Abstract
Oligomeric complexes of Trax and Translin proteins, known as C3POs, participate in a variety of eukaryotic nucleic acid metabolism pathways including RNAi and tRNA processing. In RNAi in humans and Drosophila, C3PO activates pre-RISC by removing the passenger strand of the siRNA precursor duplex using nuclease activity present in Trax. It is not known how C3POs engage with nucleic acid substrates. Here we identify a single protein from Archaeoglobus fulgidus that assembles into an octamer with striking similarity to human C3PO. The structure in complex with duplex RNA reveals that the octamer entirely encapsulates a single thirteen base-pair RNA duplex inside a large inner cavity. Trax-like subunit catalytic sites target opposite strands of the duplex for cleavage, separated by seven base pairs. The structure provides insight into the mechanism of RNA recognition and cleavage by an archaeal C3PO-like complex.
Small RNAs such as small interfering RNAs (siRNAs), microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs), mediate gene silencing in conjunction with Argonaute proteins1-3. The effector complex of RNA interference (RNAi), mediated by siRNAs, is known as RISC (RNA-induced silencing complex) and is assembled via a stepwise process4. In the first stage, siRNA precursor duplex, composed of guide and passenger RNA strands, is loaded onto Argonaute (Ago) in a process requiring ATP5-9 and Hsp90 family chaperone proteins10-12 to form pre-RISC. For Drosophila melanogaster Ago2-RISC, but not human Ago2-RISC or Drosophila Ago1-RISC, this process also requires a Dicer enzyme6-8,13-17. The loaded orientation of the duplex in Argonaute, which determines the fates of the strands (i.e. which is guide and which is passenger), is itself determined by the relative thermodynamic stabilities of the two ends of the duplex18,19. In the second stage, when pre-RISC is converted to active RISC, the passenger strand of the duplex is nicked, or ‘sliced’, by Argonaute, mirroring target mRNA cleavage20-23. Nicking, which, for a 21 nucleotide (nt) siRNA reduces the passenger to 9 and 12 nt fragments, facilitates the removal of the passenger strand, leaving the guide exposed in an active RISC complex24-27.
Recently, in both humans and Drosophila, it has been shown that a complex of Trax and Translin proteins, designated C3PO (for component 3 promoter of RISC), enhances the removal of the passenger strand in RNAi and the formation of active RISC16,17. Trax and Translin, conserved in animals, Arabidopsis thaliana and Schizosaccharomyces pombe, participate in a variety of nucleic acid metabolism pathways, in addition to RNAi28,29. However, little is known about the precise activities and mechanisms with respect to nucleic acids of Trax, Translin and C3PO. In an important breakthrough, Liu et al.16 showed that the Trax subunit, but not Translin, possesses a Glu-Glu-Asp catalytic center with the capacity to digest RNA (and DNA). This Trax-mediated catalytic activity is required for passenger strand removal and RISC activation in RNAi16,17. It is likely that Trax directly digests the passenger strand to facilitate its removal, but it is unclear how the C3PO complex engages with the siRNA precursor duplex, whilst it is associated with Argonaute, and selects one strand, the passenger, for degradation. In vitro, human or Drosophila C3PO (h/dC3PO) is inactive towards full-length (~ 21 nt) siRNA duplex16,17, but readily cleaves single-stranded siRNA16,17,30. However, human C3PO, in vitro, is able to process a ‘nicked’ siRNA duplex, mimicking one sliced by Argonaute during the second stage of RISC assembly17.
X-ray crystal and electron microscopy structures of apo human17 and Drosophila30 C3PO have been determined. They show that C3PO is oligomeric, and that human and (full-length) Drosophila C3PO adopt octameric ball-shaped structures, strikingly similar to previous structures of homo-octameric Translin31,32. The structures17 and mass-spectrometry30 reveal a predominant ratio of 2:6 for Trax to Translin in C3PO (full-length subunits). A form of Drosophila C3PO with truncated Trax and Translin subunits adopts a hexameric bowl-shaped structure30. In the octameric full-length human C3PO crystal structure17, the two Trax subunits are side-by-side and inverted relative to one another in the octameric complex, with their Glu-Glu-Asp active sites facing into the interior of the ‘ball’. It is not known how C3PO engages with nucleic acid substrate.
We set out to provide insight into the mechanism of nucleic acid recognition by a C3PO family enzyme. Here we identify a single protein from the archaebacterium Archaeoglobus fulgidus that resembles eukaryotic Translin and Trax and has the capacity to assemble into a catalytically active homo-octamer with striking structural similarity to human C3PO. Two subunits that occupy the equivalent positions to Trax in human C3PO are structurally similar to human Trax and are responsible for the primary RNA cleavage activity of the homo-octamer. Furthermore, we show that the octamer can engage duplex RNA 14 base pairs (bp) in length fully encapsulated within the octamer. The crystal structure in complex with 14 bp duplex RNA reveals that the active sites in the two Trax-equivalent subunits target opposite strands of the RNA duplex separated by seven base pairs. Four subunits, two Trax-like and two Translin-like, provide a unique and highly conserved binding surface for the RNA inside the octamer. Supported by extensive sequence and structural conservations with eukaryotic C3POs, the structure, and our assays, provide insight into C3PO-family RNA recognition and cleavage.
RESULTS
A single Trax protein in Archaeoglobus fulgidus
We identified a single protein coded in the genome of the archaebacterium Archaeoglobus fulgidus (symbol AF2260) that resembled Translin and Trax. Notably, this protein appeared to contain the four critical glutamate or aspartate residues16,17 that define the Trax catalytic site (Supplementary Fig. 1). However, there appeared to be no other protein coded in the genome that might be a separate non-catalytic Translin partner. Sequence alignment showed a greater overall resemblance to eukaryotic Trax rather than Translin (Supplementary Fig. 1) and we designated the protein AfTrax. We found that AfTrax was stable when expressed in isolation (not shown), unlike eukaryotic Trax17,33-37. To gain insight into the mechanisms of Trax-mediated RNA cleavage, we crystallized and solved the structure of AfTrax in isolation (data to 3.27 Å) and, in a different space group, AfTrax in the presence of a 14 bp siRNA-like RNA duplex (data to 2.98 Å) (Table 1).
Table 1.
Data collection and refinement statistics
| AfC3PO Selenomethionine SAD |
AfC3PO (D114A) – RNA complex |
Apo AfC3PO (wild type) |
|
|---|---|---|---|
| Data collection | |||
| Space group | P42212 | P3121 | P42212 |
| Cell dimensions a, b, c (Å) |
182.30, 182.30, 109.39 |
183.06, 183.06, 198.12 |
183.31, 183.31, 111.28 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 93.80–3.41 (3.50– 3.41) |
123.78–2.98 (3.06–2.98) |
70.75–3.27 (3.35–3.27) |
| R merge | 0.114 (0.699) | 0.110 (0.827) | 0.077 (0.752) |
| I / σI | 16.0 (4.1) | 14.6 (3.2) | 17.5 (2.8) |
| Completeness (%) | 99.9 (99.9) | 100.0 (100.0) | 99.9 (100.0) |
| Redundancy | 14.3 (14.8) | 10.1 (10.6) | 9.2 (9.5) |
| Refinement | |||
| Resolution (Å) | 83.09–2.98 | 53.56–3.27 | |
| No. reflections | 78,400 | 29,852 | |
| Rwork / Rfree | 0.208 / 0.257 | 0.221 / 0.281 | |
| No. atoms | |||
| Protein | 18106 | 11807 | |
| RNA | 1680 | Not applicable | |
| Ligand/ion | 16 | 5 | |
| Water | 86 | 8 | |
| B-factors | |||
| Protein | 54.3 | 103.6 | |
| RNA | 79.2 | Not applicable | |
| Ligand/ion | 56.9 | 85.9 | |
| Water | 44.0 | 84.9 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.021 | 0.010 | |
| Bond angles (°) | 1.242 | 1.279 |
Values in parentheses are for highest-resolution shells. A single crystal was used for each dataset.
Structural overview and RNA binding
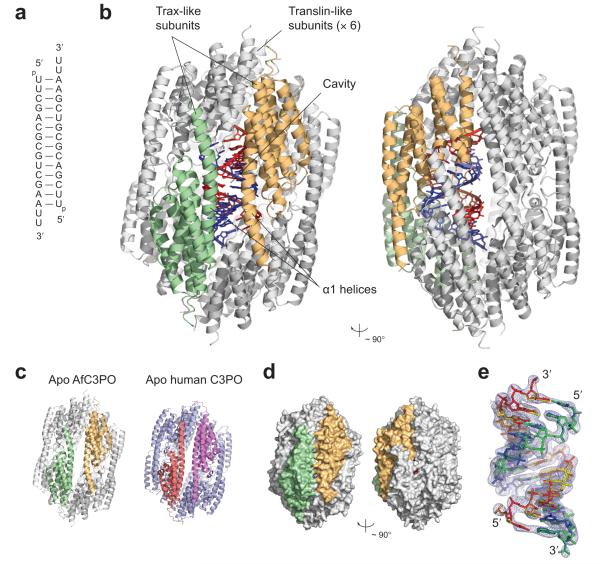
AfTrax, both in isolation and in complex with RNA, assembles into an octameric structure, resembling a cage formed by two tetramers enclosing a large inner cavity (~ 60 Å along the long axis and ~ 40 Å widest diameter) (Fig. 1a–c and Supplementary Fig. 2a). The arrangement, where each tetramer forms from four subunits related by a ~ 90° rotation and a small superhelical shift (Supplementary Fig. 2b), is similar to that observed in human C3PO17 and human32 or mouse31 homo-octameric Translin. The eight AfTrax subunits, which adopt almost identical α-helical tertiary structures (Supplementary Fig. 2c), more closely resemble human17 or Drosophila30 Trax than Translin (root mean square Cα deviation of 1.95 Å from human Trax) (Supplementary Fig. 2d, e). We analyzed AfTrax by analytical gel filtration and found that it migrated as a single peak consistent with an octamer (~ 180 kDa) (Supplementary Fig. 3a). Thus AfTrax is an archaeal Trax homologue that independently adopts a C3PO-like octameric quaternary structure.
Figure 1.
Overview of AfC3PO – duplex RNA structure. (a) Sequence and structure of the crystallized duplex. (b) ‘Front’ and ‘side’ views of the AfC3PO – RNA structure, related by a ~ 90° rotation. The two Trax-like subunits are colored green and orange, and the six Translin-like subunits are colored grey. The RNA duplex, inside the octamer, is shown in red and blue. (c) Comparison of AfC3PO (apo), colored as in panel b, with apo human C3PO (pdb code 3PJA)17, composed of two Trax subunits (salmon and violet) and six Translin subunits (blue). (d) Surface representations of the AfC3PO – RNA complex structure, colored and oriented as in panel b. (e) 2Fo – Fc electron density contoured at 1.0 σ enveloping the two 13 bp duplexes, colored red–blue or yellow–green, built overlapping in the structure. The duplexes comprise base pairs 1–13 or 2–14 of the crystallized duplex (panel a). This and other structure figures were produced using PyMOL (www.pymol.org).
Although previous studies suggest eukaryotic C3PO associates only weakly with full length (~ 19 bp) duplex siRNA17, we assessed binding of the AfTrax octamer to shorter (14 bp) siRNA-like RNA duplex. Using native electrophoretic mobility shift assays, we found that the AfTrax octamer bound 14 bp duplex RNA ~ 1,000 × more tightly than single-stranded RNA of the same length (Supplementary Fig. 3b). The octamer also bound DNA (Supplementary Fig. 3b); binding to 14 bp duplex DNA (same sequence as the RNA) was ~ 1,000 × weaker than binding to the duplex RNA (Supplementary Fig. 3b). We analyzed 14 bp duplex RNA binding by analytical gel filtration. The duplex RNA associated with AfTrax sub-stoichiometrically with respect to the AfTrax monomer (Supplementary Fig. 3a).
The X-ray crystal structure of the complex between the AfTrax octamer (mutated at catalytic aspartate 114) and the 14 bp siRNA-like RNA duplex shows that the octamer entirely encapsulates one RNA duplex (Fig. 1b, d). No part of the duplex is exposed to the outside surface of the ‘cage’. Moreover, there are no gaps between subunits large enough to accommodate a transiting duplex (Fig. 1d), indicating that the octamer must open up and then close again to incorporate the RNA. The RNA-complex subunit arrangement is almost unchanged from the apo structure. The RNA, which adopts a distorted A-form conformation (Supplementary Fig. 4), is inclined at a 24° angle to the long axis of the cage, and 8 Å away from the center, against one surface of the inner cavity (Fig. 1b). The RNA axis runs parallel to the α1 helices of two subunits (Fig. 1b), one from each of the upper and lower tetramers, which together form the most intimate contacts with the RNA. These two subunits occupy the equivalent positions in the octamer to the two Trax subunits in human C3PO17 (the ‘lowest’ positions in the rotation-and-superhelical-shift arrangements [Supplementary Fig. 2b]). We therefore designated these two subunits ‘Trax-like’, with the remaining six subunits as ‘Translin-like’ (Fig. 1b). Because of the striking composite similarity to human C3PO (e.g. Fig. 1c), we refer to the octameric form of AfTrax as AfC3PO.
Although a 14 bp duplex was used in crystallization, we observed density for only a 13 bp duplex inside AfC3PO. Therefore, we built and refined two duplexes, composed of base pairs 1–13 and 2–14, simultaneously into the electron density, each with crystallographic occupancy 0.5 (Fig. 1e). Because the 14 bp duplex is palindromic, and because the two halves of the octamer appeared to be equivalent (and are related by two-fold symmetry in one of the crystal lattices), this satisfied the symmetry requirements and provided a good match to the electron density. The pattern of FO – FC difference density between bases using a ‘minimal’ poly-uridine duplex for refinement (not shown) was consistent with this arrangement and inconsistent with either individual sequence. The figures, apart from Fig. 1e, depict just one of these (closely-overlapping) duplexes.
Positioning and detail of Trax-like subunit catalytic sites
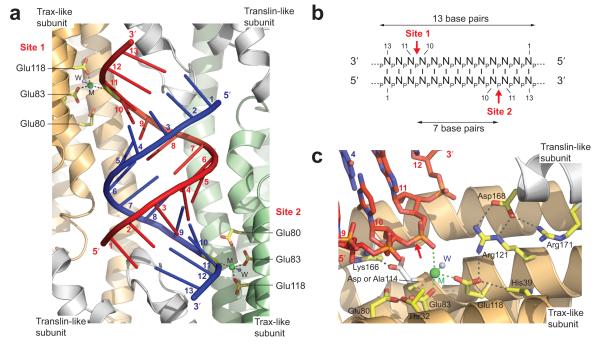
The 13 bp duplex in the octamer cavity is in close proximity to two of the AfTrax catalytic sites, located on the two Trax-like subunits (Fig. 2a). These active sites, containing metal ions (described below), target opposite strands of the RNA duplex. Measured from the 5′-end of either strand of the 13 bp visible duplex, the active sites are proximal to the phosphodiester linkages between nucleotides 10 and 11 on each strand (Fig. 2a). Thus, structurally, AfC3PO targets symmetrically opposite strands of the 13 bp RNA duplex, with an intermediate separation of seven base pairs (Fig. 2a, b).
Figure 2.
AfC3PO Trax-like subunit catalytic sites. (a) Positioning of the Trax-like subunit catalytic sites. The view shows the RNA duplex inside AfC3PO with the two RNA-proximal catalytic sites (Site 1 and Site 2), located on the Trax-like subunits. Catalytic glutamate side chains are colored yellow, metals (M) are shown as green spheres and metal-coordinated waters (W) as blue spheres. Bases on each strand are numbered 1–13 from the 5′ ends. The RNA is shown in cartoon representation for clarity. (b) Schematic representation showing the positioning of the AfC3PO Trax-like subunit catalytic sites relative to the visible RNA duplex in the structure. (c) Detailed view of a Trax-like subunit catalytic site in complex with substrate RNA. Selected side chains are shown in yellow. Catalytic Asp114, mutated to alanine in the complex with RNA, is superimposed from the apo (wild type) structure and shown in white (together with interacting Lys166). The metal (M) coordinated to the scissile phosphate is shown as a green sphere. A coordinated water molecule (W) is shown as a blue sphere. Nucleotides in the RNA (red and blue) are numbered. The putative scissile bond in the RNA is indicated by a red arrow. Parts of the subunits have been removed for visibility.
The two Trax-like subunit catalytic site regions displayed prominent, ellipsoidal, electron density between the conserved catalytic acidic residues and the proximal RNA (Supplementary Fig. 5a). Based on the density, geometry and crystallization conditions, we assigned Mg2+, octahedrally coordinated between conserved catalytic Glu83, Glu118 and the OP1 non-bridging oxygen of the phosphodiester linkage between nucleotides 10 and 11 of the RNA, with an additional coordinated water (Fig. 2c, Supplementary Fig. 5a). Previous studies17,30 showed that eukaryotic C3PO cleaves RNA generating products with 5′ phosphate and 3′-OH groups. Given the similarity to human C3PO (see below for active site comparison) and the position of the coordinated Mg2+, this suggested that the targeted scissile bond in the RNA lies between the 3′-O of nt 10 and the 5′-P of nt 11 (Fig. 2c, Supplementary Fig. 5a). In FO – FC difference density, we observed a negative peak of density over this bond in one strand (Supplementary Fig. 5b), suggesting some density depletion (data was collected after crystal growth for five weeks at 19°C), although the 2FO – FC density was entirely continuous (e.g. Supplementary Fig. 5a).
In addition to Mg2+ coordination, Glu118 interacts with Arg121 and His39 in the same subunit (Fig. 2c), both conserved in eukaryotic Trax (Supplementary Fig. 1). Interestingly, the Arg121 side chain appears to be stabilized via a bidentate interaction with Asp168 from a neighboring Translin-like subunit (Fig. 2c), notably invariantly conserved in eukaryotic Translin, but not Trax (Supplementary Fig. 1). Thus, the AfTrax monomer blends features of both eukaryotic Trax and Translin. Glu80, a third conserved catalytic glutamate, is placed adjacent to Glu83 and interacts with (non-conserved) Thr32. The fourth conserved catalytic acid residue, Asp114, was mutated to alanine in the complex with RNA, but in apo AfC3PO interacts with conserved Lys166 (Fig. 2c). Comparisons with apo human17 and Drosophila30 Trax show that the catalytic sites display very similar structural features including, where present, the positions of the four catalytic acidic side chains and side chains equivalent to His39, Arg121, Asp168 and Lys166 (Supplementary Fig. 6a, b). The similarities indicate a common catalytic mechanism.
Duplex interactions with the Tx2Tn2 subunit interface
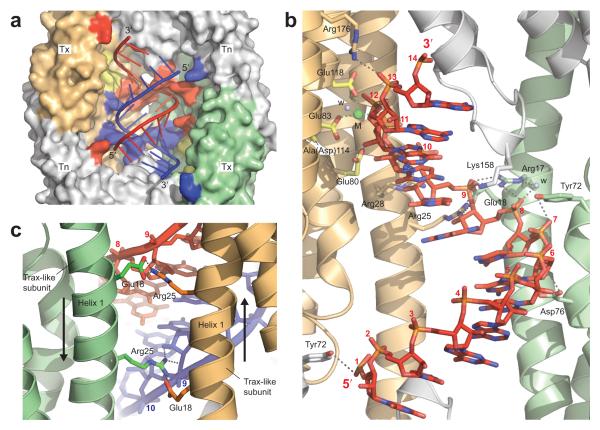
The RNA duplex beds against one wall of the inner AfC3PO cavity formed by four subunits: the two Trax-like subunits and two adjacent Translin-like subunits (Fig. 3a). We designated this face the Tx2Tn2 face. All four subunits make contributing interactions with the RNA (Fig. 3a, b). A protein surface representation (PyMOL Connolly Surface, www.pymol.org) closely enveloped the overall shape of the 13 bp duplex (Fig. 3a). Most of the interactions are with the backbone of the central part of the duplex, between nucleotides 6 and 10 of each strand (Fig. 3a, b). Residues that interact with the RNA and are conserved (Supplementary Fig. 1) include, from the Trax-like subunits, Arg17, Arg25, and Arg176, and from the Translin-like subunits, Lys158 and Arg164 (these two conserved in eukaryotic Translins but not Trax). As a result of the complex symmetry, the interactions are largely duplicated towards each strand (see Fig. 3a). Arg25 side chains from the two Trax-like subunits are particularly notable because, positioned between the α1 helices of the Trax-like subunits, they span the minor groove of the RNA duplex (Fig. 3c). Furthermore, they contact ribose 2′-hydroxyl groups (in addition to ribose 3′ oxygens), thereby participating in a dual mode of A-form RNA recognition: helical structure and ribose 2′-hydroxylation. Arg25, together with Glu18 (whose two side chains appear to stabilize the Arg25 side chains [Fig. 3c]), is invariantly conserved in eukaryotic Trax proteins (Supplementary Fig. 1). Tyr72, which interacts with the RNA from both the Trax- and Translin-like subunits (Fig. 3b), although not directly conserved, is located in the center of a motif region previously identified as critical for nucleic acid binding in both Translin and Trax (Cb-domain or MBRII)38-41.
Figure 3.
AfC3PO – duplex RNA interactions. (a) View of the RNA duplex (cartoon representation) bedded against the Tx2Tn2 surface inside AfC3PO. Trax-like subunits (Tx) are colored orange and green, and Translin-like subunits (Tn) grey. Residues that interact with the red and blue strands of the duplex are colored red and blue respectively. Catalytic residues in the Trax-like subunits are colored yellow. Opposing subunits on the opposite side of the octamer (4 × Translin-like) have been removed for visibility. (b) Detailed interactions with one strand of the duplex, viewed from inside the octamer. The interactions are largely duplicated towards the other strand (removed for clarity). (Invariantly conserved Arg164 on a Translin-like subunit makes an additional contact to the phosphate group of nt 10 of the other strand, not present in this view.) (c) View of the invariantly conserved Arg25 side chains on the Trax-like subunits bridging the RNA minor groove. The view is from the outside of the octamer.
The double-stranded-RNA specific interactions are consistent with the results of our gel shift assays, which demonstrated a strong AfC3PO affinity for 14 bp duplex RNA (Supplementary Fig. 3b). That many of these interactions involve conserved residues, within a conserved oligomeric structure, suggests that short duplex RNA recognition may be a conserved feature of C3PO enzymes. Notably, the spacing between the two catalytic Mg2+ ions in the AfC3PO Trax-like subunits (37 Å), which target opposite strands of the RNA (Fig. 2a), is almost identical to the spacing of two Mn2+ ions soaked into crystals of human C3PO17 in the two Trax catalytic sites (35 Å). This suggests a similar pattern of substrate targeting.
Because only a 13 bp duplex is visible in the structure, and the complex was co-crystallized with a 14 bp duplex (with 3′ dinucleotide overhangs), we suggest that 13 bp is the maximum base-paired length that can be accommodated within the enclosed octamer cavity. We observed no electron density for a remaining base pair at either end, even accounting for half crystallographic occupancy. Furthermore, whilst the middle 11 bp of the duplex adopt canonical Watson-Crick base pairing, the visible end base pairs do not form all the canonical Watson-Crick hydrogen bonds, and show increased separation (Supplementary Fig. 4c, d). We suggest, therefore, that the remaining base pair in the duplex is frayed open, at either end (they are equivalent) and, together with the 3′ dinucleotide overhangs, is disordered in the crystal.
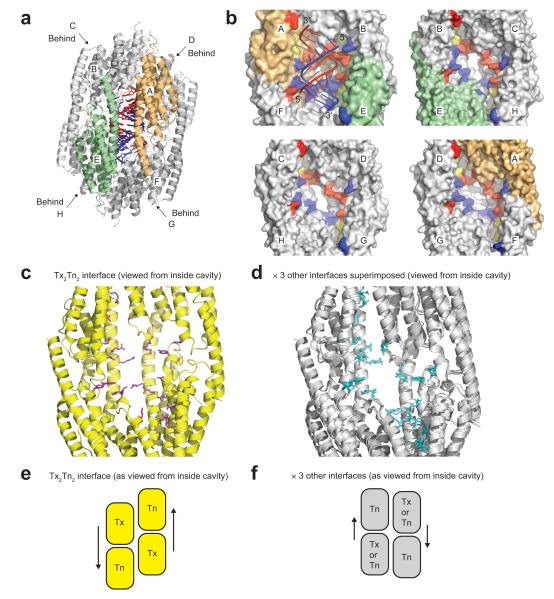
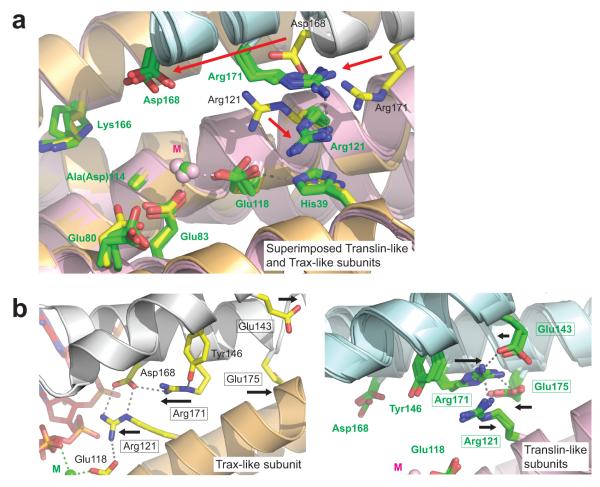
It is notable that the RNA duplex binds only one face of the inner octamer cavity, flanked by the two Trax-like subunits and two Translin-like subunits (Tx2Tn2 face), when the octamer displays quasi four-fold internal symmetry. Strikingly, if we compare the three other similar inner surfaces to the Tx2Tn2 RNA binding surface, we observe that the spatial patterning of the ‘RNA-interacting’ residues (defined from the Tx2Tn2 face) is different (but the same for the three other surfaces) (Fig. 4a–d). Thus, the Tx2Tn2 tetrameric binding surface is unique within the octamer. This is a consequence of the spiral (rotation-and-superhelical-shift) subunit arrangement within either half of the octamer (Supplementary Fig. 2b), which creates a unique subunit interface at the Tx2Tn2 junction (Fig. 4e, f). Another consequence of this arrangement is that the two Trax-like subunit active sites, which are formed at subunit interfaces, display a fundamentally different architecture to the six Translin-like subunit ‘active sites’ (Fig. 5a). In particular, invariantly conserved Arg121, which in the Trax-like subunits interacts with metal-coordinating Glu118, in the Translin-like subunits flips away, as part of a concerted inter-subunit conformational ‘switch’ (Fig. 5b). Thus, as a result of their positions in the octamer, the two Trax-like subunits are distinct and different from the six Translin-like subunits. The Tx2Tn2 subunit interface, present in human C3PO but absent from a crystallized (weakly active) hexameric form of Drosophila C3PO assembled from truncated subunits (Supplementary Fig. 6c–e), is likely to be an important catalytic sub-assembly in the C3PO octamer.
Figure 4.
The four tetrameric inner surfaces of the AfC3PO cavity. (a) Key to subunit nomenclature for panel b. Subunits A (orange) and E (green) are the Trax-like subunits, and subunits B, C, D, F, G and H (grey) are Translin-like subunits. Subunits C, D, G and H are located behind the others on the other side of the octamer. (b) Surface representations of the four tetrameric inner surfaces of the AfC3PO cavity. Subunits are colored and labeled as in panel a. The top left surface shows the tetrameric Tx2Tn2 binding surface with associated RNA, with catalytic site residues colored yellow and residues that interact with the red or blue RNA strands colored red and blue respectively (as Fig. 3a). The equivalent residues on the other three surfaces are similarly colored, but their spatial patterns differ to the Tx2Tn2 surface. The four facing subunits in each instance are removed for visibility. (c, d) RNA interacting residues (c) or their equivalents (d) on the four tetrameric inner surfaces. Subunits are colored uniformly yellow (c) or grey (d), and the four facing subunits in each view are removed for visibility. (e, f) Diagrams showing relative subunit positioning at the four tetrameric inner surfaces of AfC3PO. The diagrams correspond to the views in panels c and d. Tx and Tn refer to Trax-like and Translin-like subunits respectively. Arrows highlight relative subunit positioning.
Figure 5.
Conformational differences between AfC3PO Trax-like and Translin-like subunits. (a) Superposition of AfC3PO Trax-like and Translin-like subunit ‘catalytic sites’. The Trax-like subunit catalytic site is colored as in Fig. 2c with the RNA omitted (yellow side chains, orange and grey subunits, green metal). The Translin-like subunit ‘catalytic sites’ are colored with green side chains, pink and blue subunits, and pink metals. The superposition is of the four (independent) sites from one half of the AfC3PO octamer (one Trax-like, three Translin-like). Red arrows show notable side chains shifts, caused by altered positioning of the adjacent (grey or blue) subunits. (b) Views illustrating a concerted conformational switch adjacent to the ‘catalytic sites’ of Trax-like (left) and Translin-like (right, × 3 superimposed) subunits. Boxes highlight residues participating in the conformational switch, illustrated by black arrows.
RNA cleavage assays
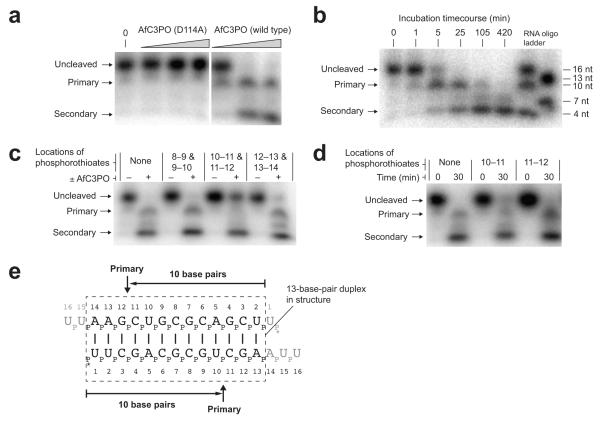
To test our structural conclusions, we performed small RNA cleavage assays. Incubation of 5′-labeled 14 bp siRNA-like duplex (sequence as crystallized) with wild type AfC3PO, but not D114A catalytic mutant AfC3PO, resulted in cleavage of the RNA (Fig. 6a). This confirmed that AfC3PO is an active, self-sufficient complex, employing the Trax-like catalytic sites. Cleavage resulted in distinct products. In a timecourse at 30°C (Fig. 6b), the first product to appear, peaking in abundance after ~ 5 min, was ~ 10 nt in length. The second major product(s) reached a peak after ~ 105 min and were short in length, ≤ ~ 5 nt. We designated the products, and the corresponding cleavage events, ‘primary’ and ‘secondary’, respectively.
Figure 6.
AfC3PO duplex RNA cleavage assays. (a) Denaturing urea-PAGE analysis of duplex RNA cleavage by increasing concentrations of D114A mutant or wild-type AfC3PO. The duplex was as crystallized (with 5′-labeling). Incubation was at 20°C for 60 min. Protein concentrations were 20 nM, 200 nM and 2 μM (AfTrax monomer concentrations), or no protein (0). Primary and secondary cleavage products are indicated. (b) Timecourse of RNA duplex cleavage (duplex as in panel a) by wild-type AfC3PO (200 nM AfTrax monomer, 30°C incubation). RNA size markers are shown at the right. (c) Cleavage of RNA substrates containing dual phosphorothioate modifications (positions between nucleotides indicated). Incubation was at 30°C for 30 min, with or without AfC3PO as indicated (200 nM AfTrax monomer). (d) Cleavage of RNA substrates containing single phosphorothioate modifications (positions between nucleotides indicated). Incubation was at 30°C for the times indicated (200 nM AfTrax monomer). (e) Schematic showing primary cleavage positions on the assay duplex. The dashed box shows a 13 bp segment as observed in the crystal structure. * indicates 5′ labeling.
To define precisely the primary cleavage position, we performed assays using RNA oligonucleotides containing phosphorothioate modifications, which can site-specifically inhibit enzymatic Mg2+-catalyzed nucleic acid cleavage42. Duplexes with modifications between nucleotides 10–11 and 11–12 were entirely blocked for primary cleavage (Fig. 6c), unlike those containing modifications either side of these positions (Fig. 6c) or individually at 10–11 or 11–12 (Fig. 6d). This is consistent with Mg2+-catalyzed cleavage of the 14 bp siRNA-like duplex positioned as observed in the crystal structure (Fig. 6e). Together, the assays and the structure indicate that the principal, most efficient, active conformation for the complex, generating primary cleavage products, is formed by an RNA duplex stably positioned at the Tx2Tn2 interface, spanning the two Trax-like subunit active sites.
Secondary cleavage occurred independently of primary cleavage (Fig. 6c) and required the AfTrax catalytic sites (Fig. 6a). In the crystal structure, none of the active sites in the octamer are positioned close to the duplex 5′ ends where they could generate short cleavage products. This implies that other conformations must also occur in solution, bringing the duplex 5′ ends in proximity with one or more active sites, to facilitate secondary cleavage. Further studies are required to understand the basis for secondary cleavage.
DISCUSSION
Here we identify a single protein from the archaebacterium Archaeoglobus fulgidus that assembles into an octameric C3PO-like complex. The structural similarities to human C3PO17, composed of two proteins, are striking. These similarities include the monomer folds, the octameric subunit arrangements (including superhelical shifts between subunits), the positions of the principal catalytic subunits, the spacing of metals coordinated in the two Trax or Trax-like catalytic sites (~ 37 Å) and the detailed structures of the principal (Trax or Trax-like) catalytic sites. Furthermore, the residues in AfC3PO involved in interaction with the RNA are well conserved in human and other eukaryotic C3POs. These similarities suggest that features of AfC3PO–RNA interactions are likely to be conserved in eukaryotes.
It is notable that, in human C3PO, catalytic activity is present only in the two subunits that in AfC3PO mediate primary cleavage of the RNA. This illustrates the importance, conserved through evolution, of the catalytic activity of these two subunits. It suggests that the AfC3PO–RNA complex reflects the core, conserved, mechanistic function of the general C3PO scaffold. Furthermore, we suggest it is possible that eukaryotic C3PO arose from a homomeric precursor, similar to AfC3PO, and over time acquired an inactive Translin subunit, for positions in the octamer where catalytic activity is unnecessary (or even detrimental). As discussed in the Results, the AfTrax monomer shares features with both Trax and Translin.
The binding and cleavage of duplex RNA is seemingly at odds with the activities of human and Drosophila C3PO, which are reported to cleave single-stranded RNAs but not siRNA precursor duplexes16,17,30. However, the AfC3PO–RNA structure reveals a crucial length-dependence for incorporation of duplex RNA inside the scaffold, less than ~ 13 or 14 bp, which is pertinent to the interpretation of the results. Longer RNA duplexes, such as siRNA precursor duplexes (~ 19 bp), would not be able to be fully accommodated inside the C3PO scaffold and must therefore bind differently (if at all). The data report on different binding modes: fully encapsulated versus non-fully encapsulated. Single-stranded RNAs, which are cleaved by human or Drosophila C3PO up to (at least) 25 nt in length16,17,30, can adopt a variety of conformations which may facilitate their cleavage by C3PO. The precise structural basis for ssRNA cleavage is unclear.
The AfC3PO–RNA structure is consistent with mutagenesis studies conducted by Ye et al. with human C3PO17. The authors mutated Lys68 and Arg200 in human Trax and Arg192 in human Translin and monitored the effects on single-stranded siRNA binding and cleavage. Mutation of Lys68 in human Trax, equivalent to Thr32 in AfTrax (Supplementary Fig. 1), severely disrupted RNA binding and cleavage. In the AfC3PO–RNA complex, Thr32 in the Trax-like subunits is located in the catalytic sites, interacting with catalytic Glu80 and in close proximity to the substrate RNA (Fig. 2c). Mutation of Arg200 in human Trax, equivalent to Arg121 in AfTrax, had a less severe effect on RNA binding but still a pronounced effect on cleavage. This is particularly interesting because we identified Arg121 as a critical residue in the AfC3PO Trax-like subunit catalytic site, bridging metal-coordinating Glu118 and conserved Asp168 from an adjacent Translin-like subunit (Fig. 2c, Supplementary Fig. 6a). Arg121 does not contact the RNA directly, but does adopt different (‘switched’) conformations in the AfC3PO Trax-like and Translin-like subunits (Fig. 5a, b). Finally, mutation of Arg192 in human Translin almost completely abolishes ss-siRNA binding and cleavage by human C3PO. This residue is equivalent to Arg164 in AfTrax (Supplementary Fig. 1), which from a Translin-like subunit in AfC3PO contacts directly a phosphate group of the RNA. Together, the mutagenesis studies conducted by Ye et al. are consistent with a similar mode of recognition towards single-stranded siRNA, by human C3PO, as observed in our duplex RNA complex.
What are the implications of the AfC3PO–RNA structure for the mechanism of RNAi activation by C3PO in humans and Drosophila? One key unresolved question is whether h/dC3PO, in degrading the siRNA passenger strand to activate RISC, binds the whole precursor duplex or just a dissociated, and isolated, passenger strand. If AfC3PO serves as a prototype, it may suggest that h/dC3PO, during RISC activation, binds, and cleaves, the duplex. This would be contrary to previous results showing that h/dC3PO is inactive against full-length precursor siRNA duplex, in vitro16,17. It is also inconsistent with the apparent length requirement (< ~ 13 or 14 bp) from AfC3PO for accommodation inside the cage (siRNA duplexes are at least ~ 19 bp). However, Ye et al. have demonstrated17 that human C3PO, in vitro, is capable of degrading the ‘passenger’ strand in a ‘nicked’ siRNA duplex (mimicking Argonaute-mediated passenger slicing) showing that, under some circumstances, human C3PO can engage and process full-length (nicked) duplex siRNA. (Nicking could perhaps provide flexibility to allow the connected duplex halves, now 9 and 10 bp, to be accommodated inside C3PO.) C3PO could also ‘open up’ to accommodate longer duplex (duplex could protrude out) in the context of Argonaute and RISC. Nonetheless, any model of RISC activation based on the AfC3PO pattern of duplex RNA cleavage would need to resolve the key issue of how C3PO processes only one strand (not both) - and the correct one - of the duplex. Further studies with eukaryotic C3PO and RISC are required to fully understand the mechanism.
AfC3PO exists in an archaebacterium with a truncated Argonaute-like protein (AfPiwi43). However, we have no evidence that they interact, or indeed participate together, in an orthologous archaeal RNAi-like pathway. C3PO in eukaryotes is not exclusively an RNAi-pathway component28 and, in Neurospora crassa, does not participate in RNAi29. Therefore, we do not anticipate that C3POs in general will display RNAi-specific properties, such as measurement of 19 bp duplex or recognition of a 3′ dinucleotide overhangs, but will rather provide versatile platforms for the processing of nucleic acids. The structure of AfC3PO in complex with duplex RNA, coupled to the extensive sequence and structure conservation with eukaryotic C3POs, provides a starting point for understanding the molecular mechanisms employed by C3PO family members.
ONLINE METHODS
Cloning, expression and purification
The AfTrax coding sequence was PCR amplified from Archaeoglobus fulgidus genomic DNA (DSMZ) and cloned into a modified version of pET-17b (Novagen) containing an N-terminal 6 × histidine tag and an intervening Prescission protease (GE Healthcare) cleavage site (vector pTwo-E, a gift from T. Oliver, University of Sussex, UK). Expression was carried out in E. coli BL21(DE3) cells induced with 1 mM IPTG for 16 h at 16°C. The protein was purified using a heat shock step (65°C for 20 min - the protein is from a thermophile) followed by Nickel-affinity chromatography, Prescission protease cleavage and Superdex 200 gel filtration (GE Healthcare) with an in-line GST-trap column (GE Healthcare). The protein was eluted from the gel filtration column in 10 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 4 mM DTT. D114A mutant AfTrax was created using a QuikChange II site-directed mutagenesis kit (Agilent Technologies), sequence verified and expressed and purified as wild type except eluted at the final stage in 5 mM Hepes pH 7.5, 200 mM NaCl, 2 mM MgCl2 and 5 mM DTT. Selenomethionine-substituted protein was produced by expressing in B834(DE3)pLysS cells grown in minimal media supplemented with selenomethionine (Molecular Dimensions) and purifying as wild type with elution from the gel filtration column in 10 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA and 10 mM DTT. Proteins were concentrated and stored in aliquots at −70°C.
Crystallization
Native and selenomethionine-substituted crystals in the P42212 crystal form were grown using the hanging drop method by mixing 1 μl of protein at 337 μM with 1 μl of 9.5–10% PEG 3K, 100 mM KCl, 200 mM MgCl2, 40 mM sodium cacodylate pH 5.5, 5 mM DTT and incubating over a 600 μl reservoir for 3–5 days at 19°C. Crystals were cryoprotected by dipping quickly into a similar solution supplemented with 17–19% MPD and frozen by plunging into liquid nitrogen.
For complex formation, RNA was ordered desalted and deprotected from Dharmacon, solubilized in 10 mM Hepes pH 7.5, 100 mM NaCl and 2 mM MgCl2, and annealed by incubation at 90°C for 1 min followed by gradual cooling from 65°C to 4°C at 1°C per 5 min. To form the complex, D114A mutant AfTrax at 752 μM was incubated with RNA at 193 μM (i.e. a ratio of 1:0.26) for 30 min at 20°C followed by 30 min at 4°C. Crystals were grown by the hanging drop method by mixing 1 μl of complex with 1 μl of 2.7 M 1,6-hexanediol, 200 mM ammonium chloride, 10 mM MgCl2, 50 mM Hepes pH 8.0, 5 mM DTT and incubating at 19°C for 5 weeks. Crystals were frozen by plunging directly (in a loop) into liquid nitrogen.
X-ray data collection and processing
Selenomethionine SAD data were collected from a single crystal on beamline I02 at Diamond Light Source, UK (DLS) at 100 K and wavelength 0.97980 Å (0.3° increments). Native RNA-complex data were collected from a single crystal on beamline I03 at DLS at 100 K and wavelength 0.97630 Å (0.15° increments). Native apo data were collected from a single crystal on beamline I04 at DLS at 100 K and wavelength 0.97630 Å (0.15° increments). In each instance, data were integrated, scaled and merged using xia2 (-3diir option)44 employing XDS45 and Scala46,47.
Structure determination and refinement
The structure was solved using selenomethionine SAD data for the P42212 crystal form, collected at the peak wavelength (0.97980 Å). ~ 48 selenium sites (six per protein chain) were located using SnB48,49 and then refined and used for phasing in autoSHARP50. The presence of eight-fold symmetry in the refined heavy atom sites (× 46) was an indication of a correct solution. Density modification exploiting the eight-fold symmetry was performed running Phenix AutoBuild51 in ‘maps_only’ mode and resulted in an interpretable density map. A preliminary model was built into the SAD density manually using Coot52 and refined using phenix.refine51 (Rw / Rfree = 0.35 / 0.38).
The preliminary model was used to solve the P3121 RNA-complex crystal form with a higher resolution dataset (2.98 Å) by molecular replacement using Phaser53. Successive rounds of model building in Coot and refinement using phenix.refine resulted in a final model (Rw / Rfree = 0.208 / 0.257) comprising twelve protein chains built from residues 1 to 187–191 (out of 196), three RNA chains each composed of two overlapping conformers of thirteen full nucleotides (discussed below), coordinated magnesium ions at the twelve catalytic sites and a number of waters, mainly involved in magnesium coordination. Non-crystallographic symmetry (ncs) restraints, secondary structure restraints, metal coordination restraints (from phenix.metal_coordination51) and Watson-Crick hydrogen bonding restraints were employed. Early on in refinement, rigid body refinement and simulated annealing in phenix.refine were used. An anomalous difference map created using the SAD dataset (re-solved with Phaser) was used to confirm and position the locations of the methionine side chains. The magnesiums at the catalytic sites were assigned based on the crystallization conditions, the results of activity assays, the electron density and the coordination geometries.
Early on in the refinement it was clear that thirteen RNA base pairs were visible in the density, whereas the crystallized RNA comprised fourteen (palindromic) base pairs. Because one of the octamer complexes in the lattice was generated by a two-fold crystallographic rotation, and because there was no clear reason why one half of the octamer should preferentially accommodate a frayed end base pair, it was deduced that two duplexes with base pairs 1–13 or 2–14 (measured from either end - the same at both ends as the duplex is palindromic) were present with equal occupancy (0.5). The conclusion was supported by the pattern of FO – FC difference density present along a refined ‘minimal’ poly-U duplex (not shown). The overlapping RNAs were constructed as separate A and B conformers of the same chains (occupancy = 0.5), which are treated independently by phenix.refine. The appropriate refinement qualifiers were used to ensure that there were no incorrect interpreted ‘clashes’ derived from ‘impossible’ duplex conformer scenarios. The resultant superimposed duplexes overlap closely, with nearly identical backbones, exhibit canonical Watson-Crick base pairing (apart from the two end base pairs) and provide a good match to the density.
The native apo structure in the P42212 crystal form was solved by molecular replacement with Phaser using a tetramer from the refined complex structure. The structure was completed through successive rounds of refinement using phenix.refine (incorporating rigid body refinement, ncs, secondary-structure and metal-coordination restraints) and manual adjustment in Coot (final Rw / Rfree = 0.221 / 0.281).
Refinement statistics are given in Table 1. For the complex, Ramachandran statistics (from phenix.refine) are: favored / allowed = 92.7% / 5.7%. For the apo structure, Ramachandran statistics are: favored / allowed = 88.8% / 9.6%.
The structure-based alignment in Supplementary Fig. 1 was formatted using ALINE54.
Analytical gel filtration
Complexes were mixed and incubated in gel filtration buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM MgCl2, 2 mM DTT) for 10 min at 20°C prior to loading. Samples were analyzed at 4°C on a Superdex 200 10/300 GL column (GE Healthcare).
Oligonucleotide labeling and annealing for assays
RNA oligonucleotides (standard and phosphorothioate-modified) were obtained desalted and 2′-deprotected from Dharmacon. DNA oligonucleotides were obtained from ATDBio. Oligonucleotides (oligos) were resuspended in 10 mM Hepes pH 7.5, 100 mM NaCl, 2 mM MgCl2, 5′-labeled using [γ-32P]ATP and T4 PNK (NEB), and purified on MicroSpin G-25 columns (GE Healthcare) equilibrated in the resuspension buffer. For annealing, oligos were treated at 90°C for 1 min followed by slow cooling from 65°C in a water bath and then a beaker of water in an ice bath.
Native gel mobility shift assays
D114A mutant AfC3PO and 5′-labeled RNA or DNA oligos were incubated in 50 mM Hepes pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM DTT, 50 ng μl−1 poly(dI-dC) (Sigma) for 1–2 h at 20°C. 10% glycerol was added to the samples which were then analyzed on 5% native PAGE gels in 1 × TBE buffer.
RNA cleavage assays
AfC3PO (wild type or D114A mutant) and 5′-labeled oligos were incubated together at 20°C or 30°C in 1 × Taq polymerase reaction buffer (NEB) supplemented with 5 mM MgCl2. Incubation times, temperatures and protein concentrations are given in the Fig. 6 legend. To stop the reactions, samples were supplemented with 52% formamide, 10 mM EDTA, 0.01% SDS. For gel analysis, samples were heated at 90°C for 5 min and run on 20% acrylamide, 7 M urea gels in 0.5 × TBE at 65°C (to prevent oligo annealing).
Supplementary Material
ACKNOWLEDGEMENTS
We thank staff at Diamond Light Source, UK for help with data collection. This work was funded by an MRC Career Development Award to J.S.P. (grant reference G0600097).
Footnotes
Accession codes. Protein Data Bank: The structure factors and coordinates for the AfC3PO–RNA complex and apo AfC3PO have been deposited with accession codes 3zc0 and 3zc1 respectively.
Note: Supplementary Information is available on the Nature Structural and Molecular Biology website.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
METHODS Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
REFERENCES
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 3.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368–76. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–21. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 6.Tomari Y, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–41. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 7.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 8.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–60. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 9.Yoda M, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasaki S, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–9. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024–6. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- 12.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–9. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–20. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–3. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye X, et al. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650–7. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 19.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–48. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 23.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–20. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–13. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–40. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–74. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elkayam E, et al. The Structure of Human Argonaute-2 in Complex with miR-20a. Cell. 2012 doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaendling A, McFarlane RJ. Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore. Biochem J. 2010;429:225–34. doi: 10.1042/BJ20100273. [DOI] [PubMed] [Google Scholar]
- 29.Li L, et al. The translin-TRAX complex (C3PO) is a ribonuclease in tRNA processing. Nat Struct Mol Biol. 2012;19:824–30. doi: 10.1038/nsmb.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y, et al. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat Struct Mol Biol. 2011;18:658–64. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pascal JM, Hart PJ, Hecht NB, Robertus JD. Crystal structure of TB-RBP, a novel RNA-binding and regulating protein. J Mol Biol. 2002;319:1049–57. doi: 10.1016/S0022-2836(02)00364-9. [DOI] [PubMed] [Google Scholar]
- 32.Sugiura I, et al. Structure of human translin at 2.2 Å resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:674–9. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
- 33.Chennathukuzhi V, et al. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol. 2003;23:6419–34. doi: 10.1128/MCB.23.18.6419-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang S, et al. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J Biol Chem. 2004;279:12605–14. doi: 10.1074/jbc.M313133200. [DOI] [PubMed] [Google Scholar]
- 35.Gupta GD, et al. Co-expressed recombinant human Translin-Trax complex binds DNA. FEBS Lett. 2005;579:3141–6. doi: 10.1016/j.febslet.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Claussen M, Koch R, Jin ZY, Suter B. Functional characterization of Drosophila Translin and Trax. Genetics. 2006;174:1337–47. doi: 10.1534/genetics.106.063016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaendling A, Ramayah S, Pryce DW, McFarlane RJ. Functional characterisation of the Schizosaccharomyces pombe homologue of the leukaemia-associated translocation breakpoint binding protein translin and its binding partner, TRAX. Biochim Biophys Acta. 2008;1783:203–13. doi: 10.1016/j.bbamcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Aoki K, Suzuki K, Ishida R, Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–6. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 39.Chennathukuzhi VM, Kurihara Y, Bray JD, Hecht NB. Trax (translin-associated factor X), a primarily cytoplasmic protein, inhibits the binding of TB-RBP (translin) to RNA. J Biol Chem. 2001;276:13256–63. doi: 10.1074/jbc.M009707200. [DOI] [PubMed] [Google Scholar]
- 40.Eliahoo E, et al. Mapping of interaction sites of the Schizosaccharomyces pombe protein Translin with nucleic acids and proteins: a combined molecular genetics and bioinformatics study. Nucleic Acids Res. 2010;38:2975–89. doi: 10.1093/nar/gkp1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta GD, Kumar V. Identification of nucleic acid binding sites on translin-associated factor X (TRAX) protein. PLoS One. 2012;7:e33035. doi: 10.1371/journal.pone.0033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heidenreich O, Pieken W, Eckstein F. Chemically modified RNA: approaches and applications. FASEB J. 1993;7:90–6. doi: 10.1096/fasebj.7.1.7678566. [DOI] [PubMed] [Google Scholar]
- 43.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. Embo J. 2004;23:4727–37. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–90. [Google Scholar]
- 45.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–92. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Blessing RH, Smith GD. Difference structure-factor normalization for heavy-atom or anomalous-scattering substructure determinations. J Appl Cryst. 1999 [Google Scholar]
- 49.Weeks CM, Miller R. The design and implementation of SnB v2.0. J Appl Cryst. 1999;32:120–4. [Google Scholar]
- 50.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–30. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bond CS, Schuttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–2. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.