Abstract
GTPases are critical molecular switches involved in a wide range of biological functions. Recent phylogenetic and genomic analyses of the large, mostly uncharacterized COG0523 subfamily of GTPases revealed a link between some COG0523 proteins and metal homeostasis pathways. In this report, we detail the bioinorganic characterization of YjiA, a representative member of COG0523 subgroup 9 and the only COG0523 protein to date with high-resolution structural information. We find that YjiA is capable of binding several types of transition metals with dissociation constants in the low micromolar range and that metal binding affects both the oligomeric structure and GTPase activity of the enzyme. Using a combination of X-ray crystallography and site-directed mutagenesis, we identify, among others, a metal-binding site adjacent to the nucleotide-binding site in the GTPase domain that involves a conserved cysteine and several glutamate residues. Mutations of the coordinating residues decrease the impact of metal, suggesting that metal binding to this site is responsible for modulating the GTPase activity of the protein. These findings point toward a regulatory function for these COG0523 GTPases that is responsive to their metal-bound state.
GTPases are molecular switches that contribute to a wide variety of critical cellular processes ranging from ribosomal protein synthesis to the cell cycle.1−3 The majority of GTPases are members of the phosphate-binding loop (P-loop) NTPase class that share a mononucleotide-binding fold composed of conserved nucleotide-binding motifs and a central, mostly parallel, β-sheet surrounded by α-helices.4,5 All P-loop NTPases contain conserved structural elements, including the P-loop (Walker A motif), the Walker B motif, and the switch I region, which are essential for binding and hydrolysis of GTP as well as for the ensuing conformational changes that trigger the downstream effects.2,4 The P-loop NTPases can be phylogenetically sorted into several families based on shared structural and sequence features.4 One such family is composed of the G3E NTPases, which possess a glutamate residue in place of the conserved aspartate in the Walker B motif, responsible for coordinating the catalytically essential magnesium ion.4 The G3E family features four main subfamilies, three of which (HypB, UreG, and MeaB/ArgK) are based on prototypical proteins with established roles in the assembly of metalloenzyme active sites.6 The fourth subfamily, COG0523, is a more diverse and ubiquitous group of proteins with mostly unknown functions and a distribution across all three domains of life.4,6
A recent phylogenetic study, which suggested that the COG0523 subfamily of P-loop GTPases can be separated into at least 15 subgroups, predicted that some members of this subfamily function in metal metabolism.6 For example, several subgroups were linked to zinc homeostasis because of promoters containing the DNA recognition sequence of Zur, a zinc-responsive transcription factor, consistent with the zinc-dependent regulation observed for COG0523 members from several organisms.6−9 In addition, there are a few reports about individual COG0523 proteins that support specific roles in transition metal pathways. The first member of this subfamily identified was Pseudomonas denitrificans CobW, a protein essential for cobalamin biosynthesis.10 The function of CobW is ambiguous, but it has been suggested that it is responsible for the delivery of cobalt to the cobaltochelatase during assembly of the metallocofactor.11−13 Another COG0523 member is the nitrile hydratase activator protein Nha3, which is involved in the biosynthesis of the iron-dependent nitrile hydratase in Rhodococcus sp. N-771 and is proposed to be responsible for trafficking the proper metal to the enzyme precursor.14 A third described COG0523 factor is Bacillus subtilis YciC. Originally designated as a low-affinity zinc transporter,7 YciC has since been proposed to serve as a metallochaperone for delivery of the metal cofactor into YciA, a backup enzyme involved in folate biosynthesis.15 Both YciC in B. subtilis(7) and the more recently reported YeiR from Escherichia coli(16) help their host bacteria survive under zinc-limited growth conditions. Furthermore, in vitro analysis of YeiR revealed a link between metal binding and the protein’s oligomerization state and GTPase activity.16
Despite the large size of this subfamily and its connection with metal homeostasis, very little is known about the biochemical properties of the COG0523 constituents. The only high-resolution structural information about this subfamily is provided by the crystal structure of apo-YjiA from E. coli,17 making this protein an attractive target for structure–activity analysis. To garner further information about the metal binding activities of the COG0523 proteins and to better understand the possible connection with metal homeostasis, we conducted a bioinorganic characterization of YjiA. Metal binding studies demonstrate that YjiA can bind stoichiometric cobalt, 2 equiv of nickel, or four zinc ions in solution, and loading the protein with metal inhibits the GTPase activity. Using X-ray crystallography and spectroscopy, we identified a unique metal-binding site that, unlike those found in the UreG or HypB subfamilies,18,19 is located in a solvent-accessible pocket formed by a cysteine and two glutamate residues. This metal-binding site is adjacent to the nucleotide-binding site and is likely responsible for the observed effects of metal on GTP hydrolysis by YjiA. This report improves our understanding of this large class of proteins, supporting a role for metal-dependent regulation, and will be useful in dissecting the function of the uncharacterized COG0523 proteins.
Materials and Methods
Materials
Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs. Primers (Table S1 of the Supporting Information) were purchased from Sigma Genosys. All chromatography media were from GE Healthcare. Kanamycin, tris(2-carboxyethyl)phosphine (TCEP), phenylmethanesulfonyl fluoride (PMSF), and isopropyl β-d-thiogalactopyranoside (IPTG) were purchased from BioShop (Toronto, ON). Nickel chloride, zinc sulfate, and cobalt sulfate salts (as a minimum, 99.9% pure) were purchased from Aldrich. All other metal stocks were atomic absorption standard solutions. All other reagents were analytical or molecular biology grade from Sigma-Aldrich. The buffers for all metal assays were treated with Chelex-100 (Bio-Rad) to minimize trace metal contamination. All samples were prepared with Milli-Q water, with an 18.2 MΩ cm resistance (Millipore).
WT and Mutant YjiA Expression Vector Construction
The yjiA gene was amplified from genomic E. coli (DH5α) DNA using primers (Table S1 of the Supporting Information) designed with restriction sites for NdeI (YjiA forward) and XhoI (YjiA reverse). The digested polymerase chain reaction (PCR) product was ligated with T4 DNA ligase into the pET24b vector (Novagen) digested with NdeI and XhoI and dephosphorylated with calf intestinal phosphatase (New England Biolabs). The mutations were introduced into the YjiA-pET24b vector using QuikChange PCR mutagenesis (Stratagene) with Pfu Turbo DNA polymerase and the primers listed in Table S1 of the Supporting Information. The template was subsequently digested with DpnI. The E42A/C66A/C67A and E37A/C66A/C67A triple mutants were constructed by using the C66A/C67A YjiA-pET24b vector as a template and the E42A and E37A primers, respectively. For production of large amounts of the WT and mutant plasmids, the plasmids were transformed into XL-2 Blue E. coli (Stratagene) and isolated using the Fermentas GeneJET plasmid miniprep kit. All plasmids were sequenced (ACGT, Toronto, ON) in the forward and reverse directions.
Protein Expression and Purification
For expression of WT and mutant YjiA, the plasmids were transformed into BL21 Star(DE3) E. coli cells (Novagen). Overnight cultures were grown, and 25 mL was used to inoculate 1.5 L of LB medium supplemented with 50 μg/mL kanamycin. The cells were grown aerobically at 37 °C until the OD600 reached 0.6, at which point they were induced with 1 mM IPTG. After incubation at 37 °C for 5 h, the cells were harvested by centrifugation and resuspended in 20 mM Tris (pH 7.5) and 100 mM NaCl supplemented with 4 mM TCEP and 200 μM PMSF. For each preparation of purified protein, a total of 9 L of cell culture was used. All subsequent steps were performed at 4 °C or on ice. The resuspended cells were sonicated and centrifuged at 25000g for 40 min. The supernatant was passed through a 0.45 μm syringe filter and then loaded onto a DEAE-Sepharose anion-exchange column (GE Healthcare) equilibrated with buffer A [20 mM Tris (pH 7.5) and 1 mM TCEP]. Fractions from a NaCl gradient were screened by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using 12.5% gels. WT and mutant YjiA eluted at approximately 250 mM NaCl. Fractions containing the protein of interest were pooled and dialyzed against buffer A. The sample was then loaded onto a HiTrapQ anion-exchange column (GE Healthcare) equilibrated with buffer A. Once again, fractions from a NaCl gradient were screened by SDS–PAGE, and fractions containing the protein of interest (eluted at approximately 150 mM NaCl) were pooled. Following concentration of the pooled fractions to 1 mL using Amicon Ultra 3K molecular weight cutoff centrifuge concentrators (Millipore), the protein was loaded onto a Superdex 200 gel filtration column (GE Healthcare) equilibrated with 25 mM HEPES (pH 7.6), 200 mM NaCl, and 1 mM TCEP. Fractions containing the protein of interest were pooled, concentrated such that the final concentration was in the range of 250–500 μM, and stored at −80 °C. The protein concentrations were calculated by using an extinction coefficient of 26930 M–1 cm–1 at 280 nm for both YjiA and the YjiA mutants in 25 mM EDTA and 4 M guanidinium hydrochloride (GuHCl).20 Unless otherwise noted, electronic absorption measurements were taken on an Agilent 8453 spectrophotometer with a 1 cm path length cuvette. Each protein was analyzed by electrospray ionization mass spectrometry (ESI-MS) (Department of Chemistry, University of Toronto), and the observed molecular masses of the proteins are listed in Table S2 of the Supporting Information. All proteins were >90% pure as estimated by Coomassie-stained SDS–PAGE quantified by using ImageJ [National Institutes of Health, Bethesda, MD (http://rsb.info.nih.gov/ij/)].
Preparation of Proteins
The reduced apoprotein was produced by incubating the protein with 10 mM EDTA and 20 mM TCEP in a Coy anaerobic glovebox at 4 °C for 48 h. The TCEP and EDTA were removed by exhaustive dialysis into protein buffer [25 mM HEPES and 100 mM NaCl (pH 7.6)]. The absence of any metal bound to the protein was confirmed by a 4-(2-pyridylazo)resorcinol (PAR) assay,21 in which the protein was denatured with 4 M GuHCl and 50 μM PAR was added to the sample. The absorbance at 500 nm, due to the formation of the (PAR)2Me(II) complex, was monitored and compared to a standard curve prepared with 50 μM PAR in 4 M GuHCl and known metal concentrations. The free thiol content of the proteins was quantified via reaction with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in the presence of 6 M GuHCl and 1 mM EDTA. β-Mercaptoethanol was used as a standard, and the absorbance of the 5-mercapto-2-nitrobenzoic acid product was measured at 412 nm. Protein samples were >95% reduced after they had been treated with TCEP.
Metal Binding and Stoichiometry
For the difference spectra, individual samples containing 20 μM apo-YjiA in protein buffer and either 20 μM NiCl2 or 20 μM CoSO4 were prepared in the glovebox and incubated overnight at 4 °C. The electronic absorption spectrum was monitored between 250 and 500 nm and corrected by background subtraction at 600 nm. The difference spectrum for each metal was generated by subtracting the apo-YjiA spectrum from that of the metal-loaded protein.
Metal stoichiometry experiments were conducted by incubating 120 μM WT or mutant apo-YjiA with either 480 μM NiCl2, 360 μM CoSO4, or 600 μM ZnSO4 overnight at 4 °C in the glovebox. Samples containing mixtures of metal were similarly prepared with 120 μM WT apo-YjiA and either 360 μM NiCl2 and 360 μM CoSO4, 600 μM ZnSO4 and 480 μM NiCl2, or 600 μM ZnSO4 and 360 μM CoSO4. Samples with GDP contained 1.5 mM GDP and 5 mM MgCl2. Excess metal was removed by passing the protein through a PD10 gel filtration column (GE Healthcare) equilibrated with protein buffer in the glovebox. The protein concentration was subsequently measured in GuHCl and EDTA, as described above, and the metal content was determined via a PAR assay. In the case of the YjiA samples containing a mixture of metals, a high-pressure liquid chromatography (HPLC)-based method for the detection and identification of metal ions in solution was utilized.22 For HPLC analysis, at least 50 μg of protein was dried by centrifugation under vacuum, reconstituted with metal-free concentrated HCl (SeaStar Chemicals), and incubated overnight at 95 °C for protein hydrolysis. The sample was once again dried to remove HCl and reconstituted in 80 μL of Milli-Q water. This sample was injected onto a Dionex IonPak CS5A column, equilibrated with 7 mM pyridine-2,6-dicarboxylic acid, 66 mM KOH, 5.6 mM K2SO4, and 74 mM HCOOH, attached to a metal-free Dionex BioLC HPLC system. The metals were detected at 500 nm following postcolumn mixing with PAR.
ESI-MS Measurements
Before mass spectrometry experiments, 120 μM WT apo-YjiA was incubated with 600 μM ZnSO4 overnight at 4 °C in the glovebox and then buffer exchanged into 10 mM ammonium acetate (pH 7.5) using two consecutive PD10 columns. Mass spectra were recorded on an AB/Sciex QStarXL mass spectrometer equipped with an ion spray source in the positive mode and a hot source-induced desolvation interface. Ions were scanned in the m/z 1000–2000 range with accumulation times of 1 s per spectrum, with no interscan time delay. The instrument parameters were as follows: ion source temperature, 200 °C; ion source gas, 50.0 psi; curtain gas, 50.0 psi; ion spray voltage, 5000 V; declustering potential, 60.0 V; focusing potential, 60.0 V; MCP (detector), 2200 V. The spectra were deconvoluted using the Bayesian protein reconstruction program included with Analyst QS (AB/Sciex) over a mass range of 30000–40000 Da, with a mass step of 1 Da.
YjiA Metal Affinities
To determine the affinity of YjiA for Ni(II), 250 nM YjiA in protein buffer was titrated with a nickel atomic absorption standard solution and the absorbance at 340 nm was monitored using a 10 cm path length cuvette and a GBC Cintra 404 spectrophotometer. The apparent Kd was calculated by determining the fractional saturation, r, and free nickel concentration, [Ni(II)]free, by using eqs 1 and 2, respectively:
| 1 |
| 2 |
where [YjiA–Ni(II)] is the concentration of the protein bound to Ni(II), [YjiA]total is the total protein concentration, A340 is the absorbance at 340 nm for a given Ni(II) concentration, Amin is the absorbance at 340 nm for apo-YjiA, Amax is the absorbance at 340 nm upon saturation, and [Ni(II)]total is the total Ni(II) concentration added to the sample. The resulting values were plotted as r versus [Ni(II)]free, and the data were fit to the Hill equation (eq 3):
| 3 |
where n is the Hill coefficient.
To determine the affinity of YjiA for Co(II), 5 μM apo-YjiA was titrated with CoSO4 and the absorbance at 350 nm was monitored. The Co(II) Kd was calculated in the same manner as for Ni(II), except that eqs 1 and 2 were adjusted to solve for a single metal site instead of two.
YjiA Zincon Zn(II) Competitions
To estimate the Zn(II) Kd of YjiA, the competitor zincon was selected because of its ability to form a 1:1 complex with Zn(II) with a reported Kd of 12.7 μM.23,24 Stocks of zincon were prepared in Milli-Q water. The affinity of zincon for Zn(II) was verified under our experimental conditions by titrating 400 nM zincon in protein buffer with increasing amounts of ZnSO4. The absorbance at 620 nm was monitored using a 10 cm path length cuvette and a GBC Cintra 404 spectrophotometer. The Kd was calculated by using the same method that was used for the Ni(II)– and Co(II)–YjiA titrations, where n = 1. Competition experiments were conducted by incubating 10 μM apo-WT or E37A/C66A/C67A YjiA with 140 μM zincon and various amounts of Zn(II). The samples were incubated overnight at 4 °C in the glovebox. The absorbance of zincon at 620 nm was monitored using a 2 mm path length cuvette.
Analytical Gel Filtration Chromatography
Samples containing 60 μM YjiA were incubated with the desired metal, GDP, or GTP at the indicated concentrations (Tables S3 and S4 of the Supporting Information) overnight at 4 °C in the glovebox. For samples containing GTP, the protein was preincubated with GTP for at least 2 h at 4 °C in the glovebox prior to injection onto the gel filtration column. All samples contained 25 mM HEPES (pH 7.6), 100 mM NaCl, and 5 mM MgCl2. Apoprotein- and metal-containing samples were loaded onto a Superdex 200 10/300 analytical gel filtration column (GE Healthcare), pre-equilibrated with chelexed and filtered 25 mM HEPES (pH 7.6), 200 mM NaCl, and 5 mM MgCl2. The column was calibrated with thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.4 kDa) from Bio-Rad. The inclusion of metal in the standards did not affect their elution profiles (data not shown). Molecular masses were determined by plotting the log molecular masses of the standards versus the partition coefficient (Kav), where Kav = (Ve – Vo)/(Vt – Vo) (Ve represents the elution volume, Vo the void volume, and Vt the total column volume).
Circular Dichroism (CD) Spectroscopy
WT and mutant YjiA samples were prepared for CD spectroscopy by diluting the protein in Milli-Q water to a final concentration of approximately 10–20 μM in the glovebox. For metal titrations, ZnSO4, CoSO4, or NiCl2 was added to the diluted samples and allowed to equilibrate overnight at 4 °C in the glovebox. All samples were analyzed on an Olis RSM 1000 spectropolarimeter with a capped 1 mm path length cuvette to minimize exposure to the air. Spectra were collected at 1 nm intervals over a spectral range of 200–260 nm with an integration time of 2 s and 2400 grating lines/nm. The final spectra obtained are averages of three scans. The observed ellipticity was converted into mean residue ellipticity ([θ]mre, in degrees square centimeters per decimole) using the following formula:25
where MW is the molecular mass of the protein in daltons, N is the number of amino acids, θ is the observed ellipticity in degrees, [protein] is the protein concentration in grams per milliliter, and l is the path length. The secondary structure content was quantitated in the 200–240 nm range using K2D3.26 The CD spectra of the mutants were similar to that of the WT protein, demonstrating that these mutations do not affect the secondary structure of the protein (data not shown).
GTPase Assay
GTPase activity was determined by the Malachite Green assay for free phosphate adapted from Lanzetta et al.27 A series of 400 μL samples containing 0.5–2 μM WT or E37A/C66A/C67A YjiA in protein buffer supplemented with 5 mM Mg(II) and concentrations of GTP between 5 and 950 μM were incubated at 37 °C in the glovebox for 2.5–5 h. Controls without protein were prepared alongside the protein samples and received the same treatment. After incubation, 100 μL of the phosphate detection reagent (2.6 mM Malachite Green, 1.5% ammonium molybdate, and 0.2% Tween 20) was added to each sample. The samples were then mixed by being vortexed and incubated at room temperature for 3 min, after which sodium citrate was added to a final concentration of 3.5%. The samples were vortexed again and incubated at room temperature for 30 min prior to being plated on a 96-well plate, and the absorbance at 630 nm was measured with a Tecan Safire2 microplate reader. The amount of phosphate released was determined via a standard curve based on a phosphate standard (Molecular Probes). The data were analyzed by fitting to the Michaelis–Menten equation using OriginPro version 7.5. Samples containing metal were preincubated with the metal overnight prior to the assay [prepared as a stock of 10–40 μM protein with 100 μM Zn(II), 100 μM NiCl2, or 250 μM CoSO4 for WT or 225 μM Zn(II), 400 μM NiCl2, or 800 μM CoSO4 for the E37A/C66A/C67A triple mutant]. These stocks were then diluted to the final protein concentration of 0.5–2 μM for the assay in a buffer that contained 100 μM Zn(II), 100 μM NiCl2, or 250 μM CoSO4 for WT or 225 μM Zn(II), 400 μM NiCl2, or 800 μM CoSO4 for the E37A/C66A/C67A triple mutant.
Crystallization and Zn(II) Soaks
WT YjiA and E37A/C66A/C67A YjiA were crystallized at 25 °C by the hanging drop vapor diffusion technique. For WT YjiA, 1.5 μL of a protein solution [13.2 mg/mL in 100 mM NaCl and 25 mM HEPES (pH 7.6)] was mixed with 0.3 μL of 100 mM CaCl2 and 1.5 μL of a precipitant solution [1.55 M (NH4)2SO4 and 0.1 M HEPES (pH 6.9)] on a glass coverslip. The coverslip was sealed with grease over a reservoir containing 500 μL of the precipitant solution. Crystals appeared overnight and grew to maximal dimensions of ∼300 μm × 50 μm × 50 μm after 3 days. Crystals of E37A/C66A/C67A YjiA were grown in the same fashion with the following modifications: the precipitant solution consisted of 1.3–1.35 M (NH4)2SO4 and 0.1 M HEPES (pH 7.5–7.7), the protein concentration was 12.4 mg/mL, and the protein solution contained 1 mM TCEP in addition to the other components.
Because cocrystallization of WT YjiA with Zn(II) was unsuccessful, Zn(II)-soaked WT YjiA crystals were generated by incubating apo-WT crystals in the precipitant solution supplemented with 3 mM ZnSO4 and 1 mM TCEP at 25 °C for 16 h. The crystals were then directly transferred to a cryogenic solution [1.5 M (NH4)2SO4, 0.1 M HEPES (pH 6.9), 3 mM ZnSO4, 1 mM TCEP, and 20% (v/v) glycerol], incubated for 5 s, and flash-frozen in liquid nitrogen. Zn(II)-soaked E37A/C66A/C67A YjiA crystals were generated in a similar fashion with the following modifications: the soaking solution contained an additional 20 mM CaCl2 to better stabilize the crystals, the cryogenic solution consisted of 1.35 M (NH4)2SO4, 0.1 M HEPES (pH 7.7), 20 mM CaCl2, 3 mM ZnSO4, 1 mM TCEP, and 20% (v/v) glycerol, and the crystals were transferred into the cryogenic solution in three steps at increasing glycerol concentrations and incubated in that solution for 20 s.
Data Collection
All Zn(II)-soaked YjiA crystals belong to space group P21 (a ≈ 56 Å, b ≈ 69 Å, c ≈ 78 Å, and β ≈ 104°) and contain two protomers in the asymmetric unit. All diffraction data were collected at 100 K.
Diffraction data for Zn(II)-soaked WT YjiA were collected at Stanford Synchrotron Radiation Laboratory beamline 9-2 in Portola Valley, CA, using a MARmosaic 325 CCD detector. An initial Zn(II)-soaked WT YjiA crystal was used for an X-ray fluorescence scan to verify the presence of bound Zn(II). On the basis of the fluorescence scan, a peak wavelength of 1.2827 Å (9665.5 eV) was determined using CHOOCH.28 A wavelength of 1.1808 Å (10500.0 eV) was chosen for the remote data set. Remote and peak data sets were collected on the same crystal in wedges of 30° in 1° oscillation steps. For each wedge, the corresponding images related by a 180° crystal rotation were collected immediately after completion of the wedge. An inflection data set was also collected on the same crystal, but not used for this study. Bijvoet mates were treated as separate reflections for both the remote and the peak data set.
Diffraction data for Zn(II)-soaked E37A/C66A/C67A YjiA were collected at Advanced Photon Source beamline 24ID-C in Argonne, IL, using a Pilatus 6M pixel detector. The crystal was annealed for 5 s prior to data collection.29 Then, a single data set was collected at a wavelength of 0.9795 Å (12658.0 eV) in 1° oscillation steps. For structure solution, Bijvoet mates were treated as symmetry-related reflections. For calculation of Zn anomalous difference Fourier maps of Zn(II)-soaked E37A/C66A/C67A YjiA, Bijvoet mates were treated as separate reflections.
All data were integrated using HKL2000 and scaled using Scalepack.30 The same reflections were marked for the free set of reflections for Zn(II)-soaked WT YjiA and for Zn(II)-soaked E37A/C66A/C67A YjiA. In addition, 5% of the reflections in the resolution ranges of 75–30 and 2.57–2.05 Å were included in the free set for the data of the triple mutant. All data collection statistics are summarized in Table S5 of the Supporting Information.
Structure Building and Refinement
The crystal structure of Zn(II)-soaked WT YjiA was determined at 2.57 Å resolution by molecular replacement with the crystal structure of native WT YjiA without water molecules [Protein Data Bank (PDB) entry 1NIJ(17)] in PHASER,31 which yielded a single solution. The output model was subjected to 20 cycles of simulated annealing in PHENIX32 to reduce model bias. The model was adjusted for any changes that had occurred through a cycle of model building in COOT33 followed by refinement in PHENIX.32 Initial difference electron density maps revealed the presence of five Zn(II) ions in the asymmetric unit. Zn(II) ions and water molecules were inserted in subsequent rounds of refinement.
The intermediate model of Zn(II)-soaked WT YjiA without water molecules or Zn(II) was used to determine the crystal structure of Zn(II)-soaked E37A/C66A/C67A YjiA to 2.05 Å resolution. The structure was determined by rigid body refinement in PHENIX followed by automated rebuilding of the structure using phenix.autobuild34 to reduce model bias. Initial difference electron density maps revealed the presence of all three mutations as well as the presence of three Zn(II) ions in the asymmetric unit. Subsequently, Zn(II) ions and water molecules were inserted, and the model was adjusted to account for the mutations as well as other changes through iterative cycles of model building in COOT33 and refinement in PHENIX.32
Because of the superior quality of the Zn(II)-soaked E37A/C66A/C67A YjiA data set (Table S5 of the Supporting Information), the model derived from this data set was then used to finalize the structure of Zn(II)-soaked WT YjiA. Following rigid body refinement of the mutant structure into the WT YjiA data in PHENIX,32 additional Zn(II) ions were inserted and the structure was completed through iterative cycles of model building in COOT33 and refinement in PHENIX.32
For both structures, the two protomers in the asymmetric unit were initially restrained by strict non-crystallographic symmetry (NCS) parameters. In advanced stages of refinement, NCS restraints were loosened for all residues and removed for residues involved in crystal contacts or Zn(II) binding. Side chains with limited electron density were truncated at the last atom with visible electron density. Zn(II) coordination was loosely restrained to average bond lengths observed in previously determined structures of proteins with Zn(II) ions bound.35 The final stages of refinement for both structures involved TLS parametrization36 using two TLS groups per protomer, as determined by the TLSMD server.37 The locations of Zn(II) ions were verified by Zn anomalous difference Fourier maps calculated from the peak data set (WT YjiA) or the remote data set (E37A/C66A/C67A YjiA) using FFT, which is part of the CCP4 program suite.38
Crystallographic refinement of both structures yielded models that possess excellent stereochemistry and small root-mean-square deviations from ideal values for bond lengths and bond angles. Because of the presence of pseudo-translational symmetry in the data, the crystallographic R factors are relatively high, but within an acceptable range. Refinement statistics are summarized in Table S5 of the Supporting Information. The final model of WT YjiA contains all 318 residues of YjiA in protomer A and residues 2–318 in protomer B. The final model of E37A/C66A/C67A YjiA contains all 318 residues in both protomers. The models were validated using simulated annealing composite omit maps calculated in CNS.39 The geometries of the final models were analyzed with Molprobity.40 Figures were generated using Pymol.41
Results
YjiA Binds Zn(II), Ni(II), and Co(II) with Micromolar Affinities
The presence of a CXCC motif in YjiA, conserved in all COG0523 proteins,6 led to the proposal that the protein can bind metals,17 but experimental evidence was lacking. Because of the homology of YjiA to G3E NTPases known to be involved in Zn(II) (YeiR16 and YciC7), Ni(II) (UreG42−44 and HypB42,44), or Co(II) (CobW10) homeostasis, these three metals were the focus of this investigation. To determine whether YjiA can bind these metals and, if so, how much, 120 μM apo-YjiA was incubated with either 480 μM NiCl2, 360 μM CoSO4, or 600 μM ZnSO4, and excess metal was removed by gel filtration chromatography. Subsequent metal analysis via a PAR assay revealed roughly stoichiometric cobalt, two Ni(II) ions, or four Zn(II) ions bound per monomer (Table 1).
Table 1. Stoichiometry of Binding of Metal to WT and Mutant YjiAa.
| Ni(II) bound | Co(II) bound | Zn(II) bound | |
|---|---|---|---|
| WT | 2.2 ± 0.2 | 1.2 ± 0.2 | 3.8 ± 0.3 |
| WT with GDP | 1.3 ± 0.1 | 0.6 ± 0.2 | 3.2 ± 0.2 |
| E37A/C66A/C67A | 0.5 ± 0.2 | 0.1 ± 0.1 | 3.9 ± 0.5 |
| E42A/C66A/C67A | 1.2 ± 0.2 | 0.2 ± 0.1 | 4.2 ± 0.3 |
Apo-YjiA (120 μM) was incubated with either 480 μM NiCl2, 360 μM CoSO4, or 600 μM ZnSO4 overnight at 4 °C in an anaerobic glovebox. Excess metal was removed by passing the proteins over a PD10 gel filtration column, and the bound metal was detected via a PAR assay. In samples containing GDP, 1.5 mM GDP and 5 mM MgCl2 were included. The data listed are average values and standard deviations of the number of metal ions bound per protein monomer from at least three independent experiments.
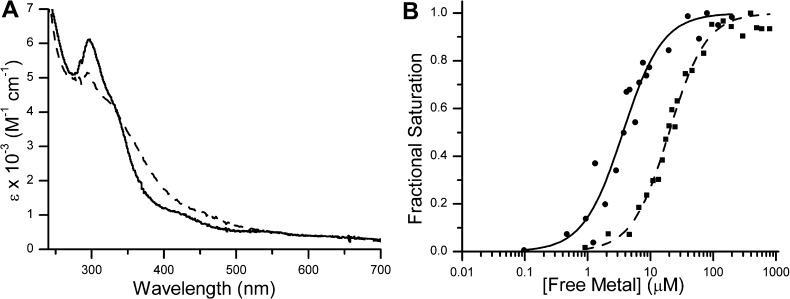
After incubation of YjiA with 1 equiv of nickel or cobalt, broad charge-transfer bands appear in the electronic absorption spectrum region of 250–400 nm (Figure 1A). The difference spectra, obtained by subtracting the signal of apo-YjiA from that of the protein loaded with 1 equiv of Ni(II) or Co(II) (Figure 1A), reveal a peak maximum at 280 nm and a broad shoulder around 340 or 350 nm for nickel and cobalt, respectively. The 280 nm and shoulder absorptions for both metals can be attributed to Cys-S– → Ni(II)/Co(II) ligand-to-metal charge transfer (LMCT),45−48 indicating the presence of at least one thiolate in the coordination sphere of both metals.
Figure 1.
Cobalt and nickel binding to WT YjiA. (A) The difference spectra of 20 μM YjiA incubated with 20 μM NiCl2 (—) or 20 μM CoSO4 (---) display ligand-to-metal charge-transfer (LMCT) signals in the region of 250–400 nm. The extinction coefficient was calculated on the basis of the protein concentration. (B) To determine the affinity of apo-WT YjiA for Ni(II), the metal was titrated into a sample of 250 nM YjiA and the increase in absorption at 340 nm was used to calculate the fractional saturation of the protein (●). Data like those shown were fit to the Hill equation to yield an apparent Kd of (3.7 ± 0.3) × 10–6 M [n = 1.2 ± 0.3 (—)] (see the text). Likewise, binding of Co(II) to 5 μM YjiA can be monitored at 350 nm (■) and fit to yield an apparent Kd of (2.0 ± 0.1) × 10–5 M [n = 1.7 ± 0.2 (---)].
The LMCTs for Ni(II) and Co(II) can be used to monitor metal binding by YjiA. A titration of 5 μM apo-YjiA with CoSO4 yields an apparent Kd of (2.0 ± 0.1) × 10–5 M and a Hill coefficient n of 1.7 ± 0.2 (Figure 1B). This Hill coefficient suggests cooperativity in cobalt binding to YjiA, a surprising result given that the protein binds only 1 equiv of cobalt. This discrepancy may be explained by the ability of cobalt to induce oligomerization in the protein (see below). The titration of YjiA with Ni(II) is more complicated because of the 2:1 Ni(II):YjiA stoichiometry as the titration may be monitoring either a single site or both Ni(II) sites simultaneously. The electronic absorption spectrum demonstrates that at least one site is spectroscopically active, but the mutants described below suggest that the second Ni(II) site does not yield a discernible electronic absorption signal. The appearance of a LMCT upon the addition of substoichiometric amounts of nickel to YjiA indicates that the second site is not significantly tighter than the spectroscopically active site. Thus, the second nickel site has either a similar or a lower affinity than the spectroscopically active site. If this site is much weaker, then the nickel titration in Figure 1B represents only the spectroscopically active site and yields an apparent Kd of (3.9 ± 0.3) × 10–6 M with a Hill coefficient n of 1.2 ± 0.2 for this one site. If the two sites have similar affinities, an apparent average Kd for the two sites can be calculated from the Ni(II) titration to be (3.7 ± 0.3) × 10–6 M (n = 1.2 ± 0.3).
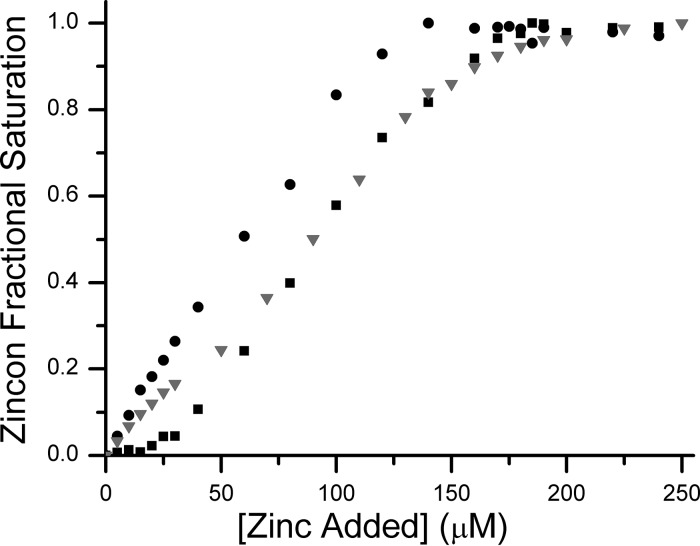
Because of the spectroscopically quiet nature of Zn(II), a metallochromic indicator, zincon, was used to estimate the affinity of Zn(II) for YjiA. The Kd of zincon for Zn(II) was determined to be 10 ± 1 μM under our buffer conditions (data not shown), in agreement with the reported Kd of 12.7 μM.23,24 In the absence of protein, metal loading of 140 μM zincon is complete upon addition of 140 μM Zn(II) (Figure 2). Upon the inclusion of 10 μM YjiA, 180 μM Zn(II) is required to observe saturation of the zincon (Figure 2), demonstrating competition by YjiA for Zn(II) and consistent with the measured Zn(II) stoichiometry of four ions per monomer. Furthermore, binding of Zn(II) to zincon is not observed until more than 20 μM metal has been added, suggesting that YjiA has two Zn(II) sites with a Kd significantly tighter than that of zincon (Kd < 10 μM), and that the remaining two Zn(II) sites have affinities comparable to that of zincon (Kd ∼ 10 μM). Finally, to confirm the Zn(II) stoichiometry, ESI-MS of YjiA incubated with excess Zn(II) was performed (Figure S1 of the Supporting Information). The mass spectrum revealed up to four Zn(II) ions bound to the protein, with two sites completely filled and two partially filled, consistent with the affinities of the sites estimated in the competition experiment.
Figure 2.
Zinc binding to YjiA. Once Zn(II) binds, the maximal absorption of zincon shifts from 488 to 620 nm. It takes 140 μM ZnSO4 to saturate 140 μM zincon (●), consistent with a 1:1 stoichiometry. In a competition between 140 μM zincon and 10 μM apo-YjiA (■), the spectrum of zincon does not change until after the addition of 20 μM ZnSO4, suggesting that there are two Zn(II) sites on YjiA that can outcompete zincon. It takes an additional 160 μM ZnSO4 to saturate the 620 nm signal, suggesting that there are two additional sites in YjiA capable of competing with zincon for Zn(II). In the case of the E37A/C66A/C67A mutant (▼), the initial plateau region is not observed, but 180 μM ZnSO4 is still required to saturate zincon, suggesting that the mutant YjiA can still bind four Zn(II) ions but with affinities weaker than that of the WT protein.
Ni(II), Co(II), and Zn(II) Share a Common Site
To determine if the binding sites of the different transition metals overlap, apo-YjiA was incubated simultaneously with two types of metal, followed by HPLC metal analysis to identify which metals are bound. When YjiA was incubated with both CoSO4 and NiCl2, 1.9 ± 0.3 Ni(II) ions and 0.15 ± 0.01 Co(II) ion were detected bound to the protein, suggesting that Co(II) shares a site with Ni(II) and that Ni(II) can outcompete Co(II) for this site. Similar experiments conducted with ZnSO4 and either NiCl2 or CoSO4 demonstrate a selectivity for Zn(II) as 3.6 ± 0.3 Zn(II) ions and 4.0 ± 0.4 Zn(II) ions per YjiA were detected, respectively, with no detectable Ni(II) or Co(II).
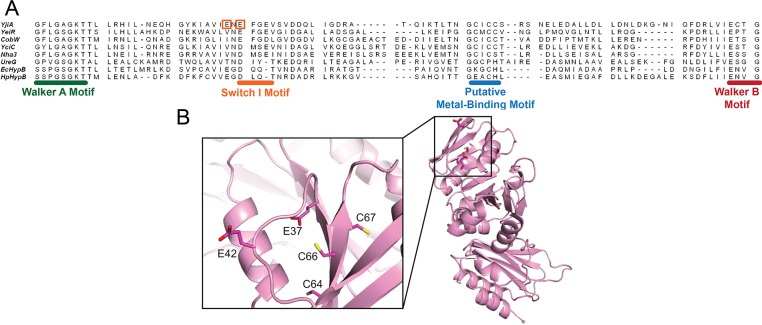
The GTPase Domain Contains the Common Metal-Binding Site
A sequence alignment of YjiA with other G3E GTPases demonstrates the presence of conserved putative metal-binding residues between the Walker A and Walker B motifs (Figure 3A). In the COG0523 subfamily, this conserved sequence corresponds to C64XCC67 of YjiA. Furthermore, examination of the published crystal structure of apo-YjiA17 revealed two nearby glutamates, Glu37 and Glu42, which could also serve as metal-binding residues (Figure 3B). To investigate the role of these glutamates as well as the CXCC motif in metal binding, we generated the E37A/C66A/C67A and E42A/C66A/C67A triple mutants of YjiA and characterized their metal binding capabilities. Upon addition of Ni(II) or Co(II) to either triple mutant, the absorbance spectra lacked the LMCTs at 340 or 350 nm, respectively, suggesting that Cys66 and/or Cys67 are the source of these signals (data not shown). Furthermore, both triple mutants exhibited weakened Co(II) and Ni(II) binding, but the Zn(II) stoichiometry was unchanged (Table 1). However, the previous observation that Zn(II) competes with the other metals (see above), suggesting overlapping sites, led us to investigate further the Zn(II) coordination of E37A/C66A/C67A YjiA by using a zincon competition. Unlike the WT protein, no initial plateau region was observed when the triple mutant was included in the zincon titration, yet 180 μM Zn(II) was still required to saturate 140 μM zincon (Figure 2). This result is consistent with the maintained Zn(II) stoichiometry of the mutant protein but indicates that the mutations have significantly weakened the affinities of two of the metal ions and suggests that there is at least one Zn(II)-binding site involving some combination of E37, C66, and C67 that has a Kd of <10 μM. Unlike WT YjiA (Figure S1 of the Supporting Information), the metalation state of the zinc-loaded mutant protein could not be observed by mass spectrometry, because of poor signal quality. Upon addition of zinc to the mutant protein, the decrease in signal was greater than that observed for WT, suggesting that it cannot just be attributed to ion suppression and is likely due to changes in the charge state of the mutant protein. Altogether, these experiments indicate that the Ni(II) and Co(II) sites and at least one Zn(II) site contain some combination of E37, E42, C66, and C67 as ligands.
Figure 3.
Structure of WT YjiA and location of the metal-binding site in the primary structure of the GTPase domain. (A) Sequence alignment of the GTPase domain regions between the Walker A and Walker B motifs of representative G3E GTPases generated by the COBALT sequence alignment program (available online at http://www.ncbi.nlm.nih.gov/tools/cobalt/).65 Located between the Walker A and B motifs is a putative metal-binding motif, the location of which is common among the G3E GTPases. The two glutamates mutated in this study are highlighted by orange boxes. The species from which the protein sequences were derived and the starting sequence positions (in brackets) are as follows: YjiA, E. coli (11); YeiR, E. coli (9); CobW, P. denitrificans (18); YciC, B. subtilis (11); Nha3, Rhodococcus sp. N-771 (13); UreG, Klebsiella aerogenes (14); EcHypB, E. coli (111); HpHypB, Helicobacter pylori (53). (B) The structure of apo-YjiA (PDB entry 1NIJ), previously published,17 features two domains. The N-terminal GTPase domain possesses a typical G3E GTPase fold with a central β-sheet core surrounded by α-helices. Located on one of the central β-strands is the conserved C64XCC67 motif. Glu37 and Glu42 are near this motif (inset).
Metal Binding Induces Oligomerization of YjiA with No Major Secondary Structure Changes
The CD spectrum of apo-WT YjiA (Figure S2 of the Supporting Information) is indicative of a mixed αβ protein, in close agreement with the published crystal structure.17 Upon addition of Co(II), Zn(II), or Ni(II), minor changes in the CD spectrum were detected, but the calculated percentage of secondary structure content was unchanged (Figure S2 of the Supporting Information), indicating that YjiA does not undergo any dramatic secondary structure changes upon binding metal.
Analytical gel filtration chromatography was used to probe the oligomeric state of apo-YjiA, as well as the metal- and nucleotide-bound species. In the apo form, the protein elutes from the column as a monomer (Figure 4 and Table S3 of the Supporting Information). The calculated molecular mass of the monomeric apo-YjiA species from the gel filtration experiments (30.0 ± 1.5 kDa) is smaller than the predicted molecular mass of 35.7 kDa, suggesting a compact protein structure. Upon the addition of 2 equiv of Ni(II), Co(II), or Zn(II), the protein oligomerizes, forming a small amount of dimer as well as a larger oligomeric species (Figure 4). The protein also dimerizes in the presence of GDP or GTP, but to a lesser degree than with metal (Figure 4). Some oligomerization was also observed upon adding metal to the E37A/C66A/C67A and E42A/C66A/C67A mutants, although the impact on quaternary structure is diminished compared to the impact in the WT protein (Table S4 of the Supporting Information).
Figure 4.
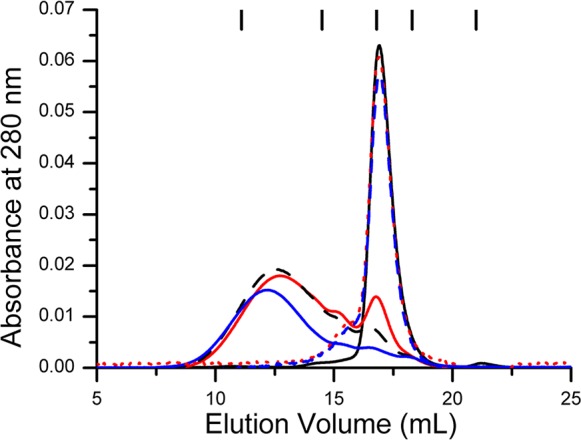
Effect of metal on the quaternary structure of YjiA. In the absence of metal, 60 μM YjiA elutes at a volume corresponding to a monomer (solid black line). The addition of 2 equiv of NiCl2 (solid red line), CoSO4 (dashed black line), or ZnSO4 (solid blue line) results in the formation of dimeric and oligomeric species. Incubation of YjiA with 2 equiv of GDP (dashed red line) or GTP (dashed blue line) followed by chromatography with 400 μM nucleotide in the running buffer resulted in only a small portion of dimeric protein. The chromatographic traces (monitored at 280 nm) are representative data sets from experiments with a Superdex 200 10/300 analytical column equilibrated with 25 mM HEPES (pH 7.6), 200 mM NaCl, and 5 mM MgCl2. The ticks at the top of the graph denote the elution volumes of the protein standards. From left to right, the identities of the standards (and their molecular masses) are thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.4 kDa), respectively.
Metal Binding Affects YjiA GTPase Activity
YjiA was previously shown to bind GTP and, on the basis of its GTPase motifs, is predicted to possess GTP hydrolysis (GTPase) activity.17 In agreement with this prediction, the apo-WT protein has low GTPase activity, with a kcat/Km of 14 ± 9 M–1 s–1 (Table 2). To determine whether metal binding modulates the GTPase activity, we incubated apo-YjiA with Ni(II), Co(II), or Zn(II) prior to analysis. The addition of Co(II) slightly disrupts GTPase activity, reducing the kcat/Km to 2.3 ± 0.8 M–1 s–1. The presence of Ni(II) significantly diminishes the GTPase activity of YjiA, such that substrate saturation was not observed with up to 950 μM GTP, indicating a significantly weaker Km versus that of the apoprotein. Attempts to use higher GTP concentrations were unsuccessful because of the departure from the the linear response region of the assay (data not shown). Zn(II) also inhibits the enzyme, and in this case, no activity was detectable in the presence of 100 μM ZnSO4 (Table 2). The results of these GTPase assays prompted experiments that aimed to examine if there was a connection between GDP binding and metal binding to YjiA. Inclusion of 1.5 mM GDP in the protein buffer lowered the metal stoichiometry of all three metals (Table 1).
Table 2. Kinetics of GTP Hydrolysis by WT and E37A/C66A/C67A YjiAa.
| metal bound | kcat (s–1) | Km (M) | kcat/Km(M–1 s–1) | |
|---|---|---|---|---|
| WT | apo | (6 ± 2) × 10–3 | (5 ± 3) × 10–5 | 14 ± 9 |
| Co(II) | (5 ± 2) × 10–4 | (2.3 ± 0.7) × 10–4 | 2.3 ± 0.8 | |
| Ni(II) | WHb | |||
| Zn(II) | NHc | |||
| E37A/C66A/C67A | apo | (5 ± 1) × 10–4 | (1.0 ± 0.7) × 10–4 | 6 ± 3 |
| Co(II) | (1.1 ± 0.1) × 10–3 | (1.7 ± 0.1) × 10–5 | 78 ± 9 | |
| Ni(II) | (6 ± 1) × 10–4 | (3 ± 1) × 10–4 | 2 ± 1 | |
| Zn(II) | NHc | |||
All GTPase assays were conducted with 0.5–2 μM WT or E37A/C66A/C67A YjiA in protein buffer supplemented with 5 mM MgCl2. Samples containing metal were preincubated with either zinc, cobalt, or nickel overnight at 4 °C in an anaerobic glovebox. The amount of released phosphate was detected using a modified Malachite Green assay. The data listed are average values from at least three independent experiments.
Weak hydrolysis (see the text for details).
No measurable hydrolysis (see the text for details).
The GTPase activity of the apo form of the E37A/C66A/C67A mutant is comparable to that of the WT protein, with a kcat/Km of 6 ± 3 M–1 s–1 (Table 2); however, unlike WT YjiA, the mutant protein was not inhibited by Ni(II), and Co(II) increased the overall catalytic efficiency of the enzyme to a kcat/Km of 78 ± 9 M–1 s–1 (Table 2). In contrast, addition of Zn(II) to the triple mutant still inhibited GTPase activity, even though it was added at lower concentrations than Co(II) and Ni(II). While the E37A/C66A/C67A triple mutant has reduced affinity for all three metals (see above), it appears that Zn(II) binds to at least some sites in YjiA with higher affinity than Co(II) and Ni(II) do, and that this affinity remains sufficiently high to cause inhibition at the concentrations used. To probe whether this inhibition of GTPase activity occurs by direct Zn(II) binding to the active site or in an allosteric fashion by Zn(II) binding to other metal sites on YjiA, we conducted a structural analysis of Zn(II)-bound YjiA.
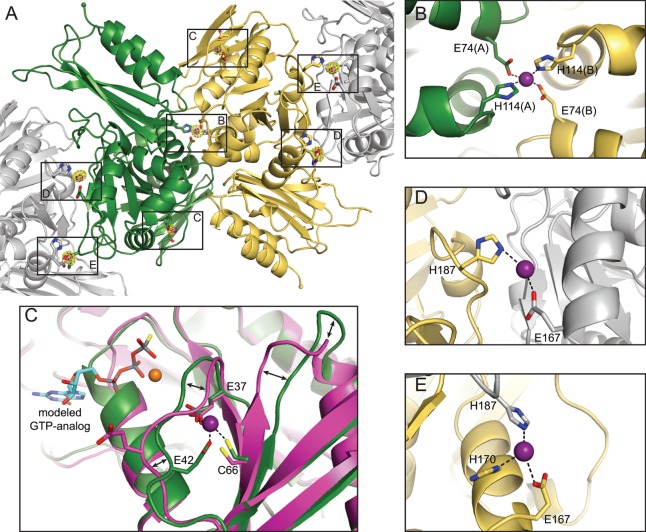
Zn(II) Binding Induces a Space Group Transition in YjiA Crystals
We determined the crystal structure of Zn(II)-bound WT YjiA to 2.57 Å resolution after soaking apo-WT YjiA crystals with ZnSO4 (Figure 5). Surprisingly, the crystals underwent a space group transition upon soaking. While unsoaked (apo) WT YjiA crystals belong to space group C2 with one protomer in the asymmetric unit,17 the space group is P21 after soaking and features two protomers in the asymmetric unit (for a more detailed discussion of space group assignment, see the Supporting Information). The arrangement of molecules in the crystal lattices of Zn(II)-soaked WT YjiA and apo-WT YjiA, however, is nearly identical despite the different space groups (Figure S3 of the Supporting Information). It seems that Zn(II) soaking induced a small shift in the crystal lattice, causing a reduction in symmetry from C2 to P21 while leaving the overall crystal lattice intact.
Figure 5.
Crystal structure of Zn(II)-bound WT YjiA. (A) Overall structure of the two YjiA protomers in the asymmetric unit (yellow and green ribbons). Symmetry-related molecules involved in Zn(II) binding are shown as gray ribbons. Four types of Zn(II)-binding sites are observed in the structure: a bridging site (labeled B), an internal site (labeled C), and two types of surface sites (labeled D and E). Bound Zn(II) ions are shown as purple spheres and coordinating residues as sticks. A Zn anomalous difference Fourier map is contoured around bound Zn(II) ions at 5σ. (B) Close-up view of the bridging Zn(II)-binding site, located on a 2-fold axis between the two protomers in the asymmetric unit. Zn(II) is coordinated by E74 and H114 from both protomers. (C) Close-up view of the internal Zn(II)-binding site. The side chains of E37, E42, and C66 (green carbons) coordinate the Zn(II), with an open coordination sphere probably occupied by a water molecule. The structure of apo-WT YjiA (PDB entry 2NIJ(17)) is superimposed and shown with magenta carbons. The region around the Zn(II)-binding site undergoes conformational changes upon Zn(II) binding, as indicated by the arrows. The Zn(II)-binding site is also located in the proximity of the nucleotide binding site, as demonstrated by modeling studies with a GTP analogue-bound HypB structure [PDB entry 2HF8,18 GTPγS shown with cyan carbons and Mg(II) shown as an orange sphere]. (D and E) Close-up views of the two types of surface sites, located at crystal contacts between YjiA protomers in the asymmetric unit and crystallographically related molecules. Zn(II) is coordinated by the side chains of E167 and/or H170 from one molecule and H187 from the other molecule.
Unfortunately, we have not been able to generate nucleotide-bound structures of YjiA itself. While YjiA exhibits the canonical GTPase motifs, these motifs are not well-ordered in the structure and the phosphate-binding Walker A motif in particular seems to exhibit multiple conformations (Figure S4 of the Supporting Information). Likely, nucleotide binding will result in a conformational change and depend on the presence of an effector protein, consistent with our unsuccessful attempts to generate nucleotide-bound structures.
YjiA Binds Zn(II) in Four Distinct Sites
Analysis of the electron density and of Zn anomalous difference Fourier maps revealed the presence of Zn(II) ions in four distinct types of sites: a “bridging site” that is located at the interface of the two YjiA protomers in the asymmetric unit (site B in Figure 5A), an “internal site” (site C in Figure 5A), and two types of “surface sites” (sites D and E in Figure 5A). In total, the two YjiA protomers in the asymmetric unit could bind seven Zn(II) ions: two in the internal sites, one in the bridging site, and four in surface sites (Figure 5A). The asymmetric unit, however, contains only five Zn(II) ions because all four surface sites are involved in crystal lattice contacts and each of those bound Zn(II) ions is shared with a neighboring asymmetric unit molecule.
The Internal Site Is Located near the GTPase Active Site
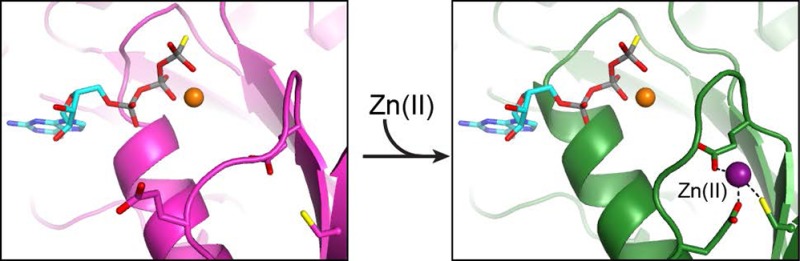
The internal site is located in the N-terminal region of a YjiA protomer, with the side chains of E37, E42, and C66 coordinating Zn(II) (Figure 5C). Although hard to resolve in the electron density at this resolution, a fourth coordination site on Zn(II) could be occupied by a water molecule, which would give rise to a tetrahedral coordination geometry. E37 and E42 are located in a loop region that undergoes a conformational change upon Zn(II) binding to bring the side chain of E42 into the proximity of the bound Zn(II) (Figure 5C). Furthermore, the region around C66 undergoes a slight shift, bringing C66 closer to the Zn(II) ion and rearranging the subsequent loop region significantly (Figure 5C).
It is clear that the internal Zn(II)-binding site is in the neighborhood of the nucleotide-binding site (Figure 5C), as demonstrated by a structural alignment with the GTP analogue-bound structure of Methanocaldococcus jannaschii HypB (PDB entry 2HF8(18)), which has the highest degree of structural homology with YjiA of the proteins in the PDB [Cα root-mean-square deviation (rmsd) of 3.1 Å over 209 residues, determined by DaliLite49]. In this model, the nucleotide-associated Mg(II) is within 5.4 Å of the side chain carboxylate of E37 and within 5.3 Å of the Cβ atom of E39 (Figure 6), which has a disordered side chain. These observations raise the possibility that E37 or E39 are involved in Mg(II) coordination upon nucleotide binding. Notably, E39 is the first residue of the switch I motif of YjiA, a common NTPase motif known to undergo conformational changes during NTPase activity, as indicated by structural and sequence alignment (Figure 3A). In M. jannaschii HypB, the first residue of the switch I motif, D75, is one of the ligands to the nucleotide-associated Mg(II) (Figure 6).18 In our structural alignment, D75 of M. jannaschii HypB aligns with E39 of YjiA (Figure 6), further suggesting that E39 could be involved in Mg(II) coordination.
Figure 6.
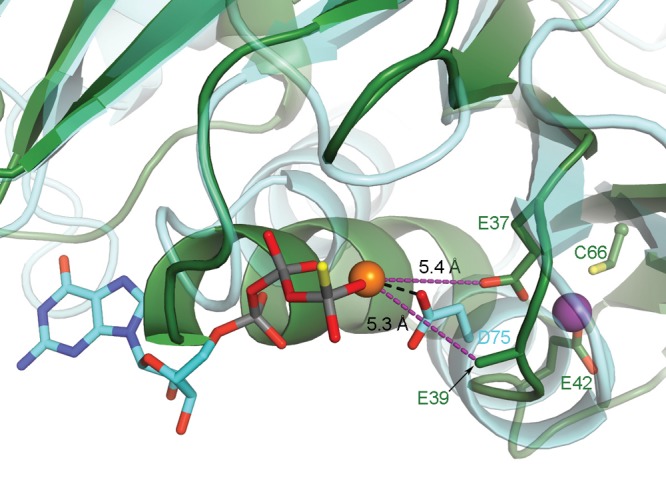
E37 or E39 of YjiA could coordinate nucleotide-associated Mg(II). GTP analogue-bound HypB (cyan carbons, PDB entry 2HF8(18)) is superimposed onto the structure of Zn(II)-bound WT YjiA (green carbons). Bound GTPγS is shown with cyan carbons, and GTPγS-associated Mg(II) is shown as an orange sphere. D75 of HypB is involved in Mg(II) coordination. Both E37 and E39 of YjiA are in the proximity of the modeled Mg(II), as indicated by the purple dashed lines, raising the possibility that these residues are involved in Mg(II) coordination in YjiA. Notably, E37 is also involved in binding Zn(II) (purple sphere). E39 was truncated to the Cβ atom because of a lack of electron density for the side chain.
The structure of this internal site is consistent with the solution studies of metal binding to WT and mutant YjiA. Given that mutagenesis of some of these ligands disrupts Co(II) and Ni(II) binding, it is likely that a variation of this internal site also binds these other metals in addition to Zn(II), with the coordination by C66 producing the LMCT absorption features observed for the Co(II)- or Ni(II)-loaded protein. Furthermore, the observation that Zn(II) can outcompete both Ni(II) and Co(II) for binding suggests that this site is one of the higher-affinity sites observed in the zincon competition assay, in agreement with the weakened affinity of the E37A/C66A/C67A mutant. As the internal site is in the proximity of the GTPase active site and involves the GTPase switch I motif, it is conceivable that the metal-induced inhibition of GTPase activity is due to metal binding at this site. Taken together, these data indicate that the internal site can be occupied by different metals with low micromolar affinity, causing subtle conformational changes that modulate the function of YjiA.
To further probe metal binding to the internal site, we determined the crystal structure of Zn(II)-bound E37A/C66A/C67A YjiA to 2.05 Å resolution. The overall structures of WT YjiA and E37A/C66A/C67A YjiA are nearly identical, with a Cα rmsd of 0.450 Å between the two structures (Figure S5A of the Supporting Information). While the bridging site and the surface sites are still occupied with Zn(II), the internal site lacks bound Zn(II) as indicated by both the electron density and the Zn anomalous difference Fourier map (Figure S5B–E of the Supporting Information). Instead, it appears that the side chain of F40 has moved into the space previously occupied by the side chains of C66 and E37 as well as the Zn(II) ion (Figure S5C of the Supporting Information).
The Bridging Site and the Surface Site Connect YjiA Protomers
As mentioned briefly above, the bridging site is located between two YjiA protomers on a noncrystallographic 2-fold symmetry axis, with Zn(II) coordinated by the side chains of E74 and H114 from both protomers in an approximate tetrahedral geometry (Figure 5B). Binding to this site likely disrupted the C2 symmetry of apo-WT YjiA crystals, as the bridging site would be located on a crystallographic axis in the C2 lattice. Analysis of the resulting dimer interface with the PISA server50 indicates that this interface alone is not as extensive as is typically observed in physiologically relevant dimers, with an average buried surface area of 1244 Å2 and a low complex formation significance score of 0.132 (on a scale of 0–1 with increasing relevance to complex formation in solution), not including the contributions from Zn(II). Thus, it seems unlikely that apo-YjiA would be a dimer in solution, in agreement with our analytical gel filtration chromatography results. Zn(II) binding, however, could strengthen this interface and thereby induce dimerization and oligomerization of YjiA.
All four surface Zn(II) sites are located at crystal contacts at the interfaces of YjiA protomers (Figure 5D,E). The Zn(II) ions in these sites are coordinated by H170 and/or E167 from one protomer and H187 from a protomer in the neighboring asymmetric unit, or vice versa. These sites exhibit a significant amount of disorder and high crystallographic B factors, indicating that the Zn(II) ions are not bound at high occupancy within the crystal lattice. There are also some differences in the exact coordination spheres of the surface sites, providing a further rationale for the asymmetry that led to the space group transition upon Zn(II) binding. Most likely, Zn(II) binding affects the individual protomers in a slightly different manner, thus causing breakdown of the crystallographic symmetry. It is unclear whether these sites would be occupied by Zn(II) ions in solution or whether binding to these sites occurred adventitiously, after the protein ligands were brought into proximity by crystallization. These sites, however, provide an additional explanation for the observed metal-induced oligomerization of YjiA, as they could stabilize higher-order oligomers.
Discussion
The mechanisms through which cells ensure the proper allocation and trafficking of metal ions are not well understood. The COG0523 subfamily of GTPases represents a large and diverse group of proteins, and on the basis of homology to known metallochaperones in the other G3E subfamilies and comparative genomics analysis, it was suggested that they play a role in metal trafficking or some other metal homeostasis process.6,51 This study of YjiA lends credence to this proposal. In particular, we demonstrate that YjiA can bind transition metals and provide biochemical and structural details about this interaction. Furthermore, the impact of metal binding on the GTPase activity of YjiA suggests that metal ions play a regulatory function.
YjiA belongs to subgroup 9 of the COG0523 subfamily and was predicted to bind metals via a conserved CXCC motif.17 As with the other members of this subgroup, YjiA does not have an assigned function, but there is a growing body of evidence that suggests a role for YjiA in the cellular response to carbon starvation.6,52−54 Within the genomic context, yjiA is found downstream of yjiY, a homologue of carbon starvation protein CstA, and yjiX, a small cytosolic protein of unknown function.6 These three genes are transcribed as a single transcript,53 which is believed to be under the control of one of the central regulators in the carbon catabolism pathway, cyclic AMP receptor protein (CRP).54 Furthermore, yjiA is downregulated by the noncoding RNA molecule Spot 42 that participates in a feed-forward loop to help enact catabolite repression in E. coli.52 Further studies will be needed to establish what role YjiA plays in the carbon starvation response and the function of metals in this pathway.
Here, we present a characterization of the metal binding capabilities of YjiA. In particular, our data identify a Zn(II)-binding site created by the side chains of E37, E42, and C66, the latter from the predicted C64XCC67 metal-binding motif.6,17 Solution studies of WT and mutant proteins suggest that Ni(II) and Co(II) also bind to at least a subset of the same ligands, although the coordination is likely to be different. Whether the other conserved cysteines in this motif have a functional role in binding metals or performing downstream effects remains to be determined. The relative metal affinities mirror the Irving–Williams series of small-molecule chelators for divalent metal ions,55 suggesting that the protein structure is not enforcing strong metal selectivity. Furthermore, at this time, it is not clear which metals are physiologically relevant for YjiA function. Many of the COG0523 subfamilies were linked to zinc pathways because of the presence of zinc-regulated genetic elements,6,51 and deletions of B. subtilis yeiR or E. coli yjiA sensitize the bacteria to zinc-deficient conditions.7,16 However, a similar phenotype was not observed in a ΔyjiA strain of E. coli, and binding sites for zinc metalloregulators were not detected in the genetic neighborhoods of subgroup 9.7,16 On the other hand, although nickel is required in E. coli as a cofactor for several [NiFe]-hydrogenase isoforms,56 the uptake of this metal is restricted to anaerobic growth44 and there is no evidence that expression of yjiA is similarly controlled. Finally, a requirement for cobalt ions has not been identified in E. coli, and this organism does not make its own cobalamin.57 It is possible, however, that the metal binding capabilities of YjiA are only called upon to meet the challenges of unusual growth conditions.
The internal metal-binding site is adjacent to the GTPase active site, as predicted by our nucleotide modeling studies (Figures 5C and 6). Most notably, the region involved in metal binding contains the switch I motif (Figure 3A), raising the possibility that metal binding restricts the conformational changes of this motif, an essential part of the GTPase cycle. In addition, structural homology suggests that E39 and E37 are candidates for Mg(II) coordination, and the ability to coordinate Mg(II) could be affected by the presence of metals in the internal site. Indeed, we observe that the presence of metals affects the GTPase activity of YjiA, with Zn(II), Co(II), and Ni(II) suppressing GTPase activity to different extents, and that GDP can modulate metal binding. Given that the CXCC motif is conserved in the COG0523 subfamily,6 it is likely that metal-dependent regulation of activity is also conserved. This hypothesis is consistent with a recent study of E. coli YeiR,16 although in this latter case the GTP hydrolysis was accelerated in the presence of metals. Perhaps structural differences in the GTPase domain metal-binding sites are responsible for the variations on this theme. YjiA and YeiR are highly homologous and are well conserved within the GTPase domain, including the CXCC motif and Glu42 (YjiA numbering), but Glu37 is not conserved (Figure 3A). Metal-responsive GTP hydrolysis was also observed for proteins from the HypB G3E subfamily,58−60 with the most dramatic impact observed with Zn(II). While these proteins are structurally similar to YjiA in their GTPase domain and also have a GTPase domain metal-binding site, there are some clear architectural differences. The YjiA internal metal-binding site is located in a small “pocket” of the enzyme with a solvent-exposed opening, whereas in the M. jannaschii HypB structure, the metal-binding site is located on the surface of the protein (Figure S6 of the Supporting Information).18
The GTPase activity of YjiA is comparable to those of other G3E GTPases such as E. coli HypB,58,61H. pylori HypB,59 and E. coli YeiR,16 none of which features a particularly high hydrolysis rate, suggesting that their in vivo rates may be different from the rates determined in vitro. In vitro, the isolated GTPases lack additional partner proteins and cofactors that may stimulate GTPase activity in vivo.62 For example, the addition of SlyD, a protein involved in the nickel insertion stage of [NiFe]-hydrogenase biosynthesis,42,44 enhances the GTPase activity of E. coli HypB.63 By analogy, it is possible that YjiA requires an additional factor to maximally activate GTP hydrolysis. A possible candidate is the small cytosolic protein YjiX that is cotranscribed with YjiA.53,64
In addition to the internal metal site, YjiA also binds multiple Zn(II) ions at interface sites between separate YjiA molecules, which may provide an explanation for the metal-induced oligomerization observed by analytical gel filtration chromatography. It is also possible, however, that rearrangements around the internal metal site could create interfaces for oligomerization. Large-scale metal-induced rearrangements would be masked in our crystal soaking experiments because the interfaces between YjiA molecules are already preformed in the crystal lattice. This scenario would explain how Co(II) can mediate changes in quaternary structure, given that the solution analysis indicates that only a single Co(II) ion binds to YjiA at the internal site. Furthermore, this model is consistent with the observation that mutation of two of the internal site ligands diminishes oligomerization induced by Ni(II) and Zn(II). Finally, there may be other metal sites on YjiA not identified by crystallography. A recent bioinformatics analysis of the apo-YjiA crystal structure suggested two surface-exposed sites with potential for metal binding, composed of H23, E27, and H29 as well as D52, D79, and D82, respectively.51 We do not observe bound metal in either of these two sites, and E27 and D52 do not appear to be in the correct positions for metal binding. We note, however, that binding to these sites could rely on structural rearrangements that are prohibited by the crystal lattice. Further studies will be necessary to probe the contribution of these residues to metal binding as well as the functional relevance of metal ions bound on the surface of YjiA.
Although characterization of the E37A/C66A/C67A mutant provides information about the structure and impact of the internal Zn(II) site, several observations require additional explanation. The ability of this triple mutant to bind four Zn(II) ions and to oligomerize in the presence of Zn(II), albeit to a lesser degree than WT YjiA, suggests that mutation of these residues is not sufficient to completely disrupt Zn(II) binding. This conclusion, supported by the zincon competition illustrating a decreased affinity for Zn(II), is surprising because the crystal structure depicts a Zn(II) coordination sphere comprised of E37, E42, and C66, such that mutation of two of these three protein ligands should be sufficient to stop metal binding. Structural rearrangement of this site to incorporate other residues as ligands, such as the nearby E39, could serve as a possible explanation. Such a rearrangement may not be observed in the E37A/C66A/C67A YjiA crystal structure because of the aforementioned preformed interfaces in the crystal prior to Zn(II) binding. Possible binding to a rearranged site in the E37A/C66A/C67A mutant may also explain the GTPase inhibition in the presence of Zn(II). An alternative explanation for the inhibition of the GTPase activity by Zn(II) is the involvement of additional allosteric sites, but evidence to support this latter explanation is needed.
Altogether, the results presented here provide a detailed biochemical and structural picture of the metal binding properties of a COG0523 GTPase. A connection between members of this subfamily of GTPases and intracellular metal pathways is emerging. This study supports the model in which metals play a role in regulating the proteins’ enzymatic activities and presents a glimpse into how this metal-mediated regulation may occur. Furthermore, as noted earlier, subgroup 9 of the COG0523 GTPases is hypothesized to be involved in carbon starvation, and the metal binding activities of YjiA offer a tantalizing hint about the interplay of metal homeostasis with other cellular processes.
Acknowledgments
We thank Andrew van Benschoten and Christine Phillips-Piro for their efforts toward a nucleotide-bound structure of YjiA, Prof. Andrew Woolley for the use of his spectropolarimeter, and Prof. Cynthia Goh for the use of her microplate reader. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Portions of this research were conducted at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the U.S. Department of Energy Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including Grant P41GM103393) and the National Center for Research Resources (Grant P41RR001209).
Glossary
Abbreviations
- CD
circular dichroism
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- ESI-MS
electrospray ionization mass spectrometry
- EDTA
ethylenediaminetetraacetic acid
- GuHCl
guanidinium hydrochloride
- HPLC
high-pressure liquid chromatography
- IPTG
isopropyl β-d-thiogalactopyranoside
- LMCT
ligand-to-metal charge transfer
- P-loop
phosphate-binding loop
- PAR
4-(2-pyridylazo)resorcinol
- PMSF
phenylmethanesulfonyl fluoride
- rmsd
root-mean-square deviation
- TCEP
tris(2-carboxyethyl)phosphine
- WT
wild type.
Supporting Information Available
A discussion of the space group transition in the YjiA crystals upon Zn(II) binding, tables with primer sequences, representative reconstructive ESI-MS results, summary of gel filtration chromatography results, crystallographic data collection and refinement statistics, circular dichroism spectra, and figures of the crystal structure of WT and mutant YjiA with Zn(II). This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The atomic coordinates for Zn(II)-soaked WT YjiA and Zn(II)-soaked E37A/C66A/C67A YjiA have been deposited in the PDB as entries 4IXM and 4IXN, respectively.
Author Present Address
⊥ K.S.R.: Department of Chemistry, University of British Columbia, Vancouver, BC, Canada V6T 1Z1.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (to D.B.Z.), National Institutes of Health Grant GM69857 (to C.L.D.), a Massachusetts Institute of Technology Poitras predoctoral fellowship (to M.J.), a Douglass College Science, Technology, Engineering, and Math Summer Research Experience Grant from Rutgers University, and HHMI-MIT Summer Research Experience in Chemical Biology Grant 52005719 (both to K.E.T.). C.L.D. is a Howard Hughes Medical Institute Investigator.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Clementi N.; Polacek N. (2010) Ribosome-associated GTPases: The role of RNA for GTPase activation. RNA Biol. 7, 521–527. [DOI] [PubMed] [Google Scholar]
- Verstraeten N.; Fauvart M.; Versées W.; Michiels J. (2011) The Universally Conserved Prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 75, 507–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R.; Sanders D. A.; McCormick F. (1990) The GTPase superfamily: A conserved switch for diverse cell functions. Nature 348, 125–132. [DOI] [PubMed] [Google Scholar]
- Leipe D. D.; Wolf Y. I.; Koonin E. V.; Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Saraste M.; Sibbald P. R.; Wittinghofer A. (1990) The P-loop: A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Haas C.; Rodionov D.; Kropat J.; Malasarn D.; Merchant S.; de Crecy-Lagard V. (2009) A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A.; Helmann J. D. (1998) Identification of a Zinc-Specific Metalloregulatory Protein, Zur, Controlling Zinc Transport Operons in Bacillus subtilis. J. Bacteriol. 180, 5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J.; Jochmann N.; Rodionov D.; Tauch A. (2010) The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano M.; Rubio M. Á.; Santamaría-Gómez J.; Olmedo-Verd E.; Robinson N. J.; Luque I. (2012) Characterization of the Response to Zinc Deficiency in the Cyanobacterium Anabaena sp. Strain PCC 7120. J. Bacteriol. 194, 2426–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J.; Levy-Schil S.; Cameron B.; Cauchois L.; Rigault S.; Rouyez M. C.; Blanche F.; Debussche L.; Thibaut D. (1991) Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J. Bacteriol. 173, 6074–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F.; Cameron B.; Crouzet J.; Debussche L.; Thibaut D.; Vuilhorgne M.; Leeper F. J.; Battersby A. R. (1995) Vitamin B12: How the Problem of Its Biosynthesis Was Solved. Angew. Chem., Int. Ed. 34, 383–411. [Google Scholar]
- Heldt D.; Lawrence A. D.; Lindenmeyer M.; Deery E.; Heathcote P.; Rigby S. E.; Warren M. J. (2005) Aerobic synthesis of vitamin B12: Ring contraction and cobalt chelation. Biochem. Soc. Trans. 33, 815–819. [DOI] [PubMed] [Google Scholar]
- Rodionov D. A.; Vitreschak A. G.; Mironov A. A.; Gelfand M. S. (2003) Comparative Genomics of the Vitamin B12 Metabolism and Regulation in Prokaryotes. J. Biol. Chem. 278, 41148–41159. [DOI] [PubMed] [Google Scholar]
- Lu J.; Zheng Y.; Yamagishi H.; Odaka M.; Tsujimura M.; Maeda M.; Endo I. (2003) Motif CXCC in nitrile hydratase activator is critical for NHase biogenesis in vivo. FEBS Lett. 553, 391–396. [DOI] [PubMed] [Google Scholar]
- Lee J. W.; Helmann J. (2007) Functional specialization within the Fur family of metalloregulators. BioMetals 20, 485–499. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas C. E.; Flood J. A.; de Crécy-Lagard V.; Zamble D. B. (2012) YeiR: A metal-binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics 4, 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil P. P.; Obmolova G.; Teplyakov A.; Howard A. J.; Gilliland G. L.; Camerini-Otero R. D. (2004) Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function. Proteins: Struct., Funct., Bioinf. 54, 371–374. [DOI] [PubMed] [Google Scholar]
- Gasper R.; Scrima A.; Wittinghofer A. (2006) Structural insights into HypB, a GTP-binding protein that regulates metal binding. J. Biol. Chem. 281, 27492–27502. [DOI] [PubMed] [Google Scholar]
- Zambelli B.; Turano P.; Musiani F.; Neyroz P.; Ciurli S. (2009) Zn(II)-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins: Struct., Funct., Bioinf. 74, 222–239. [DOI] [PubMed] [Google Scholar]
- Gill S. C.; von Hippel P. H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- Hunt J. B.; Neece S. H.; Ginsburg A. (1985) The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 146, 150–157. [DOI] [PubMed] [Google Scholar]
- Atanassova A.; Lam R.; Zamble D. B. (2004) A high-performance liquid chromatography method for determining transition metal content in proteins. Anal. Biochem. 335, 103–111. [DOI] [PubMed] [Google Scholar]
- Sadek F. S.; Schmid R. W.; Reilley C. N. (1959) Visual EGTA titration of calcium in the presence of magnesium. Talanta 2, 38–51. [Google Scholar]
- Tõugu V.; Karafin A.; Palumaa P. (2008) Binding of zinc(II) and copper(II) to the full-length Alzheimer’s amyloid-β peptide. J. Neurochem. 104, 1249–1259. [DOI] [PubMed] [Google Scholar]
- Kelly S. M.; Jess T. J.; Price N. C. (2005) How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139. [DOI] [PubMed] [Google Scholar]
- Louis-Jeune C.; Andrade-Navarro M. A.; Perez-Iratxeta C. (2012) Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins: Struct., Funct., Bioinf. 80, 374–381. [DOI] [PubMed] [Google Scholar]
- Lanzetta P. A.; Alvarez L. J.; Reinach P. S.; Candia O. A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100, 95–97. [DOI] [PubMed] [Google Scholar]
- Evans G.; Pettifer R. F. (2001) CHOOCH: A program for deriving anomalous-scattering factors from X-ray fluorescence spectra. J. Appl. Crystallogr. 34, 82–86. [Google Scholar]
- Stevenson C. E. M.; Mayer S. M.; Delarbre L.; Lawson D. M. (2001) Crystal annealing: Nothing to lose. J. Cryst. Growth 232, 629–637. [Google Scholar]
- Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology (Charles W., Carter J., and Sweet R. M., Eds.) pp 307–326, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L.-W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C.; Grosse-Kunstleve R. W.; Afonine P. V.; Moriarty N. W.; Zwart P. H.; Hung L.-W.; Read R. J.; Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames B.; Sousa S. F.; Tamames J.; Fernandes P. A.; Ramos M. J. (2007) Analysis of zinc-ligand bond lengths in metalloproteins: Trends and patterns. Proteins: Struct., Funct., Bioinf. 69, 466–475. [DOI] [PubMed] [Google Scholar]
- Painter J.; Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D62, 439–450. [DOI] [PubMed] [Google Scholar]
- Painter J.; Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111. [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G. W.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T.; Adams P. D.; Clore G. M.; DeLano W. L.; Gros P.; Grosse-Kunstleve R. W.; Jiang J. S.; Kuszewski J.; Nilges M.; Pannu N. S.; Read R. J.; Rice L. M.; Simonson T.; Warren G. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. III; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. (2010) MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.3rl (2010) Schrodinger, LLC, New York. [Google Scholar]
- Kaluarachchi H.; Chung K. C. C.; Zamble D. B. (2010) Microbial nickel proteins. Nat. Prod. Rep. 27, 681–694. [DOI] [PubMed] [Google Scholar]
- Zambelli B.; Musiani F.; Benini S.; Ciurli S. (2011) Chemistry of Ni2+ in Urease: Sensing, Trafficking, and Catalysis. Acc. Chem. Res. 44, 520–530. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zamble D. B. (2009) Nickel Homeostasis and Nickel Regulation: An Overview. Chem. Rev. 109, 4617–4643. [DOI] [PubMed] [Google Scholar]
- Stibrany R. T.; Fox S.; Bharadwaj P. K.; Schugar H. J.; Potenza J. A. (2005) Structural and Spectroscopic Features of Mono- and Binuclear Nickel(II) Complexes with Tetradentate N(amine)2S(thiolate)2 Ligation. Inorg. Chem. 44, 8234–8242. [DOI] [PubMed] [Google Scholar]
- Jenkins R. M.; Singleton M. L.; Almaraz E.; Reibenspies J. H.; Darensbourg M. Y. (2009) Imidazole-Containing (N3S)-Ni(II) Complexes Relating to Nickel Containing Biomolecules. Inorg. Chem. 48, 7280–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A. B. P. (1984) Inorganic Electronic Spectroscopy, 2nd ed., Vol. 33, Elsevier Science B.V., Amsterdam. [Google Scholar]
- May S. W.; Kuo J. Y. (1978) Preparation and properties of cobalt(II) rubredoxin. Biochemistry 17, 3333–3338. [DOI] [PubMed] [Google Scholar]
- Holm L.; Kääriäinen S.; Rosenström P.; Schenkel A. (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. (2007) Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Barnett J. P.; Millard A.; Ksibe A.; Scanlan D. J.; Schmid R.; Blindauer C. A. (2012) Mining genomes of cyanobacteria for elements of zinc homeostasis. Front. Microbiol. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C. L.; Storz G. (2011) The Base-Pairing RNA Spot 42 Participates in a Multioutput Feedforward Loop to Help Enact Catabolite Repression in Escherichia coli. Mol. Cell 41, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil P. P.; Camerini-Otero R. D. (2002) Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44, 89–105. [DOI] [PubMed] [Google Scholar]
- Novichkov P. S.; Laikova O. N.; Novichkova E. S.; Gelfand M. S.; Arkin A. P.; Dubchak I.; Rodionov D. A. (2010) RegPrecise: A database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving H.; Williams R. J. P. (1948) Order of stability of metal complexes. Nature 162, 746–747. [Google Scholar]
- Vignais P. M.; Billoud B. (2007) Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 107, 4206–4272. [DOI] [PubMed] [Google Scholar]
- Hausinger R., and Zamble D. (2007) Microbial Physiology of Nickel and Cobalt. In Molecular Microbiology of Heavy Metals (Nies D., and Silver S., Eds.) pp 287–320, Springer, Berlin. [Google Scholar]
- Cai F.; Ngu T.; Kaluarachchi H.; Zamble D. (2011) Relationship between the GTPase, metal-binding, and dimerization activities of E. coli HypB. J. Biol. Inorg. Chem. 16, 857–868. [DOI] [PubMed] [Google Scholar]
- Sydor A. M.; Liu J.; Zamble D. B. (2011) Effects of Metal on the Biochemical Properties of Helicobacter pylori HypB, a Maturation Factor of [NiFe]-Hydrogenase and Urease. J. Bacteriol. 193, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W.; Li H.; Yang X.; Wong K. B.; Sun H. (2012) Metallo-GTPase HypB from Helicobacter pylori and Its Interaction with Nickel Chaperone Protein HypA. J. Biol. Chem. 287, 6753–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T.; Jacobi A.; Sauter M.; Böck A. (1993) The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J. Bacteriol. 175, 630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y.; Sasaki T.; Matozaki T. (2001) Small GTP-Binding Proteins. Physiol. Rev. 81, 153–208. [DOI] [PubMed] [Google Scholar]
- Kaluarachchi H.; Zhang J. W.; Zamble D. B. (2011) Escherichia coli SlyD, More Than a Ni(II) Reservoir. Biochemistry 50, 10761–10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A.; Bren A.; Ronen M.; Itzkovitz S.; Kikoin I.; Shavit S.; Liebermeister W.; Surette M. G.; Alon U. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628. [DOI] [PubMed] [Google Scholar]
- Papadopoulos J. S.; Agarwala R. (2007) COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.