SUMMARY
Tropomyosins (Tm) constitute a family of ubiquitous and highly conserved actin-binding proteins, playing essential roles in a variety of biological processes. Tm isoforms produced by multiple Tm encoding genes and alternatively expressed exons along with post-translational modifications (PTMs) regulate Tm function. Therefore, to gain a better understanding of the functional role of Tm, it is essential to fully characterize Tm isoforms. Herein, we developed a top-down high-resolution mass spectrometry (MS) based targeted proteomics method for comprehensive characterization of Tm isoforms. α–Tm was identified to be the predominant isoform in swine cardiac muscle. We further characterized its sequence and localized the PTMs such as acetylation and phosphorylation as well as amino acid polymorphisms. Interestingly, we discovered a “novel” Tm isoform that does not match with any of the currently available swine Tm sequences. A deep sequencing of this isoform by top-down MS revealed an exact match with mouse β–Tm sequence, suggesting that this “novel” isoform is swine β–Tm which is 100% conserved between swine and mouse. Taken together, we demonstrated that top-down targeted proteomics provides a powerful tool for deep sequencing of Tm isoforms from genetic variations together with complete mapping of the PTM sites.
Keywords: Tropomyosin, Muscle Contraction, Actin Filament, Mass Spectrometry, Electron Capture Dissociation
Introduction
Tropomyosins (Tm) are a family of ubiquitous and highly conserved proteins found in eukaryotic cells, playing an essential role in a variety of biological processes that include but not limited to muscle contraction, cell motility, cell transformation and cytokinesis.1–4 Tm are α–helical coiled coil dimers that form a head to tail polymer along the major groove of actin filaments in all muscle fibers and many nonmuscle cells. The most well-known biological function of Tm is its role in striated muscle. Tm was originally discovered as a structural component of the skeletal muscle contractile apparatus and was believed to be a precursor of myosin, hence the name “tropomyosin”.5 Muscle contraction depends on the interaction of actin thin filaments and myosin thick filaments of the sarcomere.6–8 As one of the primary components of the actin filament, Tm interacts with actin and, together with the troponin complex, regulates muscle contraction and relaxation in a calcium-dependent manner.3, 9
Recently, Tm is becoming increasingly recognized for its function in the cytoskeleton upon the discovery of “nonmuscle contractile proteins” expressed in most tissues and cell types, regulating cell morphology, intracellular motility, cell transformation and migration among others.1, 3–4 It becomes clear that actin cytoskeleton is centrally involved in a diversity of functions in all eukaryotic cells. A plethora of Tm isoforms have been identified which significantly contribute to the diversity of actin filament function.1–4, 9–10
Tm expressed in eukaryotic cells show an increased complexity in terms of protein isoforms from yeast to human.411 The isoform diversity results from multiple Tm encoding genes and alternatively expressed exons, along with post-translational modifications (PTMs).12–14 Tm are highly conserved proteins encoded by four different genes: TPM1, TPM2, TPM3 and TPM4, while each gene utilizes alternative splicing, alternative promoters, and differential processing to encode multiple striated muscle, smooth muscle and cytoskeletal transcripts.3, 13–16 TPM1 is the most versatile and complex gene that comprises 15 exons to encode a variety of tissue specific isoforms. There are two major striated and smooth muscle Tm isoforms, α–Tm and β–Tm encoded by TPM1, TPM2 genes, respectively, whereas γ–Tm encoded by TPM3 gene is present only in slow-twitch skeletal muscle.3, 17 All four Tm genes are expressed in non-muscle cells.3–4 The production of Tm isoforms has been demonstrated to be developmentally regulated and these isoforms exhibit distinct physiological properties which are characteristic of cell type and species.1, 10, 18–19
The involvement of Tm in a variety of human diseases clearly underlines the high clinical importance of Tm.1, 3 Mutation of Tm genes are known to be associated with a number of myopathies, most notably dilated cardiomyopathy (DCM) and familial hypertrophic cardiomyopathy (FHC).3–4, 9, 20 Recently, a novel striated muscle specific isoform encoded by TPM1 gene, TPM1κ protein, was found to be significantly up-regulated in DCM and heart failure.17 Moreover, non-muscle Tm is shown to be differentially expressed in a variety of human tumors, suggesting its functional role in oncogenic signaling.4 Recent studies have demonstrated that specific isoforms of Tm may function as tumor suppressor with the ability to suppress the malignant growth of tumor cells.21–27 Hence, to better understand the functional role of Tm in both muscle and non-muscle cells, it is essential to fully characterize the sequences of Tm isoforms, which is still lacking.
Top-down mass spectrometry (MS) is emerging as a powerful tool to comprehensively characterize the protein isoforms resulting from alternative splicing and coding polymorphisms simultaneously with PTMs.28–31 Unlike the traditional bottom-up MS that requires proteolytic digestion prior to MS analysis, top-down MS measures the intact proteins without digestion, thus providing a “bird’s eye” view of all types of protein modifications including isoforms, mutations, PTMs of targeted proteins simultaneously in one MS spectrum without a priori knowledge,28–30, 32–41. Individual protein isoforms can be isolated42 and then fragmented by tandem MS techniques such as collisionally activated dissociation (CAD)43 and electron capture dissociation (ECD),44 allowing deep sequencing to detect amino acid variations and reliably map the modification sites with full sequence coverage.
Herein, we have developed a top-down mass spectrometry (MS)-based targeted proteomics approach to purify Tm from tissues and comprehensively characterize its isoforms. We have rapidly and effectively purified Tm from swine cardiac tissues without the use of antibodies and performed a deep sequencing of all swine Tm isoforms using top-down MS/MS with ECD and CAD. Moreover, PTMs including acetylation and phosphorylation, as well as amino acid polymorphisms have been characterized for the Tm isoforms. To the best of our knowledge, this is the first comprehensive characterization of Tm isoforms directly purified from tissues with full sequence coverage and PTM mapping.
EXPERIMENTAL SECTION
Chemicals and reagents
All reagents were purchased from Sigma Chemical Co. (St. Louis, MO) unless noted otherwise. Complete protease inhibitor cocktail tablets were purchased from Roche Diagnostics Corp. (Indianapolis, IN). All solutions were prepared in Milli-Q water (Millipore, Corp., Billerica, MA). Bicinchoninic acid (BCA) protein assay reagents were from Thermo Scientific (Rockford, IL).
Purification of swine Tm
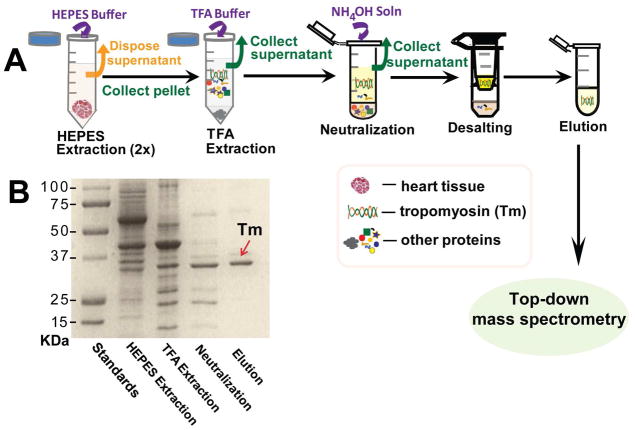
Swine cardiac tissue samples were excised from healthy juvenile Yorkshire domestic pigs, snap frozen in liquid N2 and stored in −80 °C freezer. Hearts were harvested immediately after the animals were sacrificed. All procedures were approved by UW-Madison Animal Care and Use Committee. A rapid purification method with high reproducibility was established to purify Tm from tissues (Figure 1A). Briefly, ~0.1–0.4 g of heart tissue was homogenized with HEPES extraction buffer45 (HEPES, 25 mM, pH 7.5, NaF, 50 mM, Na3VO4, 0.25 mM, PMSF (in isopropanol), 0.25 mM, EDTA, 2.5 mM, and half tablet of commercial protease inhibitor cocktail) twice using a Polytron electric homogenizer (Model PRO200, PRO Scientific Inc., Oxford, CT, USA) for 15 s. The homogenate was centrifuged at 16,100 g for 15 min at 4 °C (Centrifuge 5415R, Eppendorf, Hamburg, Germany) and supernatant was removed and saved as “HEPES extraction”. The pellet was then homogenized with TFA extraction buffer (TFA, 1%, TCEP, 1 mM) using the Polytron electric homogenizer. The homogenate was centrifuged (16,100 g, 4 °C, 15 min) to collect the supernatant. 8 VL of 10% NH4OH solution was added to the 50 VL of collected supernatant drop-by-drop to neutralize the supernatant to a pH of 7.0. As the solution was neutralized, precipitation occurred. The mixture was then centrifuged at 16,100 g for 2 min at 4 °C to remove the precipitate. Finally the supernatant was desalted using the Amicon 100 K molecular weight cutoff (MWCO) filter with 0.1 % formic acid solution as the exchange buffer. Tm was retained on top of the 100 kDa MWCO filter likely due to its elongated α–helical coiled coil polymeric structure. Fractions were analyzed for protein content by SDS-PAGE on 12.5% polyacrylamide gels stained with Coomassie Blue (Figure 1B). BCA protein assay was employed to determine the total protein concentration with a Synergy™ 2 microplate reader (Bio-Tek Instruments, Winooski, VT). A total of 200 μg Tm could be purified from 400 mg heart tissue.
Figure 1. Flow diagram (A) and SDS-PAGE (B) analysis of swine Tm purification and characterization procedures.
Tm was purified from swine heart tissue. Each lane of the SDS-gel represents different stages of the purification procedure.
Top-down MS
Approximately 2 μg of purified swine Tm was analyzed each time using a 7 T linear ion trap/Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (LTQ/FT Ultra, Thermo Scientific Inc., Bremen, Germany) equipped with an automated chip-based nano ESI source (Triversa NanoMate; Advion Biosciences, Ithaca, NY, USA) as described previously (28, 32–36). The sample was introduced into the mass spectrometer with a spray voltage of 1.2–1.6 kV versus the inlet of the mass spectrometer, resulting in a flow of 50–200 nL/min. Ion transmission into the linear ion trap and subsequently into the FT-ICR cell was automatically optimized for a maximal ion signal. The accumulated charge numbers for the full scan linear ion trap, FT-ICR cell (FT), MSn FT-ICR cell and ECD were 3×104, 8×106, 8×106, and 8×106, respectively. The resolving power of the FT-ICR was typically set at 200,000, obtaining an acquisition rate of 1 scan/s. For CAD and ECD, the protein molecular ions of the individual charge states were first isolated and then fragmented by 12–25% normalized collision energy for CAD or 2.2–3% electron energy (corresponding to 1.0–1.8 eV) for ECD with a 70 ms duration and with no additional delay, respectively. Typically, 1,000–5,000 transients were averaged to ensure high-quality CAD and ECD spectra.
All MS and MS/MS data were processed by in-house developed MASH Suite software using THRASH 46 algorithm (version 1.0) with S/N threshold of 3 and a fit factor of 60% and manually validated. The fragments in MS/MS spectra were assigned based on the protein sequence of swine Tm obtained from Swiss-Prot protein knowledgebase (Unit-ProtKB/Swiss-Prot). Allowance was made for possible PTMs such as acetylation of the N-terminus, phosphorylation and two amino acid polymorphisms using 1 Da tolerance for the precursor and fragment ions, respectively. The assigned ions were manually validated to ensure the accuracy of the assignments. For fragment ions containing possible phosphorylation site, the masses of fragment ions were manually validated for ~80 Da mass shifts to confirm or exclude the existence of phosphorylation. All the reported molecular weights (MWs) are the most abundant MWs.
Quantitative analysis of swine isoforms was performed as described previously.33, 38 All the spectra were acquired in full profile mode. Briefly, the integrated peak heights of the top five isotopomers were used to calculate the relative abundance of each detected protein form. The percentages of un- and mono-phosphorylated α–Tm, un- and mono-phosphorylated β–Tm were defined as the summed abundances of α–Tm, pα–Tm, β–Tm and pβ–Tm over the summed abundances of the entire Tm populations, respectively. Non-covalent phosphate adducts (+H3PO4) were also taken into account in the quantitation. For example, the abundance of Tm + H3PO4 peak is combined with that of the unphosphorylated Tm, whereas the abundance of pTm + H3PO4 is integrated with mono-phosphorylated Tm (pTm). Three swine hearts (three biological replicates) were investigated and each biological sample was run in triplicate (three technical replicates) to ensure the technical consistency. Data are represented as means ± S. D.
RESULTS
SDS-PAGE and High-resolution MS of Swine Tm
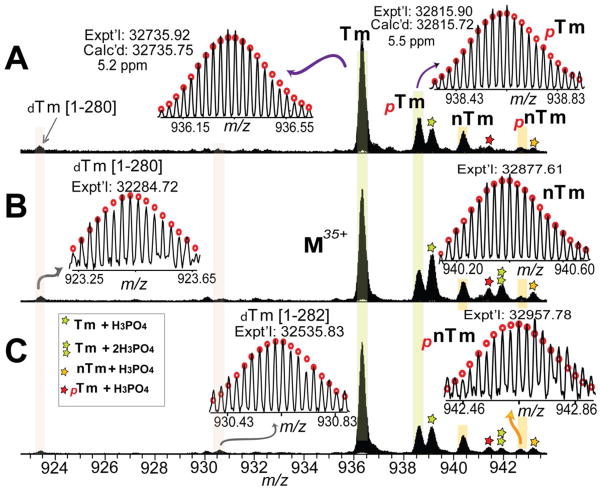
The Tm purification process is comprised of four steps (Figure 1A): 1) HEPES extraction at neutral pH (~7.5) in the presence of a variety of protease and phosphatase inhibitors to preserve the endogenous modification stage.45 This extraction contains mainly soluble cytoplasmic proteins. 2) TFA extraction of the insoluble portion from the HEPES extraction at acidic pH (~2.2) to solubilize myofilament proteins. 3) Neutralization of the TFA extraction to precipitate the majority of the myofilament proteins which are only soluble at very acidic pH. This step significantly enriched the Tm that remains soluble at the neutral pH (7.0). 4) Desalting with a membrane filter that effectively eliminates low MW species and purifies Tm that are then collected as “elution” for further MS analysis. The entire protein purification process takes less than 3 hours and was analyzed by SDS-PAGE (Figure 1B). The major band in the elution is a 32 kDa protein as shown in Figure 1B, indicating that Tm has been purified from swine heart tissue (Figure 1B). FT-ICR MS analysis with high-resolution and high mass accuracy provides an overview of all detectable protein forms present in swine Tm using the method employed here (Figure 2). The predominant peak with MW of 32735.92 likely corresponded to unphosphorylated swine Tm. The peak next to the predominant peak showed a MW of 32815.90, with a mass increment of 79.98 Da, indicating it is a mono-phosphorylated form of the predominant swine Tm peak. High-resolution MS spectra of purified Tm from three different swine hearts exhibited highly comparable profiles (Figure 2, Supplemental Figure 1) and the predominant peak in these spectra demonstrated the same experimental MW value of 32735.92 (tentatively labeled as “Tm” in Figure 2).
Figure 2. High-resolution MS analysis of intact Tm purified from three different swine hearts (A), (B) and (C).
Tm: swine α–Tm. nTm: a “novel” isoform of swine Tm. dTm[1-282] and dTm[1-280], C-terminal degradation products of swine α–Tm. pTm and pnTm represent mono-phosphorylated Tm and nTm. The peaks marked with stars represented non-covalent phosphoric acid adducts. Red circles: the theoretical isotopic abundance distribution of the isotopomer peaks corresponding to the assigned mass. Calc’d, calculated most abundant molecular weight; Expt’l, experimental most abundant molecular weight. Molecular weights were calculated based on DNA-prdicted sequence of Sus scrofa Tm (UnitprotKB/Swiss-Prot P42639, TPM1_PIG) with acetylation of the first amino acid at the N-terminus (+42.01) and two amino acid polymorphisms, P38→Q (−28.04) and P64→L (16.03).
Meanwhile, an interesting peak with MW of 32877.61 was observed with a mass increment of 141.69 compared to the predominant swine Tm peak of MW 32735.92. This mass increment indicates a possible isoform of swine Tm as it is rare to observe a PTM of 141 Da. Surprisingly, a thorough sequence homology analysis demonstrated that this isoform does not match any of the swine Tm sequences available in the Swiss-Prot and NCBI database (Supplemental Figure 2) including commonly occurring modifications such as N-terminal Met removal, acetylation and phosphorylation. Hence we tentatively labeled it as a “novel” isoform of Tm (nTm).
Minor peaks with MW of 32284.75 and 32535.83 were also detected in the spectra of swine Tm (Figure 2), likely representing trace degradation products. Other low abundance peaks with higher MW (+98 Da) than the major peak were also observed, which correspond to non-covalent phosphate adducts of swine Tm (Figure 2).
Deep sequencing of the major swine Tm isoform
A thorough sequence homology analysis among all the available swine Tm sequences in Swiss-Prot and NCBI databases found the experimental MW of the predominant swine Tm form (32735.92) matches closest with the calculated MW (32705.75) of Sus scrofa α-Tm (UnitprotKB/Swiss-Prot P42639, TPM1_PIG) encoded by TPM1 gene with a mass discrepancy of 30.17 Da (Supplemental Figure 2). Meanwhile the sequence of TPM1_PIG matches best with the MS/MS data among all available Tm sequences thus it was selected as a starting isoform for further comprehensive sequence mapping. Based on the mass discrepancy of 30.17 Da, it is reasonable to hypothesize that some modifications such as PTMs and/or amino acid polymorphisms have occurred in this predominant swine Tm isoform (defined as swine α–Tm hereafter). To confirm the possible modifications in the α–Tm chain, a single charge state of α–Tm precursor ions were isolated individually and fragmented by both ECD and CAD (Supplemental Figures 3, 4). Supplemental Figure 5 illustrated the effectiveness of a gas-phase isolation of a single charge state (M38+) of un-phosphorylated Tm, mono-phosphorylated Tm, or a mixture of both.
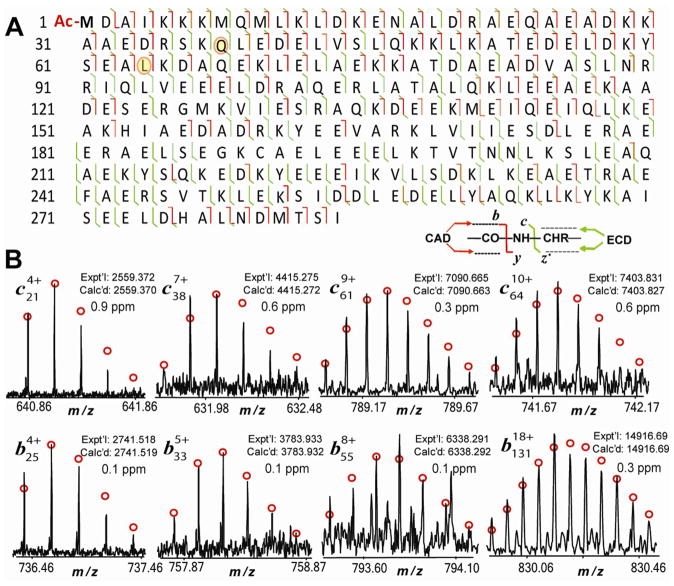
In the representative ECD experiment of swine α–Tm, only 48 z• ions matched the C–terminal ions predicted from un-modified swine α–Tm sequence in the database (UnitprotKB/Swiss-Prot-P42639, TPM1_PIG) (Supplemental Figure 3A). The fact that no single ion matched N-terminal sequence indicates possible modifications at the N-terminus of the swine α–Tm. Given that the majority of eukaryotic proteins carry N-terminal acetylation, it is highly likely that this modification occurs to the swine α–Tm. As shown in Supplemental Figure 3B, after considering acetylation of the N-terminus of swine α–Tm, 30 c ions from the N-terminus were detected, which matched the predicted value of the fragment ions of the sequence before Arg38. The acetylation of swine α–Tm would give a mass increase of 42.01 Da. Meanwhile another 19 c ions after Arg38 were detected, with a mass decrease around 28.04 Da compared with the predicted values, which suggested that a potential amino acid polymorphism occurred at Arg38. The mass decrease of 28.04 Da possibly results from the amino acid polymorphism of Arg38→Gln38 (R→Q, Δm = −28.04, Supplemental Figure 3C). Another 31 c ions after Pro64 show a mass increase of 16.03 Da compared with the predicted values, indicating that another amino acid polymorphism occurred at Pro64 (Supplemental Figure 3D). The 16.03 Da mass increase could be potentially generated from amino acid polymorphism from either Pro64→Leu64 (P/L) or Pro64→Ile64 (P/I) (P→I/L, Δm = 16.03). Indeed a homology sequence alignment indicates the presence of Gln38 and Leu64 in human, mouse, rabbit, rat, and bovine α–Tm sequences (Supplemental Figure 6). Moreover, comprehensive sequencing by ECD and CAD unambiguously identified these amino acid polymorphisms, Arg38→Gln38 and Pro64→Leu64 in swine Tm (Figure 3A, Supplemental Figure 3D).
Figure 3. MS/MS product ion map (A) and representative product ions (B) from both ECD and CAD spectra of unphosphorylated swine α–Tm.
Fragment assignments were made to the DNA-predicted sequence of Sus scrofa Tm (UnitprotKB/Swiss-Prot P42639, TPM1_PIG) with acetylation of the first amino acid at the N-terminus(42.01) and two amino acid polymorphisms, P38→Q (−28.04) and P64→L (16.03). A total of 228 out of 283 bond cleavages from five CAD and four ECD spectra are shown. Two amino acid polymorphisms were highlighted in orange circles. Circles: the theoretical isotopic distribution of isotopomer peaks. Calc’d, calculated most abundant molecular weight (MW). Expt’l, experimental most abundant MW.
One ECD experiment generated a total of 80 c ions and 48 z• ions according to the modified α–Tm sequence (Supplemental Figure 3D, Supplemental Table 1). After considering acetylation (+42.01 Da) and two amino acid polymorphisms (−28.04 Da, +16.03 Da, respectively), the calculated MW of swine α–Tm is 32735.75 (=32705.75+42.01−28.04+16.03), which matches well with the experimental MW of 32735.92 (5.5 ppm) (Figure 2). Figure 3 illustrated the representative product ions around Gln38 and Leu64 from the ECD and CAD spectra and resulting MS/MS ion map of swine α–Tm. 107 b ions and 15 y ions (118 cleavages) were generated in five CAD spectra, and 119 c ions and 78 z• ions (187 cleavages) were generated in four ECD spectra. A total of 228 out of 283 bond cleavages were obtained from the MS/MS data of swine α–Tm (Figure 3A). The top-down MS proteomics provided both the highly accurate MW measurement and effective sequencing, which unequivocally confirmed the modified sequence of swine α–Tm.
Mapping phosphorylation site in swine α–Tm
Swine α–Tm is present as both un- and mono-phosphorylated forms, at MWs of 32735.92 and 32815.90, respectively (Figure 2). The intensity of un-phosphorylated swine α– Tm is much higher than that of mono-phosphorylated α–Tm, indicating that swine α–Tm from healthy swine hearts is predominantly un-phosphorylated. No bis- or tris-phosphorylated swine α-Tm was detected, implying that higher-order phosphorylation was not present in the swine Tm samples or below the detection limit of the FTMS method applied in this study.
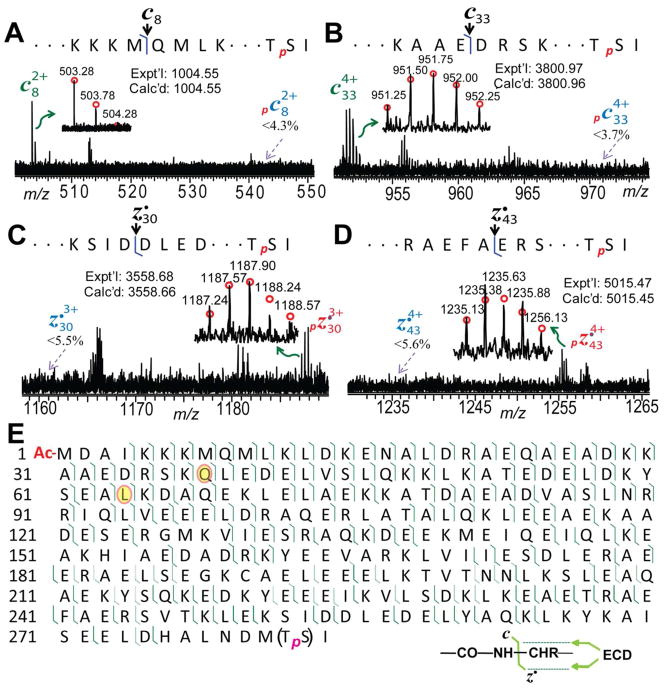
To localize the phosphorylation site of swine α–Tm, the precursor ion of monophosphorylated swine α–Tm was isolated and fragmented by ECD (Supplemental Figure 5B). 112 c ions and 79 z• ions were detected in combined four ECD spectra of mono-phosphorylated α–Tm; the representative fragment ions and the complete fragmentation map were demonstrated in Figure 4. As shown in the ECD spectra of isolated mono-phosphorylated swine α–Tm (pα–Tm), no phosphorylated product ions were detected for c8 and c33, nor other c ions detected in the ECD spectra (Supplemental Figure 7), which provided unambiguous evidence that the amino acid residues from the N-terminus of α–Tm do not contain a phosphorylation site (Figure 4A and 4B). In contrast, all 79 z• ions observed in these ECD spectra of mono-phosphorylated swine α–Tm were phosphorylated, suggesting the phosphorylation site is located near the C-terminus (Figure 4C and 4D). The smallest phosphorylated fragment ions detected was z•9 in the ECD, which suggested the phosphorylation site was localized at one of the first 9 amino acids at the C-terminus (HALNDMTSI) (Figure 4E, Supplemental Table 2). Phosphorylation occurs most commonly on serine (~90%), then on threonine (~10%) and on tyrosine (~<0.05%).47 Since there are only one serine (Ser283) and one threonine (Thr282) at this region, it is likely that Ser283 was phosphorylated as reported in the previous studies of Tm.48–49
Figure 4. ECD for mapping the phosphorylation site of swine α-Tm.
(A)–(D), Representative MS/MS spectra of c ions and z• ions from ECD spectra of mono-phosphorylated swine α–Tm. (E) MS/MS ion map from ECD spectra of mono-phosphorylated swine α–Tm. The two amino acid polymorphisms were highlighted in orange circles and p presents the mono-phosphorylation site at Ser283. A total of 173 out of 283 bond cleavages from four ECD spectra are shown.
Moreover, the representative ions from an individual ECD spectrum of un-, mono-phosphorylated swine α–Tm and their mixture were shown and compared (Supplemental Figure 7). Consistently, all the c ions in the spectra of un-, mono-phosphorylated α–Tm and their mixture were presented as un-phosphorylated form, indicating no phosphorylation site at the N-terminus. The z• ions observed were presented as un-phosphorylated, mono-phosphorylated, and a mixture of both un- and mono-phosphorylated forms in the ECD spectrum of un-, mono-, and the mixture of un- and mono-phosphorylated swine α–Tm, respectively, suggesting the phosphorylation site at the C-terminus and presumably localized at Ser283.
Sequence determination of the “novel” swine isoform
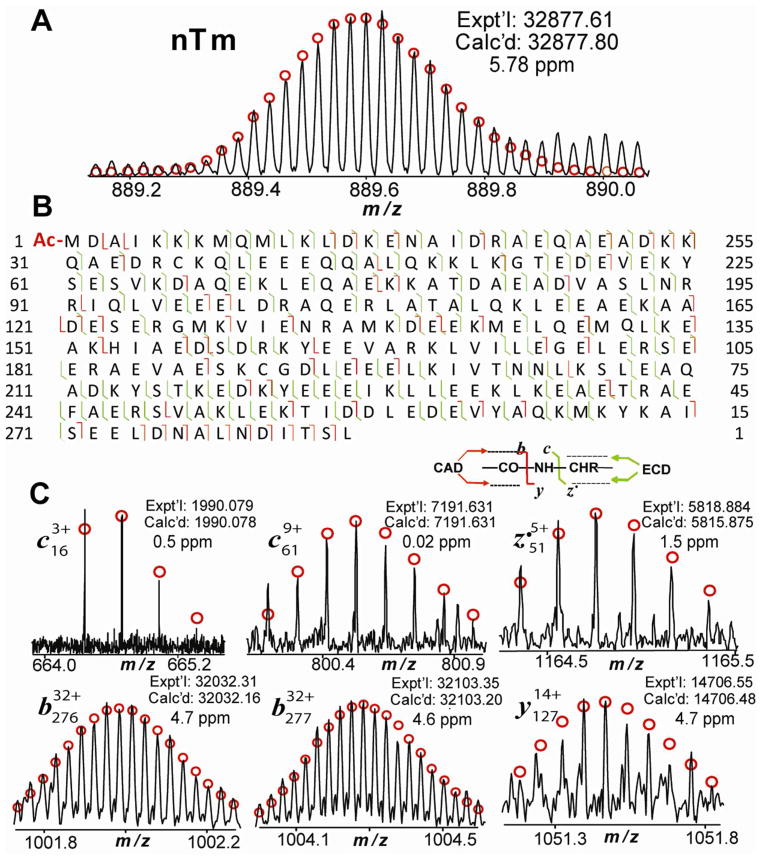
To comprehensively characterize the “novel” isoform of swine Tm (nTm in Figure 2), we isolated this swine Tm isoform and then fragmented it by ECD and CAD. A single charge state of the swine Tm isoform was isolated and demonstrated a MW of 32877.61 with highly accurate mass measurement (Figure 5A). Both ECD and CAD spectra of the nTm generated sequence tags that match part of the N-terminal ions predicted from swine α–Tm with acetylation at N-terminus. The largest ions obtained from ECD and CAD spectra that match the sequence were c29 and b29, respectively, suggesting a possible sequence difference after c29/b29. A thorough sequence alignment of both α– and β–Tm sequences among various species revealed that β–Tm among most mammalian species have one amino acid different at position 30 from N-terminus (Gln, Q) compared with swine α–Tm (Ala, A) (Figure 6, Supplemental Figures 6 and 8). Interestingly, analysis of MS/MS experiments of the swine Tm isoform discovered that this isoform matches exactly with the mouse β–Tm sequence (UnitprotKB/Swiss-Prot P58774, TPM2_MOUSE) encoded by TPM2 gene (Figure 5B). 63 b ions and 15 y ions were detected from four CAD spectra, and 79 c ions and 69 z• ions were detected from three ECD spectra, with the representative c, z•, b, y ions shown in Figure 5C and the peak list of one ECD spectrum shown in Supplemental Table 3. Moreover, the calculated MW of the modified mouse β–Tm chain (acetylation at the N-terminus: 42.01) is 32877.90 (32835.79 + 42.01), which matches exactly with the experimental MW of this swine Tm isoform 32877.61 (5.8 ppm). As we have confidently excluded the possibility of potential contamination from mouse tissue samples, the only other explanation is that this “novel” swine Tm is swine β–Tm isoform that is 100% conserved with that of mouse β–Tm.
Figure 5. MS/MS characterization of a “novel” isoform of swine Tm (nTm).
(A) High-accuracy mass measurement of nTm (M37+). (B) Fragmentation ion map of nTm from both ECD and CAD spectra. Fragment assignments were made to the DNA-predicted sequence of mouse β –Tm (UnitprotKB/Swiss-Prot P58774, TPM2_MOUSE) with acetylation of the first amino acid at N-terminus. A total of 137 out of 283 bond cleavages from 6 CAD and 2 ECD spectra are shown. (C) Representatvie ECD and CAD fragment ions from nTm.
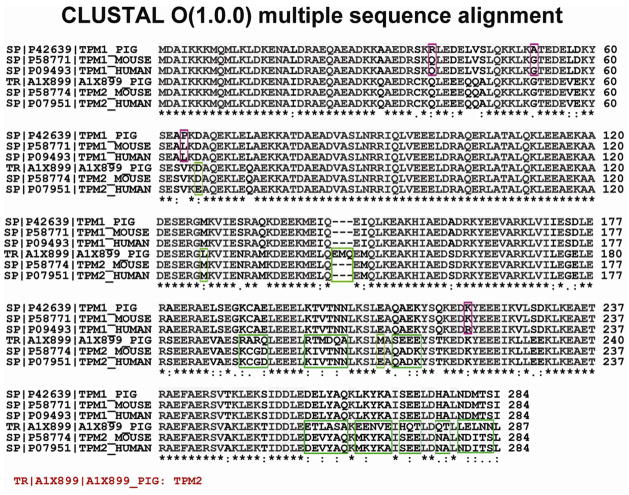
Figure 6. Protein sequence homology alignment of Tm isoforms among various species.
The α– and β–Tm isoforms presented here are detected in pig, mouse and human, encoded by TPM1 and TPM2 genes. Highlighted areas marked in purple and green indicate amino acid substitutions between α–Tm and β–Tm across differen species, respectively. Asterisks indicate the conserved sequence regions. A1X899_PIG is encoded by TPM2 gene but the sequence is marked as “unreviewed” in the Swiss-Prot database.
The sequence of swine β–Tm isoform identified here is significantly different from that of swine β–Tm chain reported in Swiss-Prot and NCBI database. The swine β–Tm sequence identified in our study has a total of 284 amino acids (Figure 3), in contrast to 287 amino acids in the hypothetical swine β–Tm sequence as reported in the database (Figure 6). Significantly, there are 39 amino acid differences between the observed (in this study) and the hypothetical swine β–Tm. Note that the hypothetical sequence for swine β–Tm encoded by TPM2 gene (A1X899 available at Swiss-Prot databases) is marked as “un-reviewed”. The sequence alignment analysis indicates that a significant sequence difference exists between un-reviewed swine β–Tm (A1X899_PIG) and reviewed mouse and human β–Tm (P58774-TPM2_MOUSE, P07951-TPM2_Human) in the Swiss-Prot database (Supplemental Figure 8a), which challenges the un-reviewed swine β–Tm sequence and supports that the sequence characterized in this study is the correct swine β–Tm isoform. More interestingly, a further investigation of β–Tm across different species found all the reviewed β–Tm sequences are highly conserved among mammals (Supplemental Figure 8b). In fact, β–Tm chains are 100% identical among mouse, rat, rabbit, pig and bovine, whereas there is only one amino acid difference in β–Tm between human and other mammals at amino acid position 66. This further supported our hypothesis that swine β–Tm sequence is 100% conserved with mouse β–Tm.
In summary, both the MS/MS data and highly accurate MW measurement unambiguously define the correct sequence for swine β–Tm isoform. The three major peaks in the spectra with MW of 32735.92, 32815.90 and 32877.61 have been defined as un-phosphorylated swine α–Tm, phosphorylated α–Tm and un-phosphorylated β–Tm, respectively. Meanwhile, low MW peaks at 32284.75 and 32535.83 were assigned as a C-terminal degradation product Tm [1-280], Tm [1-282] and their isoforms dTm [1-280], dTm [1-282], respectively (Figure 2). Due to the low intensity of these peaks, these degradation product isoforms have not been investigated further in this study.
Relative quantification of Tm isoforms and their phosphorylated form
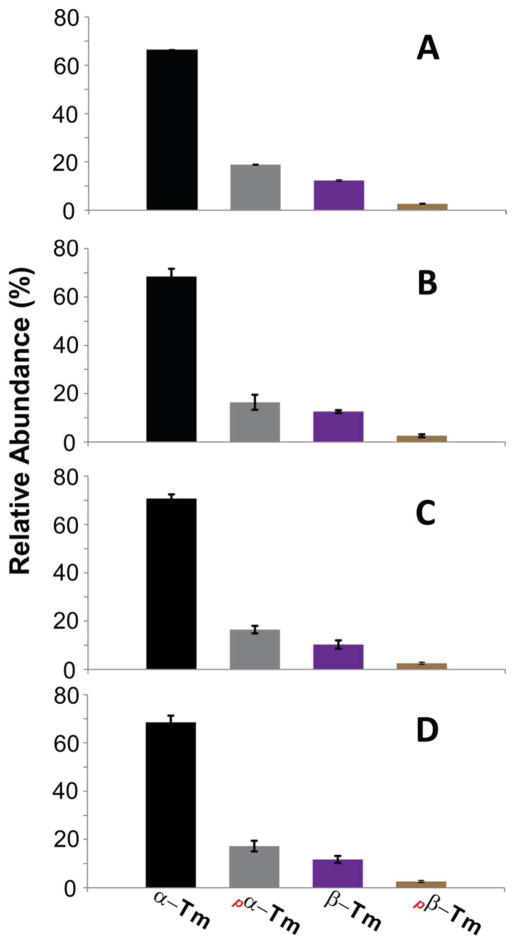
To investigate the composition of Tm isoforms in swine hearts, the percentages of swine α–Tm, β–Tm and their corresponding mono-phosphorylated forms were calculated based on the molecular ion abundances (M35+ and M36+) of Tm purified from three swine hearts. The reported results are from the three swine hearts, each with three technical replicates. Percentages of un-and mono-phosphorylated swine α–Tm and un- and mono-phosphorylated β–Tm were shown in Figure 7. The percentages of swine α-Tm are comparable among three distinct swine hearts, 66.2 ± 0.3%, 68.4 ± 3.2%, 70.7 ± 1.7% and the percentages of the mono- phosphorylated form (pα–Tm) are 18.8 ± 0.2%, 16.4 ± 3.1%, and 16.5 ± 1.5%, respectively (Figure 7A-C). The percentages of the swine β–Tm and its mono-phosphorylated form (pβ–Tm) exhibited in the three different swine hearts are 12.3 ± 0.3%, 12.6 ± 0.6%, 10.3 ± 1.7% and 2.7 ± 0.2%, 2.6 ± 0.6%, 2.6 ± 0.4 %, respectively (Figure 7A-C). The summed percentages of the Tm isoforms and their corresponding mono-phosphorylation forms over three biological replicates with triplicate measurements are 68.5 ± 2.8%, 17.2 ± 2.2%, 11.7 ± 1.4%, and 2.6 ± 0.4% for un- and mono-phosphorylated swine α–Tm and un- and mono-phosphorylated β–Tm, respectively (Figure 7D). The ratio of swine β–Tm of the total Tm population (including all Tm species regardless of isoforms and PTMs) is 14.3 ± 1.2%. The phosphorylation occupancy of α–Tm and β–Tm is 20.1 ± 1.8% and 18.3 ± 1.5%, respectively.
Figure 7. Relative quantification of two swine Tm isoforms (α–Tm and β–Tm) and their phosphorylation forms (pα–Tm and pβ–Tm) from three different swine hearts (A), (B) and (C), respectively. (D) Summary plot of Tm isoforms and their phosphorylated forms from a total of three biological replicates.
The percentage of α–Tm, pα–Tm, β–Tm and pβ–Tm was defined as summed abundances of α–Tm, pα–Tm, β–Tm and pβ–Tm over the summed abundances of the entire Tm populations, respectively. The bar graphic data were expressed with means ± S.D. Samples are run in triplicate.
DISCUSSION
The highly conserved Tm sequences
Tm are highly conserved proteins found in eukaryotic cells and encoded by a family of four alternative spliced genes: TPM1, TPM2, TPM3 and TPM4. Sequence homology alignment of α–Tm encoded by TPM1 among various mammalian species demonstrated that α–Tm is highly conserved and there are only four amino acid substitutions at positions 38, 52, 64 and 220 from the N-terminus (Supplemental Figure 6). Moreover, after considering two amino acid polymorphisms of swine α–Tm at R38→Q and P64→L, there are only two amino acid differences (positions 52 and 220) across these species. Not only α–Tm encoded by TPM1 but β-Tm chains encoded by TPM2 exhibit highly conserved sequences among different species (Supplemental Figure 8). β–Tm chains of mouse, rat, rabbit and bovine have exactly the same protein sequence and these β–Tm chains have only one amino acid discrepancy from human β–Tm at amino acid position 66 from the N-terminus (Supplemental Figure 8b). Interestingly, as discovered in this study, the corrected sequence of swine β–Tm is 100% conserved as that of mouse β–Tm, suggesting the high sequence similarity among mammalians β–Tm. Meanwhile, top-down targeted proteomics analysis of swine Tm isoforms characterized here revealed that 85.6% sequence similarity was exhibited between swine α–Tm and β–Tm from the swine heart tissue with 41 amino acid differences distributed throughout the entire sequence of 284 amino acids.
The functional significance of Tm isoforms
Tm are known to have a large number of isoforms which play important roles in regulating physiological function in various developmental stages.3–4, 9 Isoform switching accompanies morphogenesis and alterations in Tm isoforms have been detected in cancer cells.4 In striated muscle, there are three primary Tm isoforms, α–Tm, β–Tm, and γ–Tm. α–Tm is present in various tissue types and is the predominant form expressed in cardiac tissue, which is consistent with our findings here. β–Tm is expressed in both cardiac and skeletal muscles whereas γ–Tm is primarily expressed in slow twitch skeletal muscle.3, 9 The ratio of α–Tm to β–Tm present in the muscle cells is characteristic of the muscle type and species and also dependent on the developmental stages.3 Although both α– and β–Tm isoforms are expressed in fetal heart tissue, α–Tm appears to be expressed nearly exclusively in the adult myocardium of small mammals.9 Indeed, Wieczorek and co-workers showed that the endogenous β–Tm protein expression is absent (<2%) in the healthy adult mouse heart.50 In contrast, β–Tm is expressed in large animal and human myocardium in a much higher ratio (approximately 1:5 of β–Tm:α–Tm), which agrees well with our current data. The difference of α– and β–Tm ratio in cardiac muscle may be dependent on the size and contraction speed of the heart,51 with the ratio of α–Tm growing with increasing muscle speed.3 In large and slowly beating hearts such as pig and human, the ratio of β–Tm to α–Tm is much higher than that of small and faster beating hearts such as in rabbit, guinea pig and rat, where the β–Tm component appear to be nearly absent.49
Meanwhile, compelling evidence shows that different Tm isoform expressions in muscles are functionally distinct.1–4, 9,10 The Wieczorek lab has utilized transgenic mouse models to define the functional differences among these isoforms in regulating cardiac physiology. 9, 52–53 Partial replacement of α–Tm with β–Tm in the adult mouse heart significantly increased Ca2+ sensitivity of the force and decreased the rates of contraction and relaxation.52–53 Increased expression level of β–Tm (>75%) leads to significantly altered diastolic function of the myocardium and these transgenic mice die between 10–14 days postnatally.52 In contrast, γ–Tm overexpression to ~50% of total Tm population leads to decreased myofilament Ca2+ sensitivity and increased rates of contraction and relaxation.9, 15 Thus, these studies suggest Tm isoforms are not functionally equivalent and alteration of the Tm composition may directly modulate the muscle physiology.
Regulation of Tm function by PTMs
Our top-down MS/MS data has located the site of N-terminal acetylation and suggests phosphorylation at Ser283. N-terminal acetylation is a common PTM for eukaryotic proteins. The functional role of N-terminal acetylation remains enigmatic although it is typically perceived to increase the stability and half-life of proteins in the cell.54 Interestingly, Tm is among one of the few proteins in which the N-terminal acetylation has been recognized to be functionally significant. It has been demonstrated that Tm require NatB-mediated N-terminal acetylation for proper function actin-tropomyosin interaction and actomyosin regulation.55 Since Tm are coiled coil dimers which form head-to-tail polymers along the major groves in the actin thin filament, it is reasonable to believe that the modifications on the N- and C-termini are especially important for Tm polymerization. Indeed, only N-terminally acetylated Tm has been shown to have normal actin affinity and polymerization ability, underscoring the importance of Tm N-terminal acetylation.54
Here our data also suggested a single phosphorylation site at Ser283 with partial phosphorylation occupancy. This is consistent with previous studies showing the presence of phosphorylated Tm in both skeletal and cardiac muscle from various species including frog, rabbit, rat, mouse, and chicken.3, 49 Moreover, with 32P labeling and amino acid analysis, Ser283 was identified as a single phosphorylation site in α–Tm, but not β–Tm, isolated from skeletal muscle of adult frog despite of the similarity of the C-terminal sequence between β-Tm and α-Tm.49 Nonetheless, the phosphorylated β–Tm has been detected in developing striated muscles of the rat and mouse although the percentage of phosphorylation is relatively lower in β-Tm than α-Tm,3 which is in agreement with our findings. The fact that a higher extent of phosphorylation was detected in both α–Tm and β–Tm indicates a more prominent role of Tm in early development.4 Importantly, phosphorylation of Tm has been shown to regulate the function of actin filament and remodeling of the actin skeleton.3, 56 Phosphorylation of Ser283 on each of Tm chains can form salt bridges with ε–NH2 groups of Lys6 and Lys12 respectively, which may enhance the stability of the head to tail interaction involved in Tm polymerization 3, 49. In addition, phosphorylation of α–Tm may also promote the actomyosin interaction and activate myosin Mg2+-ATPase.56 A recent report shows that dephosphorylation of α–Tm mediated by p38-MAPK in murine hearts leads to depressed contractility.57
Top-down targeted proteomics for deep sequencing of protein isoforms
Top-down MS is an attractive and unique method for analysis of protein isoforms because of its inherent sensitivity, resolution and speed, providing an overview of all possible isoforms as a result of multiple gene coding, alternative splicing and polymorphisms together with PTMs of the targeted protein in one mass spectrum.28–30, 39 Moreover, top-down MS with ECD and CAD is able to provide deep sequencing to define the specific sequence variation and PTM sites to a single amino acid. In this study, we have identified two swine Tm isoforms: swine α–Tm encoded by TPM1 gene and another “novel” isoform with a mass increase of 141.67 Da from swine α–Tm in the same spectra. Interestingly, this “novel” isoform does not match any of the swine Tm sequences at NCBI or Swiss-Prot database with possible modifications. Our MS/MS with ECD and CAD allowed a deep sequencing of this “novel” isoform which suggested it was swine β–Tm and verified that it matched exactly with the mouse β–Tm sequence encoded by TPM2 gene. Apparently, the original swine β–Tm sequences deposited in the NCBI or Swiss-Prot database are incorrect although we cannot exclude the possibility that it represents a new isoform that is yet to be confirmed. Furthermore, a thorough homology sequence alignment revealed that β–Tm is highly conserved among mammals and β–Tm chains are 100% conserved among pig, mouse, rat, rabbit and bovine (Supplemental Figure 8). Moreover, the top-down MS/MS deep sequencing also mapped two amino acid polymorphisms in the sequence of swine α–Tm and localized its mono-phosphorylation site. Admittedly, most of the top-down MS studies to date have focused on the analysis of protein PTMs.29, 32–41, 58–60 Despite fewer applications, top-down MS has shown to be equally powerful for sequencing protein isoforms resulting from alternative splicing and coding polymorphism.28, 30–31, 61–63 Although protein isoforms from genetic variation can also be identified by DNA sequencing, the unique advantages of top-down proteomics are the ultimate accuracy for sequencing protein isoforms purified directly from tissue and cells as well as the concurrent characterization of protein sequence variants (e.g. alternative splicing, polymorphisms, mutations) together with PTM mapping. With the further development of advanced instrumentation,64–65 we expect that the top-down MS-based targeted proteomics will be utilized routinely for sequencing protein isoforms with high throughput and high sensitivity.
CONCLUSIONS
In this study, we have developed a top-down targeted proteomics method which effectively and highly reproducibly purified Tm from muscle tissues without the use of antibodies and deeply characterized the sequence of all Tm isoforms as well as PTMs. We can define the sequence variants and PTM sites down to a single amino acid. The ubiquitous expression of Tm in eukaryotic cells from yeast to human underscores the high importance of Tm. Indeed, the role of Tm in the regulation of actin filament is increasingly appreciated and compelling evidence exists that Tm isoforms are functionally distinct in different cell types and species. The top-down targeted proteomics method we have established here provides comprehensive characterization of Tm isoforms from cardiac tissue, which can be applied to all striated muscle tissues and other tissue/cell types (with minor modifications) to facilitate a better understanding of the functional role of Tm isoforms in muscle and nonmuscle cells.
We would like to thank Chad Dooley and Matthew Lawrence for their contribution during the very early stage of the Tm purification method development, Yi-Chen (Ivy) Chen and Albert Chen for helpful discussion, and Wei Guo for critical reading of this manuscript. Financial support was kindly provided by NIH R01HL096971 (to YG). We also would like to thank the Wisconsin Partnership Program for the establishment of UW Human Proteomics Program Mass Spectrometry Facility.
Supplementary Material
Supplemental Table 1–3. Peak list and fragment assignment of one ECD spectra of un- and mono-phosphorylated α–Tm isoform, and un-phosphorylated β–Tm isoform, respectively.
Supplemental Figure 1. Broadband ESI/FTMS spectra of swine Tm purified from three different domestic swine hearts.
Supplemental Figure 2. Protein sequence homology alignment of multiple swine Tm isoforms.
Supplemental Figure 3. MS/MS product ion map from one ECD spectrum of predominant swine Tm for assignment to un-phosphorylated swine α–Tm.
Supplemental Figure 4. (A) Isolation of single charge state (M32+) of α–Tm precursor ions, (B) ECD and (C) CAD spectra of the isolated α–Tm ions.
Supplemental Figure 5. Gas-phase separation (purification) of swine Tm.
Supplementary Figure 6. Protein sequence homology alignment of α–Tm encoded by gene TPM1.
Supplemental Figure 7. Representative ECD fragment ions for mapping the mono-phosphorylation site of swine α–Tm.
Supplementary Figure 8. Protein sequence homology alignment of β–Tm encoded by gene TPM2.
ABBREVIATIONS
- Tm
Tropomyosin
- MS
Mass spectrometry
- DCM
Dilated cardiomyopathy
- FHC
Familial hypertrophic cardiomyopathy
- CAD
Collisionally activated dissociation
- ECD
Electron capture dissociation
- PTM
Post-translational modification
- MWCO
Molecular weight cutoff
- FT-ICR
Fourier transform ion cyclotron resonance
- SDS–PAGE
Sodium dodecyl sulfate – polyacrylamide gel electrophoresis
- ESI
Electrospray ionization
References
- 1.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15(6):333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Gunning P. Tropomyosin: introduction and historical perspective. In: Gunning P, editor. Tropomyosin. Springer; New York: 2008. pp. 1–4. [Google Scholar]
- 3.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22(1):5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 4.Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88(1):1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 5.Bailey K. Tropomyosin: a new asymmetric protein component of muscle. Nature. 1946;157:368–368. doi: 10.1038/157368b0. [DOI] [PubMed] [Google Scholar]
- 6.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Ann Rev Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 8.Moss RL, Razumova M, Fitzsimons DP. Myosin crossbridge activation of cardiac thin filaments - Implications for myocardial function in health and disease. Circ Res. 2004;94(10):1290–1300. doi: 10.1161/01.RES.0000127125.61647.4F. [DOI] [PubMed] [Google Scholar]
- 9.Jagatheesan G, Rajan S, Wieczorek DF. Investigations into tropomyosin function using mouse models. J Mol Cell Cardiol. 2010;48(5):893–898. doi: 10.1016/j.yjmcc.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagatheesan G, Rajan S, Ahmed RPH, Petrashevskaya N, Boivin G, Arteaga GM, Tae HJ, Liggett SB, Solaro RJ, Wieczorek DF. Striated muscle tropomyosin isoforms differentially regulate cardiac performance and myofilament calcium sensitivity. J Muscle Res Cell Motil. 2010;31(3):227–239. doi: 10.1007/s10974-010-9228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vrhovski BTN, Thiebaud P. In: Structure and evolution of tropomyosin in tropomyosin. Peter G, editor. Spring Science; New York: 2008. pp. 6–23. [DOI] [PubMed] [Google Scholar]
- 12.Denz CR, Narshi A, Zajdel RW, Dube DK. Expression of a novel cardiac-specific tropomyosin isoform in humans. Biochem Biophys Res Commun. 2004;320(4):1291–1297. doi: 10.1016/j.bbrc.2004.06.084. [DOI] [PubMed] [Google Scholar]
- 13.Wieczorek DF, Smith CWJ, Nadalginard B. The rat alpha-tropomyosin gene generates a minimum of 6 different messenger-RNAs coding for striated, smooth, and nonmuscle isoforms by alternative splicing. Mol Cell Biol. 1988;8(2):679–694. doi: 10.1128/mcb.8.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadal-Ginard B. Muscle cell differentiation and alternative splicing. Curr Opin Cell Biol. 1990;2(6):1058–64. doi: 10.1016/0955-0674(90)90156-9. [DOI] [PubMed] [Google Scholar]
- 15.Marston SB, Redwood CS. Modulation of thin filament activation by breakdown or isoform switching of thin filament proteins - Physiological and pathological implications. Circ Res. 2003;93(12):1170–1178. doi: 10.1161/01.RES.0000105088.06696.17. [DOI] [PubMed] [Google Scholar]
- 16.Leesmiller JP, Helfman DM. The molecular-basis for tropomyosin isoform diversity. Bioessays. 1991;13(9):429–437. doi: 10.1002/bies.950130902. [DOI] [PubMed] [Google Scholar]
- 17.Rajan S, Jagatheesan G, Karam CN, Alves ML, Bodi I, Schwartz A, Bulcao CF, D’Souza KM, Akhter SA, Boivin GP, Dube DK, Petrashevskaya N, Herr AB, Hullin R, Liggett SB, Wolska BM, Solaro RJ, Wieczorek DF. Molecular and functional characterization of a novel cardiac-specific human tropomyosin isoform. Circulation. 2010;121(3):410–U116. doi: 10.1161/CIRCULATIONAHA.109.889725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittenger MF, Kazzaz JA, Helfman DM. Functional-Properties of Nonmuscle Tropomyosin Isoforms. Curr Opin Cell Biol. 1994;6(1):96–104. doi: 10.1016/0955-0674(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 19.Vrhovski B, Schevzov G, Dingle S, Lessard JL, Gunning P, Weinberger RP. Tropomyosin isoforms from the gamma gene differing at the C-terminus are spatially and developmentally regulated in the brain. J Neurosci Res. 2003;72(3):373–383. doi: 10.1002/jnr.10586. [DOI] [PubMed] [Google Scholar]
- 20.Wieczorek DF, Jagatheesan G, Rajan S. The Role of Tropomyosin in Heart Disease. In: Peter G, editor. Tropomyosin. Spring Science; New York: 2008. pp. 132–139. [DOI] [PubMed] [Google Scholar]
- 21.Franzen B, Linder S, Uryu K, Alaiya AA, Hirano T, Kato H, Auer G. Expression of tropomyosin isoforms in benign and malignant human breast lesions. Brit J Cancer. 1996;73(7):909–913. doi: 10.1038/bjc.1996.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raval GN, Bharadwaj S, Levine EA, Willingham MC, Geary RL, Kute T, Prasad G. Loss of expression of tropomyosin-1, a novel class II tumor suppressor that induces anoikis, in primary breast tumors. Oncogene. 2003;22(40):6194–6203. doi: 10.1038/sj.onc.1206719. [DOI] [PubMed] [Google Scholar]
- 23.Varga AE, Stourman NV, Zheng Q, Safina AF, Quan L, Li XR, Sossey-Alaoui K, Bakin AV. Silencing of the Tropomyosin-1 gene by DNA methylation alters tumor suppressor function of TGF-beta. Oncogene. 2005;24(32):5043–5052. doi: 10.1038/sj.onc.1208688. [DOI] [PubMed] [Google Scholar]
- 24.Pawlak G, McGarvey TW, Nguyen TB, Tomaszewski JE, Puthiyaveettil R, Malkowicz SB, Helfman DM. Alterations in tropomyosin isoform expression in human transitional cell carcinoma of the urinary bladder. Inter J Cancer. 2004;110(3):368–373. doi: 10.1002/ijc.20151. [DOI] [PubMed] [Google Scholar]
- 25.Hughes JAI, Cooke-Yarborough CM, Chadwick NC, Schevzov G, Arbuckle SM, Gunning P, Weinberger RP. High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pediatric and adult CNS tumors. Glia. 2003;42(1):25–35. doi: 10.1002/glia.10174. [DOI] [PubMed] [Google Scholar]
- 26.Galloway PG, Likavec MJ, Perry G. Tropomyosin Isoform Expression in Normal and Neoplastic Astrocytes. Lab Invest. 1990;62(2):163–170. [PubMed] [Google Scholar]
- 27.Lin JLC, Geng X, Das Bhattacharya S, Yu JR, Reiter RS, Sastri B, Glazier KD, Mirza ZK, Wang KK, Amenta PS, Das KM, Lin JJC. Isolation and sequencing of a novel tropomyosin isoform preferentially associated with colon cancer. Gastroenterology. 2002;123(1):152–162. doi: 10.1053/gast.2002.34154. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Zhang H, Ayaz-Guner S, Chen YC, Dong XT, Xu QG, Ge Y. Phosphorylation, but not alternative splicing or proteolytic degradation, is conserved in human and mouse cardiac troponin T. Biochemistry. 2011;50(27):6081–6092. doi: 10.1021/bi2006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Ge Y. Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ-Cardiovasc Genet. 2011;4(6):711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth MJ, Forbes AJ, Boyne MT, Kim YB, Robinson DE, Kelleher NL. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol Cell Proteomics. 2005;4(7):1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sancho Solis R, Ge Y, Walker JW. Single amino acid sequence polymorphisms in rat cardiac troponin revealed by top-down tandem mass spectrometry. J Muscle Res Cell Motil. 2008;29(6–8):203–12. doi: 10.1007/s10974-009-9168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayaz-Guner S, Zhang J, Li L, Walker JW, Ge Y. In vivo phosphorylation site mapping in mouse cardiac troponin I by high-resolution top-Down electron capture dissociation mass spectrometry: Ser22/23 are the only sites basally phosphorylated. Biochemistry. 2009;48(34):8161–8170. doi: 10.1021/bi900739f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong X, Sumandea CA, Chen YC, Garcia-Cazarin ML, Zhang J, Balke CW, Sumandea MP, Ge Y. Augmented phosphorylation of cardiac troponin I in hypertensive heart failure. J Biol Chem. 2012;287(2):848–857. doi: 10.1074/jbc.M111.293258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge Y, Rybakova IN, Xu Q, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc Natl Acad Sci U S A. 2009;106(31):12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y, Chen X, Sato T, Rankin SA, Tsuji RF, Ge Y. Purification and high-resolution top-down mass spectrometric characterization of human salivary alpha-amylase. Anal Chem. 2012;84(7):3339–3346. doi: 10.1021/ac300083y. [DOI] [PubMed] [Google Scholar]
- 36.Xu F, Xu Q, Dong X, Guy M, Guner H, Hacker TA, Ge Y. Top-down highresolution electron capture dissociation mass spectrometry for comprehensive characterization of post-translational modifications in Rhesus monkey cardiac troponin I. Int J Mass Spectrom. 2011;305(2–3):95–102. [Google Scholar]
- 37.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Unraveling molecular complexity of phosphorylated human cardiac troponin I by top down electron capture dissociation/electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2008;7(10):1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Guy MJ, Norman HS, Chen YC, Xu Q, Dong X, Guner H, Wang S, Kohmoto T, Young KH, Moss RL, Ge Y. Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J Proteome Res. 2011;10(9):4054–4065. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 2007;4(10):817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan SS, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation. Mol Cell Proteomics. 2010;9(5):791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: A bird’s eye view. J Proteome Res. 2006;5(2):240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 42.Guan S, Burlingame AL. High mass selectivity for top-down proteomics by application of SWIFT technology. J Am Soc Mass Spectrom. 2010;21(3):455–459. doi: 10.1016/j.jasms.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senko MW, Speir JP, Mclafferty FW. Collisional activation of large multiply-charged ions using Fourier-transform mass-spectrometry. Anal Chem. 1994;66(18):2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 44.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 45.Neverova I, Van Eyk JE. Application of reversed phase high performance liquid chromatography for subproteomic analysis of cardiac muscle. Proteomics. 2002;2(1):22–31. [PubMed] [Google Scholar]
- 46.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high-resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectrom. 2000;11(4):320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 47.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20(6):261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 48.Houle F, Poirier A, Dumaresq J, Huot J. DAP kinase mediates the phosphorylation of tropomyosin-1 downstream of the ERK pathway, which regulates the formation of stress fibers in response to oxidative stress. J Cell Sci. 2007;120(20):3666–3677. doi: 10.1242/jcs.003251. [DOI] [PubMed] [Google Scholar]
- 49.Mak A, Smillie LB, Barany M. Specific Phosphorylation at Serine-283 of Alpha-Tropomyosin from from Skeletal and Rabbit Skeletal and Cardiac-Muscle. Proc Natl Acad Sci U S A. 1978;75(8):3588–3592. doi: 10.1073/pnas.75.8.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieczorek DF, Jagatheesan G, Rajan S. The Role of Tropomyosin in Heart Disease. In: Peter G, editor. Tropomyosin. Vol. 644. Springer; 2008. [DOI] [PubMed] [Google Scholar]
- 51.Leger J, Bouveret P, Schwartz K, Swynghedauw B. Comparative-Study of Skeletal and Cardiac Tropomyosins - Subunits, Thiol-Group Content and Biological-Activities. Pflugers Arch Euro J Physiol. 1976;362(3):271–277. doi: 10.1007/BF00581181. [DOI] [PubMed] [Google Scholar]
- 52.Muthuchamy M, Boivin GP, Grupp IL, Wieczorek DF. beta-tropomyosin overexpression induces severe cardiac abnormalities. J Mol Cell Cardiol. 1998;30(8):1545–1557. doi: 10.1006/jmcc.1998.0720. [DOI] [PubMed] [Google Scholar]
- 53.Muthuchamy M, Grupp IL, Grupp G, OToole BA, Kier AB, Boivin GP, Neumann J, Wieczorek DF. Molecular and physiological effects of overexpressing striated muscle beta-tropomyosin in the adult murine heart. J Biol Chem. 1995;270(51):30593–30603. doi: 10.1074/jbc.270.51.30593. [DOI] [PubMed] [Google Scholar]
- 54.Urbancikova M, Hitchcockdegregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J Biol Chem. 1994;269(39):24310–24315. [PubMed] [Google Scholar]
- 55.Singer JM, Shaw JM. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc Natl Acad Sci U S A. 2003;100(13):7644–7649. doi: 10.1073/pnas.1232343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heeley DH, Watson MH, Mak AS, Dubord P, Smillie LB. Effect of phosphorylation on the interaction and functional properties of rabbit striated muscle alpha alpha-tropomyosin. J Biol Chem. 1989;264(5):2424–2430. [PubMed] [Google Scholar]
- 57.Vahebi S, Ota A, Li MX, Warren CM, de Tombe PP, Wang YB, Solaro RJ. p38-MAPK induced dephosphorylation of alpha-tropomyosin is associated with depression of myocardial sarcomeric tension and ATPase activity. Circ Res. 2007;100(3):408–415. doi: 10.1161/01.RES.0000258116.60404.ad. [DOI] [PubMed] [Google Scholar]
- 58.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: Human histone H4. Anal Chem. 2006;78(13):4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 59.Wu SL, Huehmer AFR, Hao Z, Karger BL. On-line LC-MS approach combining collision-induced dissociation (CID): electron-transfer dissociation (ETD): and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J Proteome Res. 2007;6(11):4230–4244. doi: 10.1021/pr070313u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang JA, Dong XT, Hacker TA, Ge Y. Deciphering Modifications in Swine Cardiac Troponin I by Top-Down High-Resolution Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 2010;21(6):940–948. doi: 10.1016/j.jasms.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitelegge JP, Zabrouskov V, Halgand F, Souda P, Bassiliana S, Yan W, Wolinsky L, Loo JA, Wong DTW, Faull KF. Protein-sequence polymorphisms and post-translational modifications in proteins from human saliva using top-down Fourier-transform ion cyclotron resonance mass spectrometry. Int J Mass Spectrom. 2007;268(2–3):190–197. doi: 10.1016/j.ijms.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Halgand F, Zabrouskov V, Bassilian S, Souda P, Loo JA, Faull KF, Wong DT, Whitelegge JP. Defining Intact Protein Primary Structures from Saliva: A Step toward the Human Proteome Project. Anal Chem. 2012;84(10):4383–4395. doi: 10.1021/ac203337s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryan CM, Souda P, Halgand F, Wong DT, Loo JA, Faull KF, Whitelegge JP. Confident Assignment of Intact Mass Tags to Human Salivary Cystatins Using Top-Down Fourier-Transform Ion Cyclotron Resonance Mass Spectrometry. J Am Soc Mass Spectrom. 2010;21(6):908–917. doi: 10.1016/j.jasms.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SMM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480(7376):254–U141. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kellie JF, Tran JC, Lee JE, Ahlf DR, Thomas HM, Ntai I, Catherman AD, Durbin KR, Zamdborg L, Vellaichamy A, Thomas PM, Kelleher NL. The emerging process of Top Down mass spectrometry for protein analysis: biomarkers, protein-therapeutics, and achieving high throughput. Mol Biosys. 2010;6(9):1532–1539. doi: 10.1039/c000896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1–3. Peak list and fragment assignment of one ECD spectra of un- and mono-phosphorylated α–Tm isoform, and un-phosphorylated β–Tm isoform, respectively.
Supplemental Figure 1. Broadband ESI/FTMS spectra of swine Tm purified from three different domestic swine hearts.
Supplemental Figure 2. Protein sequence homology alignment of multiple swine Tm isoforms.
Supplemental Figure 3. MS/MS product ion map from one ECD spectrum of predominant swine Tm for assignment to un-phosphorylated swine α–Tm.
Supplemental Figure 4. (A) Isolation of single charge state (M32+) of α–Tm precursor ions, (B) ECD and (C) CAD spectra of the isolated α–Tm ions.
Supplemental Figure 5. Gas-phase separation (purification) of swine Tm.
Supplementary Figure 6. Protein sequence homology alignment of α–Tm encoded by gene TPM1.
Supplemental Figure 7. Representative ECD fragment ions for mapping the mono-phosphorylation site of swine α–Tm.
Supplementary Figure 8. Protein sequence homology alignment of β–Tm encoded by gene TPM2.