Abstract
The RNA genome of retroviruses is encased within a protein capsid. To gather insight into the assembly and function of this capsid, we used electron cryotomography to image human immunodeficiency virus (HIV) and equine infectious anemia virus (EIAV) particles. While the majority of viral cores appeared closed, a variety of unclosed structures including rolled sheets, extra flaps, and cores with holes in the tip were also seen. Simulations of nonequilibrium growth of elastic sheets recapitulated each of these aberrations and further predicted the occasional presence of seams, for which tentative evidence was also found within the cryotomograms. To test the integrity of viral capsids in vivo, we observed that ∼25% of cytoplasmic HIV complexes captured by TRIM5α had holes large enough to allow internal green fluorescent protein (GFP) molecules to escape. Together, these findings suggest that HIV assembly at least sometimes involves the union in space of two edges of a curling sheet and results in a substantial number of unclosed forms.
Keywords: HIV-1, capsid, cryotomography, retroviruses
Introduction
Retroviruses bud from the cell as immature, noninfectious virions and then become infectious through a process of maturation. During viral maturation, the structural polyprotein Gag is cleaved in five positions by the viral protease, producing the MA, CA, and NC proteins,1,2 as well as several smaller peptides (SP1, SP2, and p6). While the myristoylated MA remains associated with the membrane, conformational changes in CA promote its assembly into a new shell (the capsid) that forms around NC, the RNA genome, reverse transcriptase (RT), integrase, and Vpr to form the “core” of the maturing virion.2–6 The HIV capsid is crucial for the delivery of the RNA genome to a newly infected cell, interactions with host cell restriction factors, reverse transcription,7–9 and transport to the nucleus10,11 and is therefore a potential drug target.12 The HIV capsid is a difficult structure to study, however, owing to its heterogeneity and instability during purification.
Direct imaging of in vitro CA assemblies (tubes, planar sheets, and cones) by electron microscopy (EM) led to the model that capsids are fullerene cones composed of CA hexamers plus 12 irregularly positioned pentamers that close the shell and give rise to the variety of three-dimensional (3-D) shapes seen.13–17 The cone shape seems to be important, since mutations that decreased the frequency of cones also decreased infectivity.12–14,18 Crystal structures have provided high-resolution details of the CA domains and the interactions that drive hexamer and pentamer formation,4,5,17,19 but it remains unclear how the various capsid shapes assemble and whether they are completely closed.
It was noted more than a decade ago that conical cores exhibit a fairly consistent cone angle and that the ribonucleoprotein (RNP) often appears concentrated in the broad base.20,21 Since then, two “end-to-end” models for capsid assembly have been proposed. In 2005, Benjamin et al. studied HIV particles using ECT, observing similarities in the size and shape of the wide end of the capsid, uniform positioning of the wide end 11 nm away from the envelope, a cone angle of 18–24° around the long axis, a frequent hole at the tip of the narrow end, and the RNP density situated inside the capsid toward the wide end.22 These observations led to the proposal that capsid assembly nucleated around the RNP density at the wide end and then extended toward the narrow end, sometimes without completely closing. In 2006, Briggs et al. confirmed these results, also reporting a cone angle of 19° around the long axis and the presence of the RNP density in the broad end.23 Noting that cores always spanned the full distance across the typically spherical membrane envelope, however, they proposed that capsid assembly begins at the narrow end and grows across the viral particle until it reaches the membrane on the opposite side, which then redirects growth toward closure of the wide end.
A third model has emerged from simulations. Early capsid-growth simulations focused on highly symmetric, spherical capsids and showed that equilibrium theories allowing switching between hexamers and pentamers can produce well-formed icosahedra.24 In an effort to understand non-icosahedral retroviruses, both Hicks and Henley and Levandovsky and Zandi tested nonequilibrium models in which once a pentamer or hexamer was formed, its polymeric nature was fixed.25,26 Levandovsky and Zandi used tapered triangular prisms to represent CA units and were able to recapitulate spherical, conical, and tubular shapes.26 Tapering caused growing sheets to curve, and as the curved sheets grew, inclusion of pentamers became necessary to relieve accumulated stress. Opposing edges of the growing sheets eventually curled around toward each other and connected and then the top and bottom ends sealed. Conical shapes therefore emerged not as a result of template interactions or membrane enclosures but through simple nonequilibrium growth of elastic sheets.
Struck by the interesting correspondence between some of the simulated structures and real viral cores, here, we have collected additional and higher-resolution cryotomograms of intact viruses and purified cores and compared them to the full range of structures produced by simulation. We find that, in addition to closed cones, spheres, and cylinders, simulations of the nonequilibrium growth of elastic sheets predict various unclosed structures including rolls and curved sheets as well as cores with tip holes and seams. Rolls and extra curved sheets are clearly seen in cryotomograms of purified virions, and more tentative but still interesting evidence of tip holes and seams is also found. Light microscopy is used to confirm that a substantial fraction (∼25%) of cytoplasmic viral cores have openings big enough to allow GFP to escape. Taken together, these findings suggest that capsid assembly sometimes involves the union in space of the edges of a curling sheet and that a substantial number of unclosed structures are produced.
Results
Simulations of capsid growth
Simulations were used to explore possible capsid assembly pathways as in Ref. 26. CA proteins were modeled as triangular prism-shaped 3-D “subunits” (Fig. 1a). Compressible spring networks were used to reflect CA's ability to occupy different, “non-equivalent” positions along the curved body of cones. To introduce curvature, we tapered the subunits so that the “top” face was larger than the “bottom”. During assembly, subunits were added one by one to a growing sheet and the spring network was allowed to relax somewhat before the next subunit was added (Fig. 1b). Line energies were used to direct the insertion of each new subunit into the position where it would have the most inter-molecular contacts. Stochasticity was introduced by choosing where the next subunit would join randomly from among equal positions along the edge of the growing sheet. As the curved sheet grew, strain within the network increased until it became energetically more favorable to close a pentameric ring than squeeze in a sixth subunit to form a hexamer (Fig. 1c).
Fig. 1.
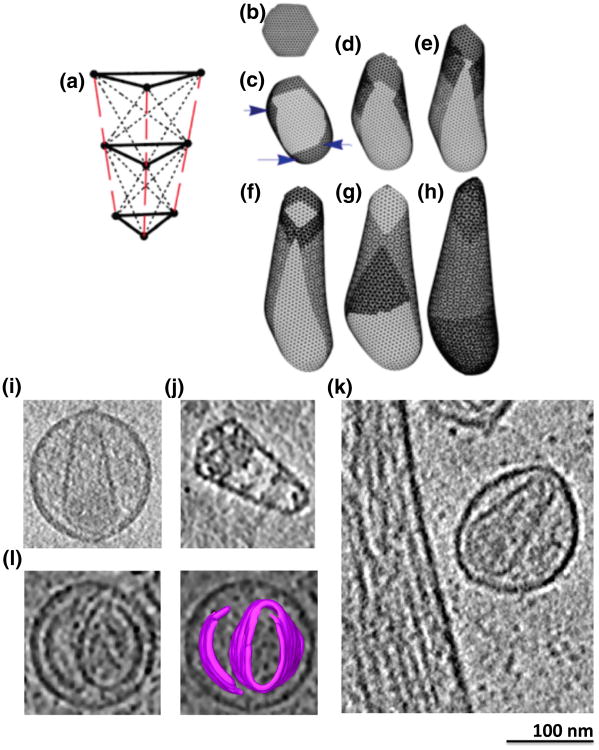
Nonequilibrium growth of elastic sheet simulations and cryotomographic reconstructions of viral samples. (a) Nine-point spring networks were used in the simulation to model CA proteins as compressible triangular prisms. (b–h) Snapshots of sheet growth and capsid completion. Prisms are added one by one to the edge where the most contacts will be formed, and then strain in the spring network is allowed to redistribute after each addition (b). Because the prisms are angled, eventually, insertion of a pentamer into the curving sheet becomes favored [blue arrows in (c) ]. Later, sheets curl over on themselves and the two edges often meet (f), followed by tip closure (h). [ (a)–(h) are reproduced from Levandovsky and Zandi26]. (i–k) Tomographic slices through a purified HIV-1 particle (i), a purified EIAV core (j), and an unpurified HIV-1 viral particle frozen next to an infected cell (k) showing typical cone-shaped cores with nucleoprotein density concentrated near the base. (l) Tomographic slice through a viral particle exhibiting an extra capsid sheet (left panel), with segmentation overlaid (right) for clarity.
The process of assembly was chosen to be irreversible in that once a pentamer or hexamer formed, it could no longer open up to add or release a subunit. In later stages of growth, the elastic sheet comprised a considerable number of hexamers and a few pentamers and curled until the edges met (Fig. 1d–f). Finally, the tops and bottoms of the resultant structures filled in even though the energetic costs of sealing the caps was high (in terms of compression and stretching of subunits) compared to building the lateral walls (Fig. 1g and h and Supplementary Movie 1).
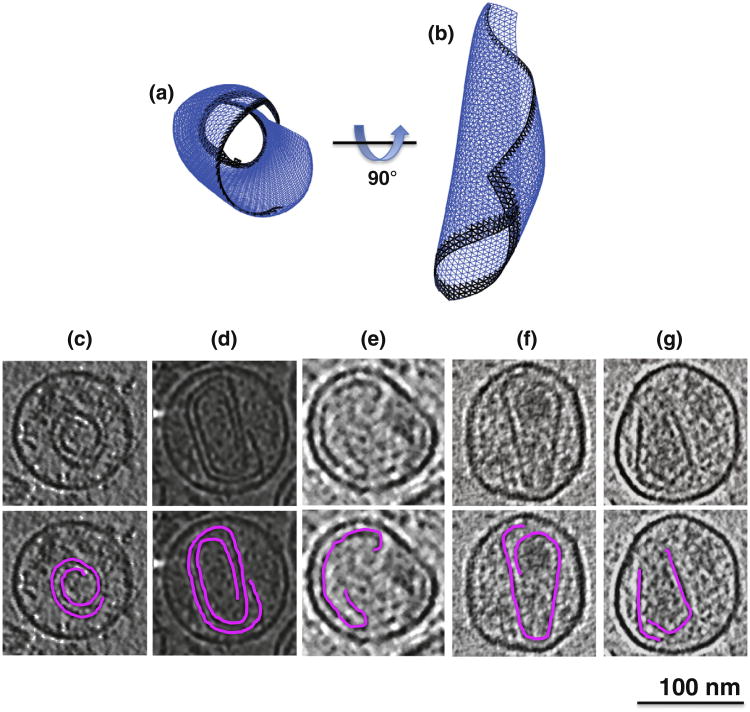
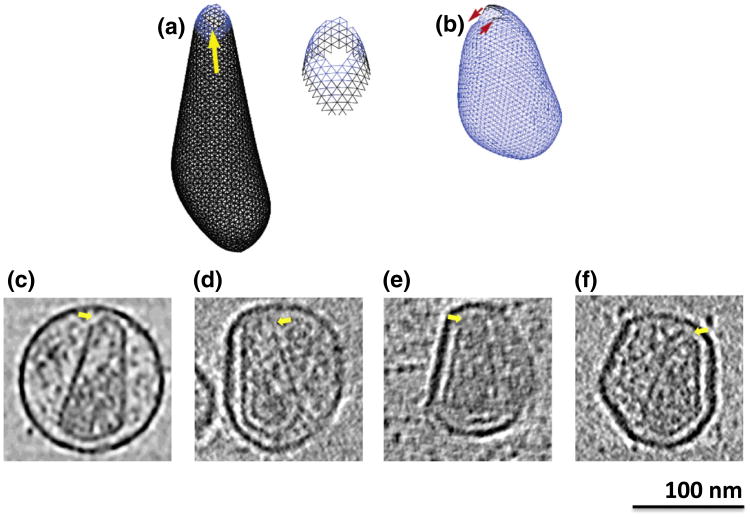
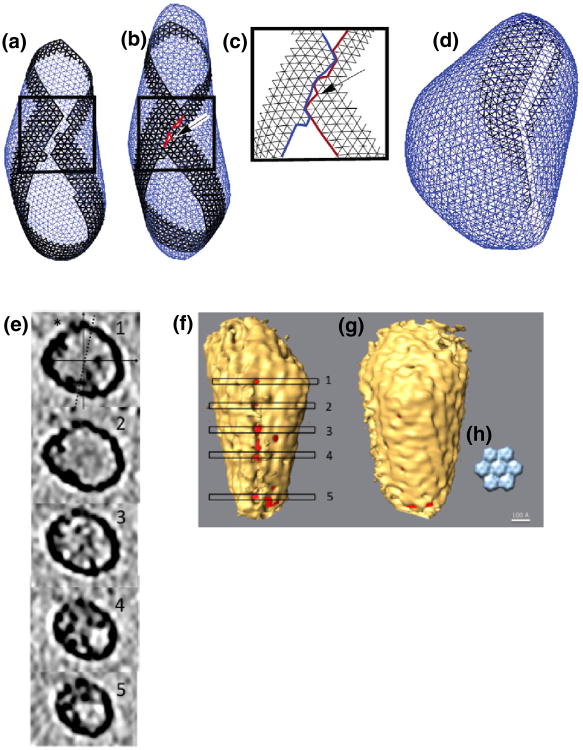
As reported in Ref. 26, depending on spring lengths and constants in the model, spherical, tubular, or conical structures form, recapitulating the various shapes exhibited by retroviral capsids. Here, we further report that unclosed structures were also frequently observed. In some cases, the edges of the curling sheet failed to unite in space, producing rolled structures (Fig. 2a). As mentioned above, high curvature tips always closed last, and in some cases, different growth rates around the edge of the closing structure led to mismatches at the tip (Fig. 3a and b). Finally, when two edges of the sheet met in the simulations, a line of defects (holes) sometimes formed (Fig. 4a–d and Supplementary Movie 2). For example, merging edges in Fig. 4a are not smooth but have small protrusions leading to the formation of a seam in Fig. 4b.
Fig. 2.
Rolled sheets. (a) Example simulated structure where the curling edges of the growing sheet failed to meet, resulting in a roll. (c–g) Tomographic slices alone (upper panels) and with segmentations overlaid (lower panels) through purified (c and d) and unpurified (e–g) HIV-1 particles exhibiting similarly rolled capsid structures.
Fig. 3.
Tip holes. (a) Example simulated structure illustrating how nonequilibrium growth can lead to tips with hard-to-close strained gaps with enlarged view of tip (right). (b) Second example illustrating how uneven growth can lead to a tip closure defect. (c–f) Tomographic slices through purified (c) and unpurified (d–f) HIV-1 particles exhibiting similarly open tips (yellow arrows).
Fig. 4.
Seams. (a and b) Before and after snapshots of a simulation where misalignment of the curling edges prevented complete closure. (c) Enlarged view of final seam. (d) Second example of a long seam of high-strain junctions. (e) Tomographic slices through an isolated EIAV core perpendicular to a putative seam. The asterisk indicates the location of the various holes along the seam. The continuous lines show the x-axis and the y-axis, and the broken line shows the tilt axis. The perpendicular orientation of the core relative to the beam and tilt axis argues that the seam is unlikely to be an artifact of the missing wedge. (f) Isosurface of the core with the seam facing the front and with the locations of the tomographic slices shown in (e) marked. The interior of the core is colored red to aid visualization. (g) Backside of the same capsid showing an otherwise mostly complete shell, including a putative pentamer (in center between levels 1 and 2). (h) Low-pass-filtered crystal structure of a CA hexamer for comparison.
Electron cryotomography
ECT is the highest-resolution imaging modality that can be used to produce 3-D reconstructions of unique biological objects such as retroviral capsids in a near-native state.27 Simulated structures were therefore compared to cryotomograms of 68 retroviral cores of three different types: (1) purified HIV-1 virions, (2) unpurified HIV-1 virions frozen next to the infected cells that produced them, and (3) purified EIAV cores. For safety, the purified HIV-1 particles were made noninfectious with single amino acid mutations in RT and RNase, but the remaining genes were wild type. Similarly, the unpurified particles were generated from a viral construct bearing a 500-bp deletion in the envelope gene. To reduce sample ice thickness and remove surrounding densities, we also imaged purified cores. EIAV cores were used because they are more stable and easier to purify than their HIV-1 analogs. Although there are subtle differences in the crystal structures of EIAV CA and HIV-1 CA, both have the same overall fold, contain the highly conserved major homology region, and assemble into capsids of very similar structure.28,29 It is thought therefore that EIAV cores are good models of HIV.30 As seen before, most cores exhibited conical, cylindrical, or “pleomorphic” shapes (among all particles, 44 were cones, 15 were cylinders, and 9 were pleomorphic; Fig. 1i–k). In addition to these well-described morphologies and in support of the idea that assembly involves the growth of curved sheets, three of the intact virions exhibited extra (presumably capsid) sheets next to the more complete cores (Fig. 1l, Supplementary Fig. 1, and Supplementary Movie 3). The extra sheets adopted curvatures similar to the cores or membranes next to them but were apparently unconnected. Such extra sheets are perhaps to be expected, since there are likely ∼3500 CA monomers in a full immature shell and only 1000–1500 are required to make a typical cone-shaped capsid. (The exact numbers of CA monomers in immature shells is uncertain: the size of immature particles and the 8-nm hexagonal Gag lattice could allow up to ∼5000 monomers,22 but tomography of immature particles revealed that the Gag lattice is incomplete 31; thus, the actual number of CA monomers is lower.32,33)
Three unclosed “rolls” without caps at the top or bottom were also clearly seen, which strongly resembled the rolls produced in the simulations (Fig. 2c–e and Supplementary Movie 4). Similarly, two cone-shaped capsids (Fig. 2f and g) also demonstrated mismatched edges at the capsid base, a feature consistent with the initiation of a rolled sheet. Weaker but still interesting evidence was also found of holes at the tips in many capsids (Fig. 3c–f and Supplementary Movie 5), though the exact number and extent could not be determined confidently due to their smaller nature and noise in the cryotomograms. The presence of holes is further supported by Benjamin et al., who aligned 16 conical cores at their narrow tips as well as possible and averaged.22 While the lateral walls of the capsid remained clear, there was a conspicuous absence of density at the tip where the alignment should have been best. This suggests that tips frequently (perhaps usually) remain unclosed.
Finally, tentative evidence of seams was found here in both purified HIV-1 virions and EIAV cores (the HIV-infected cell samples were excluded from this analysis due to their increased thickness). Isosurfaces of these cores were generated by thresholding the density of tomographic data to a level where the shape of the capsid was most clearly apparent. Typically, hexamers were discernable over parts of the surface (Fig. 4f and g), confirming the expected lattice structure of mature cores.13,17,34 When the threshold was lowered further, however, holes would usually appear uniformly over the entire surface, indicating that there were no special surfaces with lower density. In a subset of cases, however, the first holes appeared in a strikingly seam like row as the threshold was lowered. Figure 4e shows tomographic slices though one EIAV core with density missing along a line parallel with the z-axis. Figure 4f shows the isosurface of that capsid and the positions of the planes depicted in cross-sections in Fig. 4e. The seam of holes is easily detected at this threshold while the rest of the capsid is almost completely closed, exhibiting ridges that correspond to the hexameric CA lattice (backside shown in Fig. 4g). The inset (Fig. 4h) shows the low-pass-filtered, mature HIV CA hexagonal lattice as determined by electron crystallography.17 The individual CA hexamers have a diameter of 8nm, which is consistent with the individual bumps seen on these capsid segmentations.
While rows of CA hexamers therefore appear to have been resolved, whether the seams were real and their frequency was still difficult to ascertain because the holes emerged in a continuum of different configurations, ranging from randomly positioned to approximately straight rows (Supplementary Fig. 2). Nevertheless, among the 37 HIV-1 virions, 8 appeared to have seams, and among the 22 EIAV cores, 10 appeared to have seams. More seams may have been detected in the EIAV cores because their images had better signal-to-noise ratios because the samples were thinner. Putative seams were observed in many orientations with respect to the missing wedge, arguing against their being an artifact of the limited tilt range of electron microscope samples.35 Furthermore, the seams in the EIAV cores were probably not purification artifacts because seams were also observed in the intact HIV-1 virions.
Fluorescence imaging
To confirm that the closure defects seen in the cryotomograms were real and relevant to cellular infection, we developed a system in which GFP is initially present within the capsid, thereby allowing us to identify capsids with lattice gaps that are larger than GFP by loss of this fluid-phase marker. This system utilized a virus developed by Hübner et al. that contains GFP between MA and CA, flanked by HIV protease target sites.36 In this system, GFP is packaged into the immature virion as a domain of Gag and then incorporated within the capsid following viral protease cleavage. As shown in Fig. 5, a significant amount of free GFP cosediments with viral cores purified from detergent-stripped virions, revealing that a subset of the protein resides within the viral capsid.
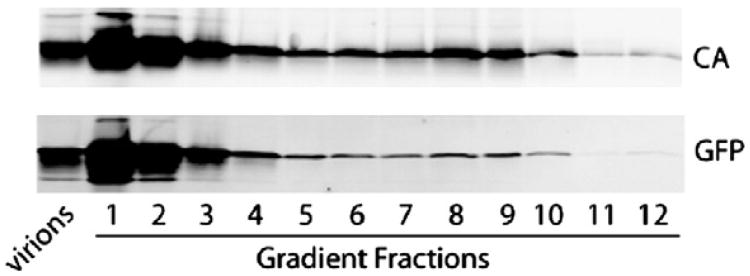
Fig. 5.
GFP comigrates with iGFP HIV cores in a sucrose gradient. Concentrated iGFP particles were subjected to equilibrium centrifugation through a layer of Triton X-100 into a linear sucrose density gradient. Fractions from the gradient were precipitated and analyzed by immunoblotting. Top panel shows detection of p24 (CA) while bottom panel shows detection of GFP. Numbers designate fractions collected from the top of the gradient. Capsids sediment to fractions 7–10. The first lane contains proteins extracted from intact iGFP virions.
To test whether these capsids retain their GFP during the early stages of viral infection, we utilized an assay in which uncoated cores are identified and captured by the restriction factor rhTRIM5α but prevented from dissociation by treatment with the proteasome inhibitor MG132. We have previously shown that, under these conditions, rhTRIM5α recognizes and sequesters intact capsid cores, thereby blocking infection.37 These captured complexes appeared to represent intact conical capsids because they contain abundant amounts of the CA protein as revealed by immunofluorescence staining.
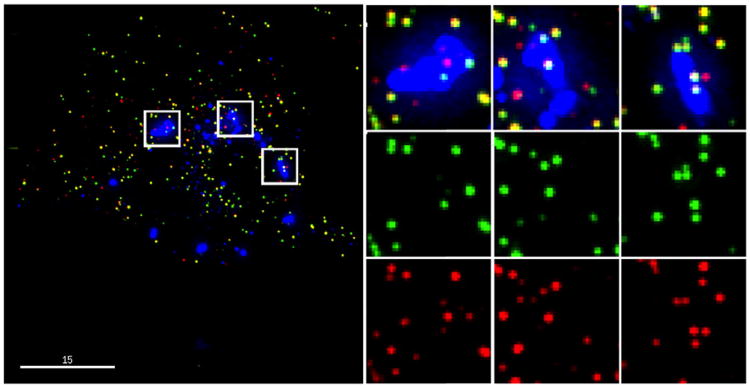
To determine whether GFP is retained within the capsids that are captured by rhTRIM5α, we infected HeLa cells that stably expressed HA-tagged rhTRIM5α with the iGFP virus that was also labeled with mCherry-Vpr. As rhTRIM5α recognizes the assembled capsid lattice, particles that label with mCherry-Vpr and rhTRIM5α must be mature viral cores that have not lost their capsids. Completely closed capsids should also contain free GFP, whereas cores with seams or other large openings would be expected to lose the free GFP while retaining mCherry-Vpr, enabling us to estimate the fraction of incompletely closed capsids. An example of this type of analysis is shown in Fig. 6, where multiple mCherry-Vpr-labeled complexes have been captured within rhTRIM5α cytoplasmic bodies. Image analysis software was used to count rhTRIM5α-associated virions and categorize them as positive for both mCherry-Vpr and GFP (closed capsids), mCherry-Vpr only (incompletely closed capsids), or GFP only (incomplete mCherry-Vpr labeling). These analyses revealed that 25.0±0.8% [s.e.m. (standard error of the mean)] of the mCherry-Vpr-labeled cores lacked detectable GFP and therefore likely represented cores that had capsid gaps large enough to lose GFP while retaining an essentially intact capsid lattice.
Fig. 6.
One-quarter of HIV capsids captured by TRIM5α in the presence of proteasome inhibition leak GFP. Captured mCherry-Vpr complexes were scored for the presence of the fluid-phase marker GFP; 25.0±0.8% (s.e.m.) of the complexes lose the GFP marker, indicating that they are unclosed.
Exactly when the GFP was lost from the structure cannot be determined from this experiment. These results are consistent, however, with the idea that a subset of authentic viral capsids has gaps that are large enough to allow loss of GFP from the core interior.
Discussion
Here, we compared high-resolution cryotomograms of retroviral cores with the predictions of a nonequilibrium growth of an elastic sheet or “curled sheet” model. As predicted by the model, curved and rolled sheet structures are present within some virions, and some evidence of other closure defects such as tip holes and seams were also found. Loss of GFP from cytoplasmic viral cores confirmed that a substantial fraction (∼25%) is indeed unclosed.
Together, these data suggest that retroviral capsid assembly frequently involves the union in space of the edges of a curling sheet, which then closes at the tips. Rolls could be explained by the two growing edges missing each other in space, extra sheets could be explained by secondary nucleation sites, unclosed tips could be explained by the energetic strains encountered by high curvature, and seams could be explained by misalignment along edge junctions.
Previous observations that led to other models can also be viewed as at least consistent with the curled sheet model. The tip holes described by Benjamin et al., for instance, could also be explained by the curled sheet model, as shown by the simulations here.22 Consistent cone angles and strong correlations between capsid length and viral diameter led Briggs et al. to propose a tip-to-base model of assembly.23 While the curling sheet model does not involve a membrane enclosure, the membrane surrounding real virions may nevertheless constrict the size and shape of the growing sheet, and at least a class of particles with a consistent cone angle would not be unexpected. It has also been proposed that the number of cores seen within one virion was dependent on the size of virion, arguing for template-mediated growth.20 Since a larger virion may contain more CA protein available for capsid assembly, more than one curled sheet may be nucleated, resulting in extra sheets as seen here.
The curling sheet model also offers an explanation for observations that challenge previous models. It is known, for instance, that CA–NC can form cones in vitro with a similar size and shape as viral cores in the complete absence of membranes and RNP.13 According to the curled sheet model, the size and shape of capsids should depend only on the elastic properties of the protein subunits, explaining why the cones observed in vitro are similar to those in vivo. The unclosed capsids observed here (rolls, extra sheets, and seams) would also not be expected to form in any strictly “end-to-end” assembly pathway.
While the support for a curling sheet model is therefore strong, it seems likely to us that capsid assembly is considerably more complicated than suggested by any of the simple models. The simulations here obviously do not take into account the stochastic release of CA proteins into the interior of the virus by protease or the decrease in the concentration of unbound protein as the capsid assembles. Nor do the simulations take into account the presence of the RNP or membrane enclosure, which must at least constrain possible assembly pathways. Other complicating factors may include Env spikes, whose presence has been seen to bias the distribution of capsid morphologies in Rous sarcoma virus.38 Different capsids may follow different assembly pathways or mixtures of pathways.
The unclosed capsids described here nevertheless suggest that, at least, some particles depend on the union in space of two edges of a curling sheet. Unfortunately, we do not know whether the unclosed particles are infectious and, thus, cannot comment on whether these defects inhibit function. Further structural studies of capsids in vivo may reveal correlations between defects in the shell and the infectivity of the capsid. If full capsid closure is essential to the HIV life cycle, small molecules that disrupt the natural curvature of growing capsid sheets or perturb the ratio of hexamers to pentamers may be effective antiretroviral agents.
Materials and Methods
Mature HIV particle preparation
We seeded 293T cells at 2.4×106 cells/10-cm plate a day before transfection. Each 10-cm plate of cells was transfected (CalPhos mammalian transfection kit; Clon-tech) with 8.1 μg of R9 ΔRT ΔRNase. After 36h, the supernatants were harvested, filtered (0.45 μm pore size), and pelleted through a 4-ml cushion of 20% sucrose in a Beckman SW-32Ti rotor (134,000g, 2h, 4°C). Each set of pelleted virions was resuspended in 50 μl of ST buffer and kept on ice till to frozen on grid.
EIAV core isolation
EIAV cores were prepared as described by Langelier et al.39 Briefly, EIAV virions were produced by co-transfection of 293T cells with an EIAV vector system. Each 10-cm plate of cells was transfected (CalPhos mammalian transfection kit; Clontech) with 7.5 μg of pEV53 (EIAV structural proteins), 7.5 μg of pSIN6.1CeGFPW (packaged GFP expression vector), and 2.2 μg of phCMV-VSV-G [vesicular stomatitis virus glycoprotein (VSV-G) envelope]. After 36h, the supernatants were removed, pooled (four plates per pool), filtered (0.45 μm pore size), and pelleted through a 4-ml cushion of 20% sucrose in a Beckman SW-32Ti rotor (134,000g, 2h, 4°C). Each set of pelleted virions was resuspended by gentle pipetting (4h at 4 °C) in 400 μl of ST buffer [10mM Tris–HCl (pH7.4) and 50mM NaCl]. Four 11.5-ml sucrose gradients [30% and 70% (weight/volume) sucrose in ST buffer] were prepared in 14mm×89mm tubes using a gradient mixer (Biocomp). The gradients were overlaid with a 0.3-ml cushion of 15% sucrose containing 1% Triton X-100 and then with a 0.3-ml barrier layer of 7.5% sucrose in ST buffer. Each tube of concentrated EIAV particles was carefully layered on top of a gradient and centrifuged in a Beckman SW-41 rotor (210,000g, 16h, 4°C). Twelve 1-ml fractions were collected from the bottom of each tube, and fraction densities were measured by using a digital refractometer (Leica); EIAV CA was assayed by Western blotting. Three 1-ml fractions of the correct density (1.22–1.26g/ml) contained intact EIAV core particles and were pooled, repelleted by centrifugation in a Beckman SW-41 rotor (210,000g, 2h, 4°C), and resuspended in 50 μl of ST buffer and kept on ice until frozen on grids.
Cryo-EM grid preparation
Quantifoil carbon grids with a hole size of 0.6 μm and spacing between holes of 1 μm were glow discharged. A 3-μl solution of 10nm nanogold was applied to the carbon film and dried in a 60°C oven. A 4-μl solution of either intact mature HIV particles or purified mature EIAV cores was applied to the pre-treated Quantifoil grids. Excess solution was blotted away with filter paper in a 100% relative humidity chamber. Grids were plunged into liquid ethane using a Vitrobot. Freezing conditions varied for each session but were optimized when cryo-grids had sample particles suspended in a thin layer of vitrified ice across the holes of carbon.
Infected cell preparation
Human umbilical vein endothelia cells (Lonza, Walkersville, MD) were seeded in 6-well plates containing carbon-coated gold EM finder grids (2 μm hole size and 2 μm hole spacing; Quantifoil, Jena, Germany) and cultured for 24h in a humidified incubator maintaining 37°C and 5% CO2. Cells were infected with the HIV-based vector NLEGFP-Delta BglIVprX (kindly provided by Dr. Irvin Chen, University of California, Los Angeles) pseudotyped with the VSV-G envelope [obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH): pHEF-VSV-G from Dr. Lung-Ji Chang]. Infection was carried out by spinoculation at 800g for 1h at 16°C. Spinoculated samples were washed two times with fresh media prewarmed to 37°C. Two milliliters of fresh media was then added to each well, and samples were incubated for 36h to allow for virus expression before cryopreservation. EM grids on which virus-producing cells were growing were removed from the culture plate using forceps and treated with 3 μl of warm media containing 10nm gold fiducials. Forceps and grid were transferred to the environment chamber of a Vitrobot Mark III (FEI) maintained at 37°C and 80% relative humidity. Excess liquid was manually blotted from the grids on one side before the grids were plunged into liquid ethane.
Electron cryotomography
Cryo-grids were transferred to a 300-kV FEI G2 Polara transmission electron microscope equipped with a field emission gun and energy filter. Data were collected on a Gatan Ultracam 4k×4k lens-coupled charge-couple device in the energy-filtered transmission electron microscopy mode with an energy slit width of 20eV. Tilt series were collected within a range from −65° to +65° with a step size of 2°, 41,000× magnification, a total dose of 80–100e/Å2, and a defocus of −6 to 8 μm. The software package used to collect the tilt series was UCSF Tomo, and the 3-D reconstruction was performed with IMOD using a weighted back-projection algorithm. Tomographic slices of capsids with seams were created by fitting an ObliqueSlice in Amira to be perpendicular to the seams or to the long axis of the cones in the case of the control.
Segmentation
The segmentations were performed manually using the Amira package by selecting the darkest density through several slices. To generate the isosurface, we cut subvolumes of tomograms containing regions of interest and generated an isosurface with UCSF Chimera. The threshold was chosen to minimize noise while maintaining the capsid density before holes appeared throughout. A red cylinder was fit inside the core using UCSF Chimera to highlight the holes in the capsid.
Simulations
The simulations were performed as in Ref. 26.
HIV Gag-iGFP core purification and analysis
Viral cores were isolated from HIV-1 particles generated by transfection of 293T cells with the HIV-1 Gag-iGFP viral clone36 as previously described.40 Fractions were collected from the top of the gradient, and the proteins were precipitated with trichloroacetic acid. The precipitated proteins were dissolved in Laemmli buffer and separated on 4–20% polyacrylamide gradient gels and blotted to nitrocellulose. The blots were probed with a mixture of affinity-purified rabbit polyclonal antibodies against GFP-derived peptides (Clontech), followed by a monoclonal antibody to HIV-1 CA (clone 183-H12-5C) with appropriate IR dye-conjugated secondary antibodies. Bands were detected with a LI-COR Odyssey.
HIV Gag-iGFP fluorescent imaging and analysis
For fluid-phase virus imaging experiments, HIV-1 Gag-iGFP virus was generated by transfection of 293T cells with HIV Gag-iGFP ΔEnv, mCherry-Vpr, and CMV-VSV-G. Ninety percent of the viral stocks used for these studies were doubly labeled. Cells stably expressing HA-tagged rhTRIM5α (provided by Joseph Sodroski) were seeded onto coverslips treated with fibronectin (Sigma-Aldrich) and infected in the presence of 10 μg/ml diethylaminoethanoldextran and 1 μg/ml MG132 (Sigma-Aldrich). Cells were incubated at 37°C for 6h before fixing in 3.7% formaldehyde (Polysciences) in PIPES buffer. Coverslips were stained with rabbit-anti-HA (Sigma-Aldrich) at a 1:300 dilution to detect rhTRIM5α, followed by AMCA donkey anti-rabbit (Jackson ImmunoResearch) at a 1:400 dilution, then mounted onto slides (VWR) using Fluoro-Gel (Electron Microscopy Sciences). Images were acquired and deconvolved on a DeltaVision microscope equipped with soft-WoRx software (Applied Precision). Images were analyzed using algorithms written for Interactive Data Language to identify virions containing mCherry-Vpr (tetramethylrhodamine isothiocyanate-positive) associated with rhTRIM5α (4′,6-diamidino-2-phenylindole, dihydrochloride-positive) and determine whether they were also positive for fluid-phase GFP (fluorescein isothiocyanate-positive). Three independent experiments were performed, and a total of seven coverslips were analyzed. Twenty-five images were taken for each coverslip. A total of 3242 mCherry-Vpr-labeled viral complexes associated with TRIM5α were quantified. The proportion of incompletely closed virions (TRITC-positive, DAPI-positive, FITC-negative) in the text represents the mean±s.e.m. (N=7).
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grants 2P50GM082545-06 (to T.J.H. and G.J.J.), R01 AI076121 (to C.A.), and R01 AI407770 (to T.J.H.); National Science Foundation Grant DMR-06-45668 (to R.Z.); and gifts to Caltech from the Gordon and Betty Moore Foundation and to Northwestern from the James B. Pendleton Charitable Trust. The following was obtained from the NIH AIDS Research and Reference Reagent Program: 183-H12-5C hybridoma from Dr. Bruce Chesebro. We thank Dr. Ben Chen for providing the HIV Gag-iGFP plasmid, Dr. Joseph Sodroski for cells expressing rhTRIM5α, Dylan Morris for help with the supplementary movies, Drs. Jordan Schooler and Elizabeth Wright for help with data collection, and Drs. Wesley Sundquist and Edward Campbell for critically reading the manuscript.
Abbreviations used
- HIV
human immunodeficiency virus
- EIAV
equine infectious anemia virus
- GFP
green fluorescent protein
- RT
reverse transcriptase
- EM
electron microscopy
- 3-D
three-dimensional
- RNP
ribonucleoprotein
- ECT
electron cryotomography
- NIH
National Institutes of Health
Footnotes
Supplementary Data: Supplementary materials related to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2012.10.006
References
- 1.Pettit S, Sheng N, Tritch R, Erickson---Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 2.Vogt VM. Retroviral virions genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 27–69. [PubMed] [Google Scholar]
- 3.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, H SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 263–334. [PubMed] [Google Scholar]
- 4.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 5.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 6.Bartonova V, Igonet S, Sticht J, Glass B, Habermann A, Vaney MC, et al. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem. 2008;283:32024–32033. doi: 10.1074/jbc.M804230200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Dornadula G, Pomerantz RJ. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage or viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arhel N, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, et al. HIV-1 DNA Flap formation promotes uncoating of the preintegration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mcdonald D. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundquist WI, Hill CP. How to assemble a capsid. Cell. 2007;131:17–19. doi: 10.1016/j.cell.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 13.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 15.Cardone G, Purdy JG, Cheng N, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganser BK, Cheng A, Sundquist WI, Yeager M. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 2003;22:2886–2892. doi: 10.1093/emboj/cdg276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 18.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, et al. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 20.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Krausslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briggs JA, Grünewald K, Glass B, Förster F, Kräusslich HG, Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Zandi R, Reguera D, Bruinsma RF, Gelbart WM, Rudnick J. Origin of icosahedral symmetry in viruses. Proc Natl Acad Sci USA. 2004;101:15556–15560. doi: 10.1073/pnas.0405844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks SD, Henley CL. Irreversible growth model for virus capsid assembly. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2006;74:031912. doi: 10.1103/PhysRevE.74.031912. [DOI] [PubMed] [Google Scholar]
- 26.Levandovsky A, Zandi R. Nonequilibirum assembly, retroviruses, and conical structures. Phys Rev Lett. 2009;102:198102. doi: 10.1103/PhysRevLett.102.198102. [DOI] [PubMed] [Google Scholar]
- 27.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2011;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egberink HF, Ederveen J, Montelaro RC, Pedersen NC, Horzinek MC, Koolen MJ. Intracellular proteins of feline immunodeficiency virus and their antigenic relationship with equine infectious anaemia virus proteins. J Gen Virol. 1990;71:739–743. doi: 10.1099/0022-1317-71-3-739. [DOI] [PubMed] [Google Scholar]
- 29.Goudsmit J, Houwers DJ, Smit L, Nauta IM. LAV/HTLV-III gag gene product p24 shares antigenic determinants with equine infectious anemia virus but not with visna virus or caprine arthritis encephalitis virus. Intervirology. 1986;26:169–173. doi: 10.1159/000149697. [DOI] [PubMed] [Google Scholar]
- 30.Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 31.Wright E, Schooler J, Ding H, Kieffer C, Fillmore C, Sundquist WI, Jensen G. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briggs JA, Simon MN, Gross I, Kräusslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 33.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Implications for viral capsid assembly from crystal structures of HIV-1 Gag1–278 and CAN133–278. Biochemistry. 2006;45:11257–11266. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 34.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arslan I, Tong J, Midgley P. Reducing the missing wedge: high-resolution dual axis tomography of inorganic materials. Ultramicroscopy. 2006;106:994–1000. doi: 10.1016/j.ultramic.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Hübner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol. 2007;81:12596–12607. doi: 10.1128/JVI.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J Cell Biol. 2008;180:13. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content: implications that nucleation controls morphogenesis. J Mol Biol. 2008;376:1168–1181. doi: 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, et al. Biochemical characterization of a recombinant TRIM5α protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah VB, Aiken C. In vitro uncoating of HIV-1 cores. J Visualized Exp. 2011 doi: 10.3791/3384. http://dx.doi.org/10.3791/3384. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.