FRA10E, an APH-induced common fragile site co-located at a BrdU-induced rare fragile site FRA10B. However, a literature search revealed that FRA10E was initially classified as a provisional common fragile site (37) and has not been verified since. Therefore, due to its overlap with a rare fragile site and poor classification, this region may not be an APH-induced common fragile site. Together, these results show that APH-induced common fragile sites on chromosome 10 possess more sequences with potential to form highly stable secondary structures, and these structures cluster together more often than non-fragile DNA.
Refining of cytogenetically defined fragile sites
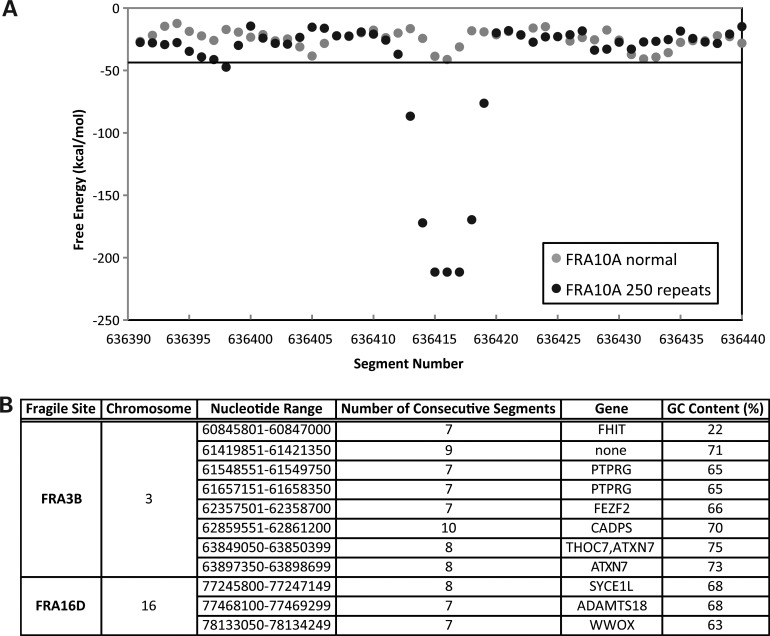
Most fragile sites are identified by G-banding of metaphase chromosomes (38,39), and therefore, not all regions within these sites are indeed fragile due to the limited resolution of determining fragile site boundaries cytogenetically. Therefore, if highly stable secondary structure-forming ability is a contributing factor to fragile site instability, this property could be used to narrow down sites of fragility within cytogenetically defined fragile sites. Next, utilizing the Mfold DNA secondary structure data, we investigated the development of a threshold to predict regions susceptible to fragile site breakage. The formation of alternative DNA secondary structures corresponding to disease-associated trinucleotide repeat expansions has been well established at rare, folate-sensitive fragile sites (6). All folate-sensitive fragile sites sequenced to date consist of a CGG repeat, including FRA10A, the most prevalent folate-sensitive rare fragile site in the human genome (40). FRA10A has been characterized at 10q23.3 (40) and 10q24.2 (37,41,42). Sequence analysis of FRA10A at 10q23.3 suggested a minimum of approximately 250 CGG repeats for fragile site breakage (40). The reference sequence (hg build 37.2) that we used in this study has eight CGG repeats within the FRA10A allele at 10q23.3. Mfold predictions of the reference sequence containing the non-expanded repeats and surrounding sequence (7650-nt) do not show any significant DNA secondary structure-forming potential (Fig. 2A). This is not surprising since the large repeat expansions necessary for fragile site breakage are not present in the reference sequence. To determine a threshold of predicted DNA secondary structure-forming potential necessary to result in fragile site breakage, we artificially inserted an additional 242 CGG repeats into the reference sequence, resulting in a total of 250 repeats. Mfold analysis then revealed that the segments corresponding to the expanded CGG repeat exhibit low free-energy values, where using a 95% confidence interval, we found seven consecutive 300 nt segments below −43.61 kcal/mol (Fig. 2A). Since expanded CGG repeats form stable secondary structures in vitro and at least 250 repeats at FRA10A results in fragile site breakage, we established a threshold for potential fragile site breakage as having at least seven consecutive segments with a free-energy value of below −43.61 kcal/mol.
Figure 2.
Establishment and validation of a threshold for Mfold prediction of chromosomal fragility. (A) The free-energy values for the most favorable Mfold-predicted DNA secondary structure for each 300 nt segment with 150 nt increments are shown for FRA10A at 10q23.3 with eight CGG repeats and surrounding sequence found in the reference sequence (gray dots) and artificially inserted CGG repeats to total 250 repeats (black dots). The (CGG)8-containing sequence is not predicted to form significant DNA secondary structures. A minimum of 250 CGG repeats is necessary for FRA10A breakage, and this sequence is predicted to form stable DNA secondary structures. When using a 95% confidence interval for chromosome 10, these expanded repeats exhibit seven consecutive segments below −43.61 kcal/mol (horizontal black line). (B) Applying the fragility prediction threshold to Mfold-predicted DNA structures for FRA3B and FRA16D, eight and three regions were identified, respectively. Overlap with gene sequences and the percentage of GC content are indicated for each of the regions.
Next, we examined the validity of this threshold by analyzing common fragile sites characterized on the molecular level. The two most active common fragile sites in the genome, FRA3B and FRA16D (18) display fragility in the regions of 4.5 (43) and 2.8 Mb (44), respectively. The DNA sequence corresponding to these characterized regions was analyzed for DNA secondary structure-forming potential, using the Mfold program in the same manner as the chromosome 10 sequence. Upon applying the DNA fragility threshold, we identified eight regions within FRA3B and three within FRA16D, encoding for six and three genes, respectively (Fig. 2B and Supplementary Material, Fig. S3). Together, these regions account for 0.23 and 0.13% of the defined FRA3B and FRA16D sequences, respectively. One FRA3B region is within a previously defined ‘active’ region located within tumor suppressor gene FHIT (45,46) and corresponds to APH-induced hybrid deletions (16), a hereditary renal cell carcinoma translocation breakpoint (47) and deletions found in gastric (48), lung (49), cervical (50,51), breast, acute lymphoblastic leukemia, esophageal, liver, brain, skin and prostate cancers and cancer cell lines (Tumorscape). Within FRA16D, we identified a region corresponding to the tumor suppressor gene WWOX, which contains heterozygous deletions found in gastric and colorectal adenocarcinomas (52) and frequent loss-of-heterozygosity in breast cancer (53). Additionally, all the three regions identified in FRA16D correspond to deletions observed in colorectal, breast, lung, brain, skin and prostate cancers (Tumorscape). Therefore, these findings indicate the ability of the DNA fragility threshold to predict regions with the potential of fragile site breakage.
We next applied the DNA fragility threshold to the chromosome 10 sequence, and identified a total of 615 potential fragile regions (Supplementary Material, Table S4), with an average of 9.6 ± 3.4 consecutive segments below −43.61 kcal/mol and a range of 7–39. These regions account for 0.74% of the overall chromosome 10 sequence. Approximately 31% of the predicted fragile regions are located within the cytogenetic boundaries of previously detected common fragile sites, and the remaining regions are located within the non-fragile regions. The latter group may represent potential new fragile sites not detected previously or too small to be detected on the cytogenetic level. The former group, including 172 regions within APH-induced sites and 15 within a BrdU-induced site, can be used to refine potential sites of fragility within previously known fragile sites. For example, within FRA10G, the largest (∼10 Mb) of all four APH-induced sites on chromosome 10, there are 74 regions with at least seven consecutive segments below −43.61 kcal/mol (Supplementary Material, Table S4 and Fig. S4), and their distribution revealed variations in the presence of the predicted fragile regions (Supplementary Material, Fig. S5), with a cluster of regions possessing a high number of consecutive segments below −43.61 kcal/mol located at the 5′ portion of the sequence.
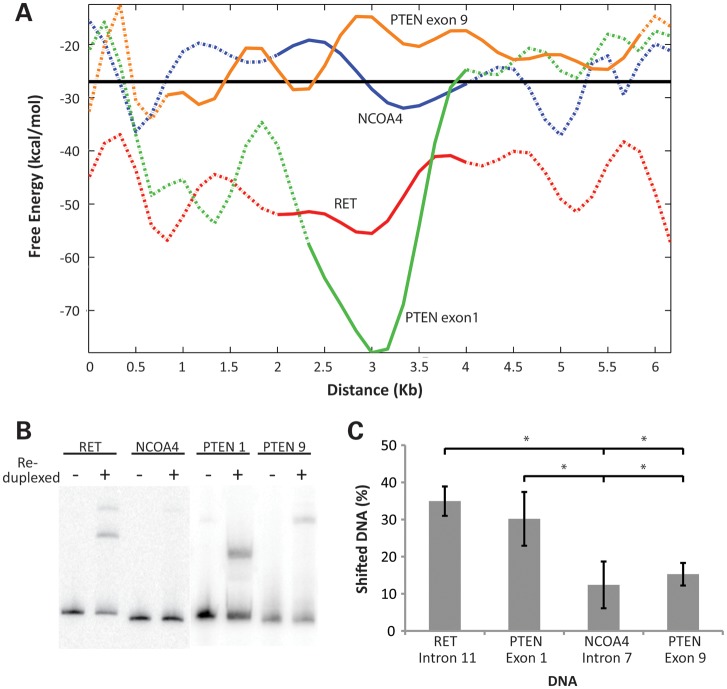
RET, an oncogene involved in 12 translocations that result in papillary thyroid carcinoma, known as RET/PTC rearrangements (54), is located within the cluster at the 5′ portion of the FRA10G sequence (Supplementary Material, Fig. S5). The average free energy of the entire RET sequence is −41.0 ± 11.9 kcal/mol, which is significantly lower than the remaining chromosome 10 sequence (−27.2 ± 9.3 kcal/mol) (P = 4.77E−68), and the RET gene contains five predicted fragile regions with a range of 9–16 consecutive segments below −43.61 kcal/mol (Supplementary Material, Table S4 and Fig.S4). In each of the reported RET/PTC rearrangements, RET fuses with a different partner gene. NCOA4, one genetic partner of RET which participates in the RET/PTC3 translocation (55), is also located in FRA10G. In contrast, NCOA4 is located within a region devoid of predicted fragile regions identified by the threshold (Supplementary Material, Fig. S5). The average free energy of the NCOA4 sequence is −26.8 ± 8.2 kcal/mol, comparable with the value for the overall chromosome 10 sequence.
Studies of RET/PTC tumors indicate that patient breakpoint cluster regions are located in intron 11 of RET and intron 7 of NCOA4 (55). As with the overall free-energy values for RET and NCOA4, intron 11 of RET has a significantly more energetically favorable free-energy value (−48.5 ± 7.9 kcal/mol) compared with the average predicted free-energy value of chromosome 10 (−27.2 ± 9.3 kcal/mol) (P = 1.60E−17), and is within the predicted fragile regions, while intron 7 of NCOA4 (−25.9 ± 6.7 kcal/mol) exhibits a value similar to the chromosome 10 average (Fig. 3A). Further supporting its secondary structure formation predictions and its involvement in APH-induced DNA fragility, RET, but not NCOA4, exhibits high levels of chromosomal breakage following APH treatment in the HTori-3 human thyroid epithelial cell line (17). In these experiments using fluorescence in situ hybridization assays, we observed RET breakage at a rate of 6% of chromosomes with 0.4 μm APH treatment compared with only 0.62% of chromosomes for NCOA4. DNA breakage within intron 11 of RET has also been confirmed on the nucleotide level following APH treatment. We detected 0.024 ± 0.015 breaks per 100 cells within intron 11 with 0.4 μm APH treatment, compared with 0.004 ± 0.009 breaks per 100 cells without treatment (P = 0.010) (17).
Figure 3.
DNA secondary structure prediction and in vitro detection within the regions predicted to exhibit fragile site instability. (A) The computed lowest free-energy value of the predicted DNA secondary structures from segments analyzed by the Mfold program was fit to a curve for regions isolated by the threshold for potential fragility within RET (red) and PTEN exon 1 (green), and regions not isolated by the threshold within NCOA4 (blue) and PTEN exon 9 (orange). The Matlab function polyfit found coefficients of a polynomial P(X) of degree N that fit the raw data best in a least-squares sense. Intron 11 of RET (solid line), exon 1 of PTEN (solid line), intron 7 of NCOA4 (solid line) and exon 9 of PTEN (solid line) plus flanking sequences (dashed) are depicted. The mean free energy of chromosome 10 (−27.2 kcal/mol) is depicted by the horizontal black line. The x-axis indicates the size of the sequences and the y-axis displays the free energy of the predicted structure. (B) Representative gel electrophoresis analysis of re-annealed RET intron 11, PTEN exon 1, NCOA4 intron 7 and PTEN exon 9 DNAs. DNA fragments were denatured and subjected to re-duplexing in the presence of 0.1 m NaCl and analyzed by native 4% PAGE. When compared with untreated samples, slower-migrating products suggest the formation of a secondary structure during re-duplexing of these DNAs. (C) Average percentages of DNA secondary structures for DNA fragments of RET (n = 12), PTEN exon 1 (n = 7), NCOA4 (n = 12) and PTEN exon 9 (n = 5) are shown with ±SD. The intensity of all bands was measured by densimetric analysis, and the percentage of shifted bands were calculated and averaged over 5–12 individual experiments performed over a range of NaCl concentrations (50 mm–1 m). Statistical analysis was performed using a Student's t-test (two-tailed), where * refers to P < 2E−3.
These data suggest that the ability to potentially form a highly stable DNA secondary structure contributes to the observed fragile site-induced breakage at RET and to some extent chromosomal breakage in patients leading to RET/PTC rearrangements, which ultimately result in papillary thyroid carcinoma. Using DNA secondary structure predictions and the established threshold, we have refined regions of fragility within the cytogenetic boundaries of FRA10G and other common fragile sites, as well as predict potential new fragile sites.
Validating the formation of secondary structures
To validate the prediction of the Mfold program, we investigated secondary structure formation within the breakpoint cluster regions of RET and NCOA4 by subjecting these DNA fragments to re-duplexing with various concentrations of NaCl to allow re-annealing of the single strands following denaturation. This in vitro re-duplexing assay has been used to analyze the formation of DNA secondary structures generated by CTG (56,57) and CGG (58) repeats, as well as the FRA16B AT-rich repeat sequence (27). Here, in addition to the two sequences derived from FRA10G, two regions (exon 1 and exon 9) within the tumor suppressor gene PTEN were also examined. PTEN, one of the most highly mutated tumor suppressor genes (59), contains one predicted fragile region with eight consecutive segments below −43.61 kcal/mol (Supplementary Material, Table S4). Mutations within PTEN are found throughout the gene and its promoter region, with few mutations located after exon 8 (60). The region identified in this study contains exon 1 and the promoter sequence of PTEN, where high levels of mutations have been observed. The average free energy for exon 1 of PTEN (−58.4 ± 21.3 kcal/mol) is significantly more energetically favorable compared with the average free energy of chromosome 10 (−27.2 ± 9.3 kcal/mol) (P = 1.25E−3) (Fig. 3A). Exon 9 of PTEN is located within a region lacking mutations (60), and like NCOA4, has an average free energy (−21.2 ± 4.0 kcal/mol) similar to chromosome 10 (Fig. 3A).
The 366 bp RET intron 11 and 317 bp PTEN exon 1 DNA fragments predicted to form highly stable secondary structures (with free-energy values −74.5 and −73.2 kcal/mol, respectively), and the 348 bp NCOA4 intron 7 and 289 bp PTEN exon 9 DNA fragments predicted to form less stable structures (−20.1 and −23.2 kcal/mol, respectively) (Supplementary Material, Fig. S6), were subjected to the re-duplexing reaction to observe the formation of DNA secondary structures. We observed slower migrating products in the re-duplexed samples, but not in the untreated samples (Fig. 3B), suggesting the formation of secondary structures during re-duplexing of these DNAs. RET, PTEN exon 1, NCOA4 and PTEN exon 9 all exhibited the formation of secondary structures to varying degrees (Fig. 3B). While the concentration of NaCl (0.05–1 m) had no impact on the amount of the secondary structure formation, DNA sequence did. RET and PTEN exon 1 showed significantly greater secondary structure formation compared with NCOA4 (P = 4.8E−10 and 3.0E−5, respectively) and PTEN exon 9 (P = 5.7E–8 and 1.6E–3, respectively) (Fig. 3C). No significant difference was observed in the level of secondary structure formation between RET and PTEN exon 1 or between NCOA4 and PTEN exon 9. Therefore, these data confirm that an energetically favorable free-energy value predicted by Mfold program corresponds to greater DNA secondary structure formation, validating the differential folding propensity of the Mfold program predictions. These results demonstrate the validity of Mfold secondary structure predictions.
Analysis of regions capable of forming highly stable secondary structures
The co-localization of these predicted fragile regions with genes was next compared. Of these 615 regions, 426 (69%) are located within regions encoding for 258 genes (Supplementary Material, Table S4). Among them, 47 genes (possessing 71 predicted fragile regions) have known mutations of insertions, deletions, translocation, or point mutations that result in cancer or other diseases in humans (Table 3; Supplementary Material, Table S5). Specifically, six of the 22 genes with mutations consisting of insertions, deletions, or translocations are specifically found in cancer (Fig. 4). Included within these genes are RET and PTEN, which form significant levels of DNA secondary structure in vitro. CCDC6, the partner gene of RET in the RET/PTC1 rearrangement in PTC (54), was also identified. Located within the BrdU-induced common fragile site FRA10C, CCDC6 exhibits chromosomal breakage in HTori-3 cells at a rate of 2.72% of chromosomes following BrdU treatment (17). Additionally, the region predicted in this study to form high levels of secondary structure corresponds to intron 1 of CCDC6, the breakpoint cluster region isolated in RET/PTC patient tumors (55). Also among these genes are ZMIZ1 (61), ADD3 (62) and NFKB2 (63), all known to be disrupted in chromosomal translocations resulting in leukemia. In B-cell acute lymphoblastic leukemia, ZMIZ1 rearranges with ABL1 (61), and translocations of ADD3 with NUP98 (62) are detected in T-cell acute lymphoblastic leukemia. NFKB2 rearranges with INA, resulting in B-cell chronic lymphocytic leukemia and cutaneous T-cell lymphoma (64); with TBXAS1, resulting in multiple myeloma and with IGH@, resulting in B-cell non-Hodgkin's lymphoma (65). NFKB2 also displays deletions, point mutations and gene amplification found in B- and T-cell leukemias and lymphomas (63).
Table 3.
Genes located in regions capable of forming highly stable secondary structures and disease associations
| Gene | Chromosomal position | Fragile site | Insertion, deletion, translocationa | Point mutationa |
|---|---|---|---|---|
| PHYH | 10p13 | + | ||
| NMT2 | 10p13 | + | ||
| VIM | 10p13 | + | ||
| CACNB2 | 10p12.33 | + | ||
| NEBL | 10p12.31 | + | ||
| BMI1 | 10p12.2 | + | ||
| PTF1A | 10p12.2 | + | ||
| MAP3K8 | 10p11.23 | + | ||
| ZEB1 | 10p11.22 | + | ||
| RET | 10q11.21 | FRA10G | + | + |
| CXCL12 | 10q11.21 | FRA10G | + | |
| ALOX5 | 10q11.21 | FRA10G | + | |
| CHAT | 10q11.23 | FRA10G | + | |
| CCDC6 | 10q21.2 | FRA10C | + | |
| RHOBTB1 | 10q21.2 | FRA10C | + | |
| STOX1 | 10q21.3 | FRA10C | + | |
| HK1 | 10q22.1 | FRA10D | + | |
| NEUROG3 | 10q22.1 | FRA10D | + | |
| PCBD1 | 10q22.1 | FRA10D | + | |
| CDH23 | 10q22.1 | FRA10D | + | |
| CHST3 | 10q22.1 | FRA10D | + | |
| KCNMA1 | 10q22.3 | + | ||
| ZMIZ1 | 10q22.3 | + | ||
| CDHR1 | 10q23.1 | + | ||
| LDB3 | 10q23.2 | + | ||
| BMPR1A | 10q23.2 | + | + | |
| GLUD1 | 10q23.2 | + | ||
| PTEN | 10q23.31 | FRA10A | + | + |
| RBP4 | 10q23.33 | FRA10A | + | |
| HPS1 | 10q24.2 | FRA10A | + | |
| ABCC2 | 10q24.2 | FRA10A | + | + |
| PAX2 | 10q24.31 | + | + | |
| PDZD7 | 10q24.31 | + | ||
| FGF8 | 10q24.32 | + | ||
| HPS6 | 10q24.32 | + | + | |
| PITX3 | 10q24.32 | + | + | |
| NFKB2 | 10q24.32 | + | ||
| CNNM2 | 10q24.32 | + | + | |
| COL17A1 | 10q24.33 | + | + | |
| ADD3 | 10q25.1 | + | ||
| ADRB1 | 10q25.3 | + | + | |
| BAG3 | 10q26.11 | FRA10F | + | + |
| FGFR2 | 10q26.13 | FRA10F | + | + |
| PLEKHA1 | 10q26.13 | FRA10F | + | |
| HTRA1 | 10q26.13 | FRA10F | + | |
| OAT | 10q26.13 | FRA10F | + | + |
| ADAM12 | 10q26.2 | + |
aDetailed descriptions of diseases and references are found in Supplementary Material, Table S5.
Figure 4.
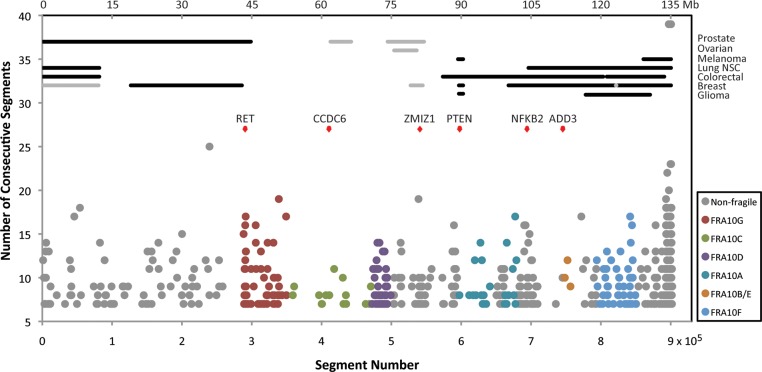
Location of regions predicted to exhibit fragile site instability and correlation with cancer-associated chromosomal aberrations. Regions on chromosome 10 with at least seven consecutive segments below −43.61 kcal/mol are presented. The lower x-axis depicts the number of the first 300 nt segment of the consecutive segments identified by the threshold, the upper x-axis displays the corresponding chromosome 10 nucleotide number and the y-axis depicts the number of consecutive segments within the each identified region. The locations of the six genes with insertion, deletion or translocation mutations in cancer that coincide with regions isolated by the threshold are marked by red diamonds. Regions of copy number alterations (Tumorscape) across various tumor types that overlap with regions identified by the threshold are depicted in horizontal lines. Black lines represent deletions and gray lines represent amplifications.
The location of these predicted fragile regions was also compared with copy number alterations observed in various tumors and cancer cell lines (66) (Tumorscape). We found that the regions identified by the fragility prediction threshold mostly overlap with the regions containing deletions in breast, colorectal, non-small-cell (NSC) lung and prostate cancers; glioma; melanoma; and amplifications in breast, ovarian and prostate cancers (Fig. 4; Supplementary Material, Table S6). Deletions in breast, colorectal, glioma and melanoma are found in the region identified to contain PTEN exon 1, in which deletions, insertions and point mutations have been sequenced in various cancers (59), and stable DNA secondary structures were observed in vitro (described above). The region identified within NFKB2 is included in the locations of deletions found in colorectal and breast cancers, and the predicted fragile region within ADD3 overlaps with deletions found in NSC lung, colorectal and breast cancers. Deletions found in melanoma, NSC lung, colorectal and breast cancers, and glioma span the identified regions within the APH-induced common fragile site FRA10F, and sequences located near the telomere on the q-arm of chromosome 10 (10q26.2-qter), which constitutes 33% of the predicted fragile regions but was not previously identified as a fragile site. Amplifications found in breast, ovarian and prostate cancers include regions of predicted DNA secondary structure that cluster within 10q22.2–3, which contains the ZMIZ1 gene, and borders the cytogenetic boundaries of the APH-induced common fragile site FRA10D (10q22.1). Additionally, 86 significant regions were identified on the p-arm of chromosome 10, where no chromosomal fragile site was previously defined. These regions on the p-arm coincide with deletions found in breast, prostate, NSC lung and colorectal cancers, as well as amplifications found in breast cancer.
Therefore, using the ability to form highly stable secondary structures within a given threshold defined by the analysis of molecularly defined fragile sites, we have narrowed down cytogenetically defined fragile sites; identified possible new fragile sites in previously unidentified regions on chromosome 10 and correlated these sites both with known mutations in human disease and regions of copy number alterations in cancer. We suggest that the secondary structure-forming/replication stalling mechanism for DNA breakage could occur within these regions to generate the respective cancer-specific gene rearrangements.
DISCUSSION
In this study, we analyzed the differences in multiple stem loop DNA secondary structure-forming potential between fragile and non-fragile DNA on chromosome 10 using the DNA secondary structure prediction program, Mfold. APH-induced common fragile sites, FRA10G, FRA10D and FRA10F, had a greater potential to form stable DNA secondary structures compared with non-fragile DNA (Table 1). Additionally, DNA capable of forming these stable secondary structures clustered more densely in these APH-induced common fragile sites compared with non-fragile DNA (Table 2). FRA10E, although classified as an APH-induced common fragile site, did not have a greater potential to form stable DNA secondary structures compared with non-fragile DNA (Table 1, Table 2). However, the original classification of this fragile site was listed as provisional (67), and further experiments to confirm its fragility have not been performed. Therefore, we suggest that this region may not be a true APH-induced common fragile site. To our knowledge, this is the first unbiased demonstration supporting secondary structure formation at proven APH-induced common fragile sites as a distinguishing feature of these regions from non-fragile sites.
We also confirmed the validity of the secondary structure predictions using an in vitro re-duplexing assay of RET, PTEN and NCOA4 sequences. Intron 11 of RET and exon 1 of PTEN are located within the regions identified by the fragility prediction threshold (Supplementary Material, Table S4), and are predicted to form secondary structures with energetically favorable free-energy values significantly lower than the average value of the chromosome 10 sequence (Fig. 3A). In contrast, intron 7 of NCOA4 and exon 9 of PTEN were not isolated in our study and are predicted to form secondary structures with free-energy values similar to the overall chromosome 10 sequence (Fig. 3A). The in vitro re-duplexing assay showed that RET intron 11 and PTEN exon 1 DNA form a significantly higher amount of stable secondary structures than NCOA4 intron 7 and PTEN exon 9 DNA (Fig. 3B and C), agreeing with the differential folding propensity predicted by the Mfold program. Further, we previously found that RET, but not NCOA4, exhibits high levels of chromosomal breakage following APH treatment (17), and the formation of DNA breaks within intron 11 of RET (the major patient breakpoint cluster region found in human tumors) was also detected at the nucleotide level following APH treatment (17). Interestingly, NCOA4 is located within the same APH-induced fragile site, FRA10G, as RET. However, NCOA4, due to its inability to form highly stable secondary structures and, more importantly, the absence of chromosomal breakage in response to APH, does not fit the criteria for a bona fide APH-induced site, unlike the RET sequence. Rearrangements in NCOA4 are most frequent in patients with a history of radiation exposure, ranging from 63 to 76% of the total RET/PTC rearrangements in pediatric tumors (68–71), suggesting that radiation, not DNA secondary structures, causes DNA breakage at this site. Mutations within the PTEN gene, including deletions, are found in many types of cancer. While mutations are commonly found in exon 1 of PTEN, they are fairly devoid within exon 9 (60), agreeing with our secondary structure predictions and observations by the in vitro re-duplexing assay. These results validate the Mfold analysis as a method for predicting potential regions of DNA fragility, and support the role of alternative DNA secondary structure in the mechanism of APH-induced common fragile site instability.
The boundaries of fragile sites have traditionally been defined cytogenetically, and to date only 23 common fragile sites have been cloned and characterized. Rare fragile sites are the result of CGG trinucleotide repeats or AT-rich minisatellite repeats occupied only a small area of the cytogenetically defined regions (72). Unlike rare fragile sites, no consensus sequence has been identified for common fragile sites to narrow down regions of fragility, but it is unlikely that these regions are fragile throughout the currently defined cytogenetic boundaries. Additionally, cloning and characterization of fragile sites is time-consuming. Here, through Mfold analysis of FRA10A, we observed a signature of fragility when comparing the reference sequence to a fully expanded repeat sequence known to produce chromosomal breaks, thereby establishing a threshold for predicting potential fragile sites (Fig. 2A). This threshold was demonstrated by the analysis of the two most active fragile sites in the genome, FRA3B and FRA16D, which have been defined on the molecular level. Within FRA3B and FRA16D, FHIT and WWOX, respectively, exhibits highly frequent fragile site breakage as identified previously, and each contains one of the predicted fragile regions. Other predicted regions of FRA3B and FRA16D correspond to the gene location of deletions and amplifications found in cancer (73)(Supplementary Material, Fig. S3C). Although breakage of these regions is not highly induced by APH, they may contribute to the fragility of regions within FHIT and WWOX, even if they themselves do not show much direct chromosome breakage. Using this threshold, we could pinpoint sites of potential fragility within the cytogenetic boundaries of fragile sites on chromosome 10 (Fig. 4; Supplementary Material, Table S4). We isolated a total of 172 regions within APH-induced common fragile sites, effectively narrowing down fragility within these regions, as in the example of FRA10G described above. We also identified 15 regions within the BrdU-induced common fragile site FRA10C. Interestingly, in a global screening study of APH-induced fragile sites, FRA10C showed levels of chromosomal breakage in peripheral blood lymphocytes in the presence of APH, at least half that of the APH-induced common fragile sites on chromosome 10 (74). Therefore, FRA10C may have characteristics of both APH and BrdU-induced fragile site breakage, and the regions that we identified in this study may be responsible for the APH breakage (74).
Another drawback of defining DNA fragility cytogenetically at metaphase chromosomes is that some fragile regions are too small to be detected or are difficult to locate by G-banding. Therefore, using the ability to form highly stable secondary structures, we can predict ‘micro’-fragile sites or previously unidentified fragile sites within non-fragile regions. Using the same threshold as used to narrow down cytogenetically defined fragile sites, we predicted an additional 428 potential regions of fragility within non-fragile regions (Fig. 4; Supplementary Material, Table S4). A total of 86 of these regions are located on the p-arm, where no fragile sites have been well established. Interestingly, 75 of these regions correspond to the locations of chromosome breaks observed by Mrasek et al. in their global study (74).
The sequence composition of the 615 predicted fragile regions (Supplementary Material, Table S4), analyzed by RepeatMasker program (http://www.repeatmasker.org), contains 13.5% of repetitive elements, including LINEs (3%), SINEs (2.5%), LTR retrotransposons (4.5%), DNA transposons (0.8%) and simple sequence repeats (2.7%). Comparison of the analyzed sequences with the human genome showed no over-representation of any elements at these predicted fragile regions (human genome: LINEs—21%, SINEs—13%, LTRs—8%, DNA transposons—3% and simple repeats—3%, (75)). This suggests that no obvious repeat sequences can account for these fragile regions. The nucleotide composition of these regions is generally GC rich (average GC content 65.1 ± 6.9%, range 8–79%). Interestingly, only a few regions were AT rich. Previous analyses of several common fragile sites revealed AT-rich sequences with an inherent higher flexibility (21,22). Although most sequences we identified are GC rich, those that are not are instead AT rich. The GC richness of the sequences was also found in the regions identified within FRA3B and FRA16D (Fig. 2B), where one sequence was AT rich and all the rest were GC rich. In a global analysis of sequence content and DNA flexibility, Tsantoulis et al. found common fragile site sequences to be on average more GC rich and less flexible than non-fragile sequences (76). Our results agree with those observations, and further support DNA secondary structure-forming potential rather than DNA flexibility as an important feature of common fragile sites.
Extensive studies (21–31) have suggested that the ability of fragile sites to form stable secondary structures during DNA replication likely contributes to their breakage by stalling replication fork progression. In addition to replication fork stalling, paucity of replication initiation in fragile site regions (77) and the presence of transcription-derived R-loops during DNA replication of fragile sites (78), are also suggested to be involved in the mechanism of fragility. However, it is not clear whether the potential of fragile site sequences to form highly stable secondary structures participates in the latter two mechanisms. Interestingly, the formation of R-loops promotes trinucleotide repeat instability. The ability of trinucleotide repeats to form stable secondary structure may stabilize the presence of R-loops by adopting hairpin structure on the non-template DNA strand, and therefore, favoring hybrid formation between RNA transcripts and the DNA template strand (79–82). Further investigation of the specific role of secondary structure-forming ability of fragile sites in the mechanism of fragility is needed.
This study is the first unbiased investigation of DNA secondary structure-forming potential at fragile sites versus non-fragile regions, and supports the role of DNA secondary structure formation in the mechanism of APH-induced common fragile site instability. Additionally, we have established a method whereby sites of fragility can be predicted, allowing systematic identification of common fragile sites. The co-localization of fragile sites and genes deleted, amplified or rearranged in cancer is well documented (12). Also, genomic and epigenomic instability in neuropsychiatric diseases such as schizophrenia and autism has been linked to fragile sites (15). Therefore, the ultimate goal is to create a list of legitimate sites that are prone to DNA breakage caused by the secondary structure-forming mechanism(s), to evaluate genomic stress caused by endogenous and exogenous insults. To create a panel of such regions and measure their fragility could tailor diagnosis and therapy based on direct knowledge of DNA damage.
MATERIALS AND METHODS
DNA secondary structure prediction
The nucleotide sequence of chromosome 10 was obtained from the Human Genome Project build 37.2 (hg37.2) as FASTA text files. The downloaded contig sequences were assembled sequentially, in the p- to q-arm direction, with unsequenced gap nucleotides between contigs assigned a sequence of ‘N’. A list of known fragile sites on chromosome 10 and their location was obtained from NCBI GeneBank (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene using "fragile site" as a search term). For example, FRA10G is located at cytogenetic position 10q11.2, corresponding to nucleotides 42 300 001–52 900 000. Fragile sites were then assigned based on common or rare, and mode of induction (APH, BrdU or folic acid). Non-fragile regions were defined by removing all known fragile sites, centromeric (32), subtelomeric (33) and gap sequences.
Using the Mfold program (34), the potential of single-stranded DNA to form a stable secondary structure can be predicted along with its free-energy value. The secondary structure-forming potential of the entire chromosome 10 was analyzed by inputting 300 nt segments with a 150 nt shift window into the Mfold program. We chose the 300 nt length because it equals the Okazaki initiation zone of the DNA replication fork in mammalian cells, which possesses a single-stranded property during DNA replication (35,36). The default [Na+], [Mg2+] and temperature used were 1.0, 0.0 m and 37°C, respectively. The free-energy value of the most stable predicted secondary structure for each segment was used in the analyses. A section was arbitrarily defined as 50 consecutive segments for evaluation whether segments with a low predicted free-energy value clustered within a region.
Analyses of DNA secondary structure within FRA3B and FRA16D were performed using the Mfold program in the same manner as above. The FRA3B sequence, defined by Becker et al. (43), is located between nucleotides 59 619 291 and 64 206 880 of chromosome 3. The FRA16D sequence, defined by Krummel et al. (44), is located between nucleotides 76 517 028 and 79 359 665 on chromosome 16.
Statistical analysis
The mean and SD of the predicted free-energy values were first calculated for the entire chromosome to generate the threshold values significantly deviating from the overall sequence (<1 SD or 2 SD). The average predicted free-energy value of the entire chromosome 10 is −27.25 ± 9.35 kcal/mol. Therefore, we defined two threshold values of −40 and −50 kcal/mol that correspond to ∼1 and 2 SD below the average predicted free energy. A free-energy value of each segment below these two thresholds indicates theoretically that it is energetically more favorable to fold into secondary structures compared with the overall sequence. The mean and SD were then calculated for each fragile site as well as the non-fragile regions, using the predicted free-energy values for the 300 nt segments contained within those regions. Segments containing sequences overlapping with other regions or ‘N’ gap sequence were not used in analyses. To compare the overall ability of each fragile or non-fragile region to form stable secondary structures, the percentage of segments within each region with free-energy value less than −40 or −50 kcal/mol was calculated. Using a chi-square test, the percentage of segments below each threshold was compared between the non-fragile region and each fragile site.
To investigate whether low free-energy segments form clusters, 50 consecutive 300 nt segments were grouped into one section and numbered consecutively from the p- to q-arm direction. The proportion of segments with a free-energy value less than −40 kcal/mol within each section was calculated. A high proportion suggests that the low free-energy segments tend to cluster together in that section when compared with other sections. The mean and SD of the proportion of segments were calculated for the sections contained within each fragile site and the non-fragile regions. The sections containing sequences overlapping with other regions or ‘N’ gap sequence were not used in analyses. We first assessed for overall difference in proportion of low free-energy segments among the seven non-fragile and fragile regions by analysis of variance. Then, we performed post-hoc comparisons for the mean difference in the proportion of segments per section for each fragile site versus the non-fragile DNA using a Student's t-test (two-tailed). The P-values were calculated after Bonferroni correction for multiple comparisons. All statistical analyses were performed using IBM SPSS Statistics v.19 (SPSS, Chicago, IL, USA).
Plasmids
The plasmid containing intron 11 of human RET sequence, pRET3, was constructed by first generating a 2177 bp fragment produced by polymerase chain reaction (PCR) of human genomic DNA using primers (5′-TACCCTGCTCTGCCTTTCAGATGG-3′ and 5′-CTGTCCTCTTCTCCTTCATC-3′) flanking intron 11 of RET, and then cloning into the Smal site of the pGEM3zf(+) vector (Promega Co.).
The plasmid containing intron 7 of human NCOA4 sequence, pELE1, was constructed by first generating a 2262 bp fragment produced by PCR of human genomic DNA using primers (5′-AGACCTTGGAGAACAGTCAG-3′ and 5′-CAGAGCCTCCTTCTCACAATTC-3′) flanking intron 7 of NCOA4, and then cloning into the pGEM3zf(+) vector.
Re-duplexing assay
Re-duplexing reactions were performed as previously described (27,56,57) with slight modifications. The 366 bp RET fragment (corresponding to 43 610 126–43 610 492 of chromosome 10 from hg37.2) was obtained by digesting pRET3 with restriction enzymes PstI and NaeI. The 348 bp NCOA4 fragment (corresponding to 51 583 022–51 583 371) was obtained by digesting pELE1 with restriction enzymes SwaI and HincII. The 317 bp PTEN exon 1 fragment (corresponding to 89 623 390–89 623 693) was obtained by digesting a PTEN cDNA clone (Open Biosystems, Catalog # MHS1011–61 412) with restriction enzymes EcoRI and BanI. The 289 bp PTEN exon 9 fragment (corresponding to 89 725 202–89 725 489) was obtained by digesting the PTEN cDNA clone with DpnI and AseI. The fragments were then gel-purified, dephosphorylated at the 5′ end with calf intestinal alkaline phosphatase, and end-labeled with [α-32P] ATP (PerkinElmer) using T4 kinase. End-labeled DNA (1 ng) was added to 10 μl denaturing solution (0.5 m NaOH, 1.5 m NaCl) and incubated at room temperature for 5 min. Following denaturation, 490 μl 5× TE buffer containing NaCl (ranging from 0.05 to 1 m) was added to the reaction mixture and incubated at 37°C for 3 h. DNA was ethanol-precipitated in the presence of glycogen (Roche). The DNA pellets were then air-dried and resuspended in TE buffer. DNA samples were electrophoresed in a 4% polyacrylamide gel cast in tris-borate-EDTA at 150 V for 3 h at room temperature. The gel was dried and visualized by phosphorimaging (GE Healthcare). All enzymes were purchased from New England Biolabs.
Copy Number Alterations
Regions of copy number alteration on chromosome 10 were obtained from Tumorscape (www.broadinstitute.org/tumorscape).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (RO1CA113863 to Y.-H.W. and T32GM095440 to L.W.D.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rafael Diaz-Garcia for generating the plasmids, pRET3 and pELE1.
Conflict of interest. None declared.
REFERENCES
- 1.Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. doi:10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J., Bacolla A., Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. doi:10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wells R.D. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. doi:10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. doi:10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 5.Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. doi:10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz M., Zlotorynski E., Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. doi:10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka H., Bergstrom D.A., Yao M.C., Tapscott S.J. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 2005;37:320–327. doi: 10.1038/ng1515. doi:10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 8.Abeysinghe S.S., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. doi:10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- 9.Chuzhanova N., Abeysinghe S.S., Krawczak M., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer II: potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum. Mutat. 2003;22:245–251. doi: 10.1002/humu.10253. doi:10.1002/humu.10253. [DOI] [PubMed] [Google Scholar]
- 10.Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. doi:10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards R.I. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17:339–345. doi: 10.1016/s0168-9525(01)02303-4. doi:10.1016/S0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 12.Popescu N.C. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. doi:10.1016/S0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 13.Burrow A.A., Williams L.E., Pierce L.C., Wang Y.H. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. doi: 10.1186/1471-2164-10-59. doi:10.1186/1471-2164-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. doi:10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith C.L., Bolton A., Nguyen G. Genomic and epigenomic instability, fragile sites, schizophrenia and autism. Curr. Genomics. 2010;11:447–469. doi: 10.2174/138920210793176001. doi:10.2174/138920210793176001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durkin S.G., Ragland R.L., Arlt M.F., Mulle J.G., Warren S.T., Glover T.W. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl Acad. Sci. USA. 2008;105:246–251. doi: 10.1073/pnas.0708097105. doi:10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi M., Dillon L.W., Pramanik S., Nikiforov Y.E., Wang Y.H. DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene. 2010;29:2272–2280. doi: 10.1038/onc.2009.502. doi:10.1038/onc.2009.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durkin S.G., Glover T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. doi:10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 19.Cheng C.H., Kuchta R.D. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993;32:8568–8574. doi: 10.1021/bi00084a025. doi:10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- 20.Glover T.W., Berger C., Coyle J., Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984;67:136–142. doi: 10.1007/BF00272988. doi:10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 21.Mishmar D., Rahat A., Scherer S.W., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl Acad. Sci. USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. doi:10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. doi:10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. doi:10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gacy A.M., Goellner G., Juranic N., Macura S., McMurray C.T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. doi:10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 25.Usdin K., Woodford K.J. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. doi:10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samadashwily G.M., Raca G., Mirkin S.M. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. doi:10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 27.Burrow A.A., Marullo A., Holder L.R., Wang Y.H. Secondary structure formation and DNA instability at fragile site FRA16B. Nucleic Acids Res. 2010;38:2865–2877. doi: 10.1093/nar/gkp1245. doi:10.1093/nar/gkp1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Freudenreich C.H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cervisiae. Mol. Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. doi:10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah S.N., Opresko P.L., Meng X., Lee M.Y., Eckert K.A. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 2010;38:1149–1162. doi: 10.1093/nar/gkp1131. doi:10.1093/nar/gkp1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamath-Loeb A.S., Loeb L.A., Johansson E., Burgers P.M., Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. doi:10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 31.Ozeri-Galai E., Lebofsky R., Rahat A., Bester A.C., Bensimon A., Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell. 2011;43:122–131. doi: 10.1016/j.molcel.2011.05.019. doi:10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 32.Jackson M.S., Rocchi M., Thompson G., Hearn T., Crosier M., Guy J., Kirk D., Mulligan L., Ricco A., Piccininni S., et al. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. doi:10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- 33.Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. doi:10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. doi:10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson S., DePamphilis M.L. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J. Biol. Chem. 1979;254:11495–11504. [PubMed] [Google Scholar]
- 36.Hay R.T., DePamphilis M.L. Initiation of SV40 DNA replication in vivo: location and structure of 5′ ends of DNA synthesized in the ori region. Cell. 1982;28:767–779. doi: 10.1016/0092-8674(82)90056-3. doi:10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- 37.Muleris M., Dutrillaux A.M., Lombard M., Dutrillaux B. Noninvolvement of a constitutional heritable fragile site at 10q24.2 in rearranged chromosomes from rectal carcinoma cells. Cancer Genet. Cytogenet. 1987;25:7–13. doi: 10.1016/0165-4608(87)90153-1. doi:10.1016/0165-4608(87)90153-1. [DOI] [PubMed] [Google Scholar]
- 38.Hecht F. Fragile sites, cancer chromosome breakpoints, and oncogenes all cluster in light G bands. Cancer Genet. Cytogenet. 1988;31:17–24. doi: 10.1016/0165-4608(88)90005-2. doi:10.1016/0165-4608(88)90005-2. [DOI] [PubMed] [Google Scholar]
- 39.Mishmar D., Mandel-Gutfreund Y., Margalit H., Rahat A., Kerem B. Common fragile sites: G-band characteristics within an R-band. Am. J. Hum. Genet. 1999;64:908–910. doi: 10.1086/302299. doi:10.1086/302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarafidou T., Kahl C., Martinez-Garay I., Mangelsdorf M., Gesk S., Baker E., Kokkinaki M., Talley P., Maltby E.L., French L., et al. Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84:69–81. doi: 10.1016/j.ygeno.2003.12.017. doi:10.1016/j.ygeno.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 41.Dutrillaux B., Muleris M., Prieur M. Exact localization of several fragile sites remains uncertain. The example of fra(10) sensitive to folate. Ann. Genet. 1985;28:161–163. [PubMed] [Google Scholar]
- 42.Giraud F., Ayme S., Mattei J.F., Mattei M.G. Constitutional chromosomal breakage. Hum. Genet. 1976;34:125–136. doi: 10.1007/BF00278880. doi:10.1007/BF00278880. [DOI] [PubMed] [Google Scholar]
- 43.Becker N.A., Thorland E.C., Denison S.R., Phillips L.A., Smith D.I. Evidence that instability within the FRA3B region extends four megabases. Oncogene. 2002;21:8713–8722. doi: 10.1038/sj.onc.1205950. doi:10.1038/sj.onc.1205950. [DOI] [PubMed] [Google Scholar]
- 44.Krummel K.A., Roberts L.R., Kawakami M., Glover T.W., Smith D.I. The characterization of the common fragile site FRA16D and its involvement in multiple myeloma translocations. Genomics. 2000;69:37–46. doi: 10.1006/geno.2000.6321. doi:10.1006/geno.2000.6321. [DOI] [PubMed] [Google Scholar]
- 45.Paradee W., Wilke C.M., Wang L., Shridhar R., Mullins C.M., Hoge A., Glover T.W., Smith D.I. A 350-kb cosmid contig in 3p14.2 that crosses the t(3;8) hereditary renal cell carcinoma translocation breakpoint and 17 aphidicolin-induced FRA3B breakpoints. Genomics. 1996;35:87–93. doi: 10.1006/geno.1996.0326. doi:10.1006/geno.1996.0326. [DOI] [PubMed] [Google Scholar]
- 46.Wilke C.M., Hall B.K., Hoge A., Paradee W., Smith D.I., Glover T.W. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum. Mol. Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. doi:10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 47.Boldog F.L., Gemmill R.M., Wilke C.M., Glover T.W., Nilsson A.S., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A. Positional cloning of the hereditary renal carcinoma 3;8 chromosome translocation breakpoint. Proc. Natl Acad. Sci. USA. 1993;90:8509–8513. doi: 10.1073/pnas.90.18.8509. doi:10.1073/pnas.90.18.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huiping C., Kristjansdottir S., Bergthorsson J.T., Jonasson J.G., Magnusson J., Egilsson V., Ingvarsson S. High frequency of LOH, MSI and abnormal expression of FHIT in gastric cancer. Eur. J. Cancer. 2002;38:728–735. doi: 10.1016/s0959-8049(01)00432-4. doi:10.1016/S0959-8049(01)00432-4. [DOI] [PubMed] [Google Scholar]
- 49.Sozzi G., Alder H., Tornielli S., Corletto V., Baffa R., Veronese M.L., Negrini M., Pilotti S., Pierotti M.A., Huebner K., et al. Aberrant FHIT transcripts in Merkel cell carcinoma. Cancer Res. 1996;56:2472–2474. [PubMed] [Google Scholar]
- 50.Corbin S., Neilly M.E., Espinosa R., 3rd, Davis E.M., McKeithan T.W., Le Beau M.M. Identification of unstable sequences within the common fragile site at 3p14.2: implications for the mechanism of deletions within fragile histidine triad gene/common fragile site at 3p14.2 in tumors. Cancer Res. 2002;62:3477–3484. [PubMed] [Google Scholar]
- 51.Mimori K., Druck T., Inoue H., Alder H., Berk L., Mori M., Huebner K., Croce C.M. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc. Natl Acad. Sci. USA. 1999;96:7456–7461. doi: 10.1073/pnas.96.13.7456. doi:10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finnis M., Dayan S., Hobson L., Chenevix-Trench G., Friend K., Ried K., Venter D., Woollatt E., Baker E., Richards R.I. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum. Mol. Genet. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. doi:10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- 53.Chen T., Sahin A., Aldaz C.M. Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res. 1996;56:5605–5609. [PubMed] [Google Scholar]
- 54.Nikiforova M.N., Nikiforov Y.E. Molecular genetics of thyroid cancer: implications for diagnosis, treatment and prognosis. Expert Rev. Mol. Diagn. 2008;8:83–95. doi: 10.1586/14737159.8.1.83. doi:10.1586/14737159.8.1.83. [DOI] [PubMed] [Google Scholar]
- 55.Smanik P.A., Furminger T.L., Mazzaferri E.L., Jhiang S.M. Breakpoint characterization of the ret/PTC oncogene in human papillary thyroid carcinoma. Hum. Mol. Genet. 1995;4:2313–2318. doi: 10.1093/hmg/4.12.2313. doi:10.1093/hmg/4.12.2313. [DOI] [PubMed] [Google Scholar]
- 56.Pearson C.E., Tam M., Wang Y.H., Montgomery S.E., Dar A.C., Cleary J.D., Nichol K. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. doi:10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson C.E., Wang Y.H., Griffith J.D., Sinden R.R. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. doi:10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearson C.E., Sinden R.R. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. doi:10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 59.Keniry M., Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. doi:10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Escudero I., Oliver M.D., Andres-Pons A., Molina M., Cid V.J., Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum. Mol. Genet. 2011;20:4132–4142. doi: 10.1093/hmg/ddr337. doi:10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 61.Soler G., Radford-Weiss I., Ben-Abdelali R., Mahlaoui N., Ponceau J.F., Macintyre E.A., Vekemans M., Bernard O.A., Romana S.P. Fusion of ZMIZ1 to ABL1 in a B-cell acute lymphoblastic leukaemia with a t(9;10)(q34;q22.3) translocation. Leukemia. 2008;22:1278–1280. doi: 10.1038/sj.leu.2405033. doi:10.1038/sj.leu.2405033. [DOI] [PubMed] [Google Scholar]
- 62.Lahortiga I., Vizmanos J.L., Agirre X., Vazquez I., Cigudosa J.C., Larrayoz M.J., Sala F., Gorosquieta A., Perez-Equiza K., Calasanz M.J., et al. NUP98 is fused to adducin 3 in a patient with T-cell acute lymphoblastic leukemia and myeloid markers, with a new translocation t(10;11)(q25;p15) Cancer Res. 2003;63:3079–3083. [PubMed] [Google Scholar]
- 63.Courtois G., Gilmore T.D. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. doi:10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 64.Migliazza A., Lombardi L., Rocchi M., Trecca D., Chang C.C., Antonacci R., Fracchiolla N.S., Ciana P., Maiolo A.T., Neri A. Heterogeneous chromosomal aberrations generate 3′ truncations of the NFKB2/lyt-10 gene in lymphoid malignancies. Blood. 1994;84:3850–3860. [PubMed] [Google Scholar]
- 65.Neri A., Chang C.C., Lombardi L., Salina M., Corradini P., Maiolo A.T., Chaganti R.S., Dalla-Favera R. B cell lymphoma-associated chromosomal translocation involves candidate oncogene lyt-10, homologous to NF-kappa B p50. Cell. 1991;67:1075–1087. doi: 10.1016/0092-8674(91)90285-7. doi:10.1016/0092-8674(91)90285-7. [DOI] [PubMed] [Google Scholar]
- 66.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M., et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. doi:10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sutherland G.R., Ledbetter D.H. Report of the committee on cytogenetic markers. Cytogenet. Cell Genet. 1989;51:452–458. doi: 10.1159/000132804. doi:10.1159/000132804. [DOI] [PubMed] [Google Scholar]
- 68.Fugazzola L., Pilotti S., Pinchera A., Vorontsova T.V., Mondellini P., Bongarzone I., Greco A., Astakhova L., Butti M.G., Demidchik E.P., et al. Oncogenic rearrangements of the RET proto-oncogene in papillary thyroid carcinomas from children exposed to the Chernobyl nuclear accident. Cancer Res. 1995;55:5617–5620. [PubMed] [Google Scholar]
- 69.Klugbauer S., Lengfelder E., Demidchik E.P., Rabes H.M. High prevalence of RET rearrangement in thyroid tumors of children from Belarus after the Chernobyl reactor accident. Oncogene. 1995;11:2459–2467. [PubMed] [Google Scholar]
- 70.Nikiforov Y.E., Rowland J.M., Bove K.E., Monforte-Munoz H., Fagin J.A. Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 1997;57:1690–1694. [PubMed] [Google Scholar]
- 71.Rabes H.M., Demidchik E.P., Sidorow J.D., Lengfelder E., Beimfohr C., Hoelzel D., Klugbauer S. Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-chernobyl papillary thyroid carcinomas: biological, phenotypic, and clinical implications. Clin. Cancer Res. 2000;6:1093–1103. [PubMed] [Google Scholar]
- 72.Lukusa T., Fryns J.P. Human chromosome fragility. Biochim. Biophys. Acta. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. doi:10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 73.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. doi:10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mrasek K., Schoder C., Teichmann A.C., Behr K., Franze B., Wilhelm K., Blaurock N., Claussen U., Liehr T., Weise A. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int. J. Oncol. 2010;36:929–940. doi: 10.3892/ijo_00000572. [DOI] [PubMed] [Google Scholar]
- 75.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 76.Tsantoulis P.K., Kotsinas A., Sfikakis P.P., Evangelou K., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. doi:10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 77.Letessier A., Millot G.A., Koundrioukoff S., Lachages A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. doi:10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 78.Helmrich A., Ballarino M., Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. doi:10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 79.Grabczyk E., Mancuso M., Sammarco M.C. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. doi:10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. doi:10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reddy K., Tam M., Bowater R.P., Barber M., Tomlinson M., Nichol Edamura K., Wang Y.H., Pearson C.E. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. doi:10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin Y., Wilson J.H. Transcription-induced DNA toxicity at trinucleotide repeats: double bubble is trouble. Cell Cycle. 2011;10:611–618. doi: 10.4161/cc.10.4.14729. doi:10.4161/cc.10.4.14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.