Summary
Given a randomized treatment Z, a clinical outcome Y, and a biomarker S measured some fixed time after Z is administered, we may be interested in addressing the surrogate endpoint problem by evaluating whether S can be used to reliably predict the effect of Z on Y. Several recent proposals for the statistical evaluation of surrogate value have been based on the framework of principal stratification. In this paper, we consider two principal stratification estimands: joint risks and marginal risks. Joint risks measure causal associations of treatment effects on S and Y, providing insight into the surrogate value of the biomarker, but are not statistically identifiable from vaccine trial data. While marginal risks do not measure causal associations of treatment effects, they nevertheless provide guidance for future research, and we describe a data collection scheme and assumptions under which the marginal risks are statistically identifiable. We show how different sets of assumptions affect the identifiability of these estimands; in particular, we depart from previous work by considering the consequences of relaxing the assumption of no individual treatment effects on Y before S is measured. Based on algebraic relationships between joint and marginal risks, we propose a sensitivity analysis approach for assessment of surrogate value, and show that in many cases the surrogate value of a biomarker may be hard to establish, even when the sample size is large.
Keywords: Estimated likelihood, Identifiability, Principal stratification, Sensitivity analysis, Surrogate endpoint, Vaccine trials
1. Introduction
1.1 Surrogate endpoints
The identification and evaluation of surrogate endpoints is a major goal of many clinical studies. In vaccine trials, for example, pinpointing a biomarker which reliably predicts protection from infection could provide an invaluable biological target for vaccine development and allow researchers to predict vaccine efficacy in new populations without the need to conduct a full-fledged trial. Several statistical approaches to surrogate endpoint assessment are summarized in Weir and Walley (2006). While the common scientific goal of these approaches is to predict the effect of treatment on the outcome in a future setting given information about the effect of treatment on the biomarker, Joffe and Greene (2009) (henceforth JG) distinguish between two paradigms based on the quantities used to make these predictions. In the “causal effects” (CE) paradigm, treatment effects on the outcome are predicted by combining the effect of treatment on the biomarker with knowledge of the effect of the biomarker on the outcome. The “causal association” (CA) paradigm predicts future treatment effects on the outcome based on the (previously observed) association between treatment effects on the biomarker and treatment effects on the outcome. While prototypical CA methods (eg.Buyse et al. (2000);Gail et al. (2000)) are based on meta-analysis of multiple trials, we focus on the case where the data arise from a single trial. We consider the counterfactual-based principal stratification approach proposed by Frangakis and Rubin (2002) (henceforth FR) and extended by Gilbert and Hudgens (2008) (henceforth GH), which allows the CA paradigm to be applied in the single-trial setting.
A major aim of this paper is to illustrate the inherent statistical difficulty of the surrogate endpoint assessment problem. We begin with a brief introduction to the problem and describe the vaccine trial setup and basic assumptions under which we will operate. Next, we introduce joint and marginal risks, the two estimands central to the paper. In Section 3, we describe the degree to which joint and marginal risks are statistically identifiable under a variety of scenarios. Section 4 outlines an estimated likelihood procedure for estimation of joint and marginal risks. Section 5 presents a sensitivity analysis approach to assessing surrogate value wherein we assume known functions for the unidentified distributions, indexed by fixed sensitivity parameters. Section 6 presents simulation results. We conclude with some final thoughts on the challenges of surrogate endpoint assessment.
1.2 Framework
In what follows, we consider a vaccine trial where subjects i = 1,…, n are randomly assigned at time 0 to either vaccine (Zi = 1) or placebo (Zi = 0) and followed for infection (Yi = 1). At some time Τ > 0, biological samples are collected from all subjects who remain uninfected at Τ (denoted by ). We assume that Τ is the same for all subjects. Let Si be the biomarker of interest which is measured from the collected samples. If , due to infection in the time interval [0, Τ], then Si is undefined, and we set Si = *. Lastly, we assume that a vector of baseline covariates Wi is available for each subject.
Prentice (1989) defined a surrogate as a replacement endpoint such that a test of the null hypothesis of no treatment effect on this endpoint (S) provides a valid test of the corresponding null hypothesis for the clinical endpoint (Y). Mathematically, this definition can be formulated as
| (1) |
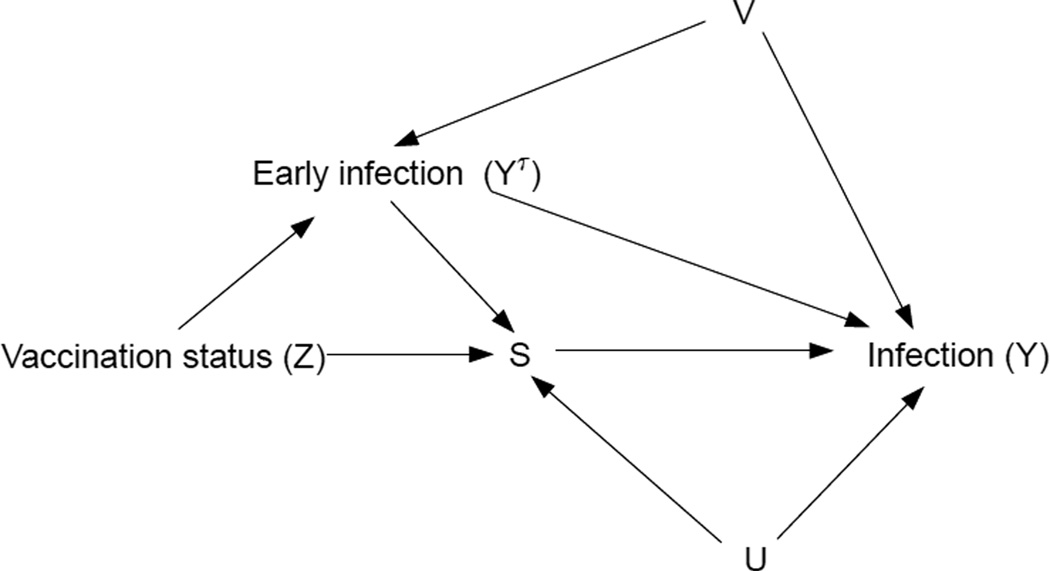
According to Prentice, the two major criteria which S must satisfy to qualify as a surrogate for Y, conditional on W, are 1) S has some predictive value for Y, and 2) Conditional on S, Y is statistically independent of Z. Approaches to surrogate endpoint assessment which aim to verify these criteria belong to JG's CE paradigm. Figure 1 of their paper illustrates the well-known result from directed acyclic graph theory (see, eg., Pearl (2000)) that conditioning on the post-randomization event S may induce statistical dependence between Z and Y if the set of simultaneous predictors of S and Y (say U) is not accounted for. A further complication arises when the outcome of interest may occur before S is measured, a case not considered by JG. Our Figure 1 illustrates this situation: YΤ, the early infection indicator, is affected by treatment and affects both S (since YΤ = 1 ⇒ S = *) and Y. The same graph theoretic arguments as above imply that the appropriate criterion 2) for evaluating the Prentice definition is statistical independence between Z and Y conditional on S and on both U and V, where V are factors that simultaneously predict YΤ and Y.
Figure 1.
Directed acyclic graph showing relationships between variables in a hypothetical vaccine trial.
Since the determinants of S, YΤ, and Y are often poorly understood during the trial design stage, it is unlikely that all of the important information about (U, V) will be captured by conditioning on the available covariates W alone. Hence, as pointed out in FR, CE estimands designed to verify criterion 2) may mislead about a biomarker's true surrogate value. To address this problem, FR proposed to assess surrogate value via potential outcomes (i.e. counterfactuals). Expanding on FR's work, GH suggested ways to gauge surrogate value via contrasts of the estimands
| (2) |
| (3) |
where are the potential values of (Y, YΤ, S) under treatment assignment Z = z, for z ∈ {0,1}.
GH proposed to judge the value of S as a surrogate by the degree to which it satisfies the following two criteria, conditional on W:
Average causal necessity (ACN): R1(S0; S1;W) = R0(S0; S1;W) for all S1 = S0.
Average causal sufficiency (ACS): There exists a constant C ≥ 0 such that R1(S0, S1, W) ≠ R0(S0, S1, W) for all |S0–S1|>C
ACN and ACS are checked by estimating contrasts of Y1 and Y0 within a principal stratum . Since the stratum to which an individual belongs is unaffected by treatment, these contrasts represent causal effects and are not subject to post-randomization selection bias arising from unmeasured confounding (see FR, page 23 and Lauritzen (2004)). We note that ACN is equivalent to the definition given by Vanderweele (2008) of “no average principal strata direct effects”, while ACS implies the existence of some “average principal strata indirect effects” in the sense of Vanderweele.
While the principal stratification estimands R1 and R0 have appealing properties, we pay a price for their “robustness” to unmeasured confounding in terms of statistical non-identifiability. In what follows, we investigate how different assumptions affect the identifiability of (2), (3) and related counterfactual estimands.
2. Estimands
2.1 Joint risks
We will henceforth refer to the quantities R1(S0, S1,W) and R0(S0, S1,W) as joint risks, to indicate that they depend on the joint values (S0, S1). We focus on identifiability and estimation in the special case of “constant biomarkers” considered in GH:
[CB] Constant biomarkers under placebo Provided YΤ = 0, S0 = c for some constant c
Though condition [CB] may not be satisfied in all situations, it often applies in vaccine trials where subjects have not previously been infected by the pathogen targeted by the vaccine under study. In HIV vaccine trials, for example, enrollment criteria generally require that subjects be HIV-uninfected at the start of the trial. Subjects who are not vaccinated, therefore, are not “primed” to recognize the HIV epitopes present in the vaccine, and hence are unlikely to show a measurable immune response when exposed to these epitopes via an immunological assay (Gilbert et al. (2005)). In this case, c might represent the lower limit of detection of the relevant assay. We note that, unlike some of the untestable assumptions introduced later, [CB] can be directly verified in the lab by measuring the biomarker values of placebo recipients who remain uninfected at time Τ.
When [CB] holds, we have (S0, S1) = (c, S1), and hence for ease of notation we can drop the dependence of R1 and R0 on S0, yielding
| (4) |
| (5) |
(the reader should recall, however, that these quantities remain “joint” risks since they are equivalent to R1(c, S1, W) and R0(c, S1, W)). The surrogate value of S can then be assessed, conditional on W, by considering the magnitude of contrasts of R0 and R1 for a variety of values of S1. We refer the reader to GH's Figure 2, which plots R0(S1, W) — R1(S1, W) as a function of S1 for biomarkers having no, low, moderate, and high surrogate value.
Four assumptions allowed GH to identify the joint risks under [CB]:
-
[A1] Stable Unit Treatment Value Assumption (SUTVA)
[No interference] The potential outcomes for one subject are independent of the treatment assignments of other subjects, i.e. there is no “interference” between experimental units.
[Consistency] For an individual receiving treatment Z = z and with observed outcome Y, we have Y = Yz, i.e. the observed outcome is equal to the potential outcome under the treatment actually received.
-
[A2] Ignorable Treatment Assignments
Conditional on W, Z is independent of
-
[A3] Equal Individual Clinical Risk prior to time Τ
if and only if , i.e. the vaccine has no causal effect on the risk of infection before the immune response is measured at Τ.
-
[A4] Risk restriction
For the case where S1 has J distinct levels 1,…, J and W has K distinct levels , for z ∈ {0, 1}.
Assumptions [A1] and [A2] are standard in most work on causal inference. The validity of the “No interference” part of [A1] may be questioned when the disease of interest is infectious, but is defensible if the vaccine trial enrolls a small fraction of the at-risk population. Recent work by Hudgens and Halloran (2008) discusses relaxation of SUTVA. [A2] will hold in a randomized trial where blinding is maintained.
The validity of [A3], however, is doubtful in many contexts. Since most current vaccine trials are designed to measure peak immune response, an effective vaccine may offer at least partial protection to vaccine recipients before their immune responses are measured, thereby violating [A3]. [A4] requires that the effect of S on the clinical endpoint not vary within levels of W. Both [A3] and [A4] are untestable, although [A3] has the testable implication that the population vaccine effect up to time Τ is zero. A major goal of this paper is to explore the consequences of relaxing [A3] and [A4].
2.2 Marginal Risks
In the Identifiability section below, we show that if assumptions [A3] and [A4] are relaxed, then joint risks are statistically non-identifiable. We might therefore ask whether any useful information can be obtained from the family of estimands which are identifiable from the available data when these assumptions fail.
We begin by noting that [A3] implies the equalities
| (6) |
| (7) |
The left-hand sides of (6) and (7) correspond to the expressions for R1 and R0, and from the expressions on the right-hand sides we define the marginal risks
| (8) |
| (9) |
Since and do not condition on , S0 may be undefined and hence S1 does not specify the basic principal stratum to which an individual belongs: an individual with S1 = s and may belong to either the stratum {S1 = s, S0 = *} or {S1 = s, S0 = c}. As a result, without [A3] marginal risks do not allow an assessment of how causal clinical effects vary with contrasts of S1 and S0, and hence cannot be used to check ACN and ACS. Nevertheless, contrasts of the marginal risks are causal effects and may be used to predict treatment effects in a new setting. In particular,
| (10) |
where is the overall causal treatment effect for level-W subjects uninfected at Τ under active treatment, which is a useful summary of treatment efficacy. Suppose in a new setting (e.g., for a refined version of the treatment to be tested in a follow-up efficacy trial in the same study population), is distributed according to Fnew(s, W). Assuming the marginal risk functions are the same for the efficacy trial and the new setting (an untestable assumption whose plausibility may be defended based on biological and mechanistic knowledge), the predicted TEpost−Τ(W) for the new setting is . A biomarker valuable for treatment/vaccine development will be one such that certain manipulations of its distribution lead to a large predicted gain in overall treatment efficacy (from TEpost−Τ(W) to TEnewpost−Τ(W;Fnew)). This amount of predicted gain is a useful summary measure of surrogate value in its own right; though we stress that it quantifies a different type of surrogate value than a measure of association between biomarker and clinical treatment effects. Follmann (2006) considered this idea under assumption [A3], with bivariate normal (S, W) and probit risk models. Further discussion of the interpretation of marginal and joint risk estimands appears in Gilbert et al. (2008).
3. Identifiability of Joint and Marginal Risks
In this section, we explore the identifiability of joint and marginal risks in a variety of scenarios. We restrict attention to the case where S has J discrete levels, 1,…,J, and omit baseline covariates W (they are assumed fixed and do not affect the identifiability arguments). With this setup, the joint and marginal risks are considered as functions of S1 only, and have J possible values. The propositions below describe how many of these J values are identifiable from observed data under different sets of assumptions. Proofs of these propositions appear in Web Appendix A.
Proposition 1 (Identifiability under [CB], [A1]–[A2] (Base case)): When [CB] and [A1]–[A2] hold, is identifiable from the available data but R1, R0, and are non-identifiable and each requires J parameters to be specified for their J possible values.
Since R1, R0, and are non-identifiable under [CB] and [A1]–[A2], it is natural to consider the question: Are there additional plausible assumptions which can help to identify these estimands? One possible additional assumption is a weakened version of [A3]:
[A3′] Pre-Τ monotonicity
The pre-Τ monotonicity assumption asserts that a participant's clinical outcome prior to Τ is no worse under assignment to treatment than under assignment to placebo. [A3′] is implied by [A3], but unlike [A3] it allows for a pre-Τ treatment effect. In the context of blinded vaccine trials where the treatment is generally safe, [A3′] will often hold, but it must be considered carefully: data from the Step trial (which was stopped prematurely in late 2007 because the vaccine product did not appear to be effective) suggested the possibility of vaccine enhancement, whereby subjects who received active vaccine were more likely to be HIV-infected than those who received the placebo (Buchbinder et al. (2008)). Beyond the vaccination context, [A3′] may fail in trials comparing two active treatments, or when a treatment is highly toxic to some but of benefit to others.
When [A3′] holds in addition to [CB] and [A1]–[A2], we have the following result:
Proposition 2 (Identifiability under [CB], [Al]–[A3′] (Monotonicity)): Under [CB], and [A1]–[A3′], the number of parameters required to describe R0 is reduced by 1, to J–1. Identification of R1 and still requires J parameters each.
It can be shown (see the proof of Proposition 3, Web Appendix A) that would be identifiable from the observed data if we could identify the distribution of S1 among subjects who would have remained uninfected over the entire trial if they had received the placebo. An idea due to Follmann (2006) suggests a way to identify this quantity: an augmented study design called closeout vaccination, where placebo recipients who remain uninfected at the end of the trial are given the active vaccine (“closed out”) and have their immune response measured Τ time-units later. Denote this closeout measurement by , and the indicator that an individual is infected during the duration-Τ closeout period by . Under [A1]–[A3′], closeout vaccination yields information about . The following two assumptions, taken together, guarantee that this distribution is equivalent to the distribution of interest, :
[A5] Time constancy of the immune response distribution: For subjects with almost surely
[A6] No infections during the closeout period:
The time constancy assumption implies that the immune responses measured from closed out subjects would not have been different if these subjects had been assigned to the vaccine at the beginning of the trial and had their immune response measured at time Τ. [A5] may be evaluated from knowledge of the immune system; ) recent research (eg. Goronzy and Weyand (2005), Lacroix-Desmazes et al. (1999)) suggests that several key markers of T cell production/diversity and antibody reactivity remain relatively constant in adults under age 65, and hence [A5] may reasonably apply to these biomarkers in the context of vaccine trials of relatively short duration. If a substantial proportion of study subjects are co-infected with another pathogen during the study period, however, the validity of [A5] may be questioned. When [A5] fails, closeout vaccination does not provide the information necessary to identify the marginal risk .
Unlike [A5], [A6] is directly testable; one need only check whether any infections occurred during the closeout period. Since placebo recipients who remain infection-free for the duration of the trial (and hence are candidates to be closed out) are likely to have a lower risk of infection than the general placebo population, [A6] may be reasonable in certain circumstances. If there is evidence showing that [A6] is violated, however, the relationship between is non-identifiable. In the Sensitivity Analysis section, we describe this relationship explicitly in terms of non-identifiable sensitivity parameters.
The following proposition summarizes the identifiability of the joint and marginal risks when [A5] and [A6] additionally hold:
Proposition 3 (Identifiability under [CB], [Al]–[A3′], [A5]–[A6] (Monotonicity + Closeout)): Under [CB], [Al]–[A3′] and [A5]–[A6], marginal risks and are identifiable from the observed data. R1 remains non-identifiable and one of the J possible values of R0 is identified.
Table 1 summarizes the results derived in this section, along with some others which follow using similar arguments. Note the dramatic simplification achieved when [A3] replaces [A3′]: the joint risks R0 and R1 are equivalent to the (identifiable) marginal risks and hence ACN and ACS can be verified non-parametrically, allowing surrogate value to be assessed from the observed data.
Table 1.
For a discrete biomarker with J levels, number of values (from a total of J possible values) of marginal and joint risk which are statistically non-identifiable from observed data under various assumptions. ECR refers to [A3] (Equal Clinical Risk prior to τ ).
| Assumptions | Marginal risks | Joint risks | ||
|---|---|---|---|---|
| R1 | R0 | |||
| [CB],[A1]-[A2] (Base case) | 0 | J | J | J |
| [CB],[A1]-[A3′] (Monotonicity) | 0 | J | J | J–1 |
| [CB],[A1]-[A2],[A5]-[A6] (Closeout) | 0 | J | J | J |
| [CB],[A1]-[A3′],[A5]-[A6] (Monotonicity + Closeout) | 0 | 0 | J | J–1 |
| [CB],[A1]-[A3],[A5]-[A6] (ECR + Closeout) | 0 | 0 | 0 | 0 |
4. Estimation
Following the notation presented above, the iid observed data are , where we reintroduce the vector Wi of baseline covariates assumed to be available for all study subjects. Note that in the definition of Oi and the conditional likelihood (11) below, subscripts refer to individual subjects and not potential outcomes.
Only subjects with contribute to the conditional likelihood, which takes the form
| (11) |
See Web Appendix B for details of the individual terms which make up the likelihood. L is a function of parameters of interest Θ, which specify and , and nuisance parameters Γ. We propose to make inference about Θ based on the estimated likelihood method described in Pepe and Fleming (1991), which consists of plugging in estimates of the elements of Γ into L and maximizing over Θ. In this application, we suggest using the bootstrap to obtain standard errors; previously derived asymptotic variance results for estimated likelihood rely on the assumption that each subject has a non-zero probability of having S1 observed, which does not apply in our situation since infected placebo recipients cannot have S1 measured.
5. Sensitivity Analysis
Since the joint risks are not statistically identifiable unless certain potentially implausible assumptions hold, we suggest a sensitivity analysis approach to assessing surrogate value. Our strategy relies on the fact that one can re-express the joint risks in terms of marginal risks and non-identifiable quantities π and π* as
| (12) |
where (a proof of (12) is given in Web Appendix C). Different values of π and π* correspond to different degrees of post-randomization selection bias induced by the unmeasured simultaneous predictors (U, V) from Figure 1. Sensitivity analysis proceeds by estimating and and then varying the values of π(j) and π*(j) to observe their effect on contrasts of R1 and R0. Certain features of the observed data, along with the requirement that probabilities lie in [0,1], restrict the range of allowed sensitivity parameter values. These constraints are described in Web Appendix D; they may be further tightened based on additional assumptions and/or subject matter knowledge.
If [A6] fails and some subjects are infected during the closeout period, is no longer identifiable from observed data. In this case, we suggest introducing additional nonidentifiable parameters η(j) defined by so that . η can be viewed as an analog of π for the closeout period. For the sake of clarity, in this paper we assume that [A6] holds throughout so that η(j) = 1 for all j.
6. Simulations
We simulate a vaccine trial where n participants are randomized to vaccine or placebo in a 1:1 ratio. is generated as 1 + Bernoulli(0.5), so that S1 ∈ {1,2}; we do not consider covariates W. [CB] is assumed to hold with S0 = 1 for all subjects with , so that ACN can be assessed by checking whether Δ1 = R1(1) – R0(1) = 0 and ACS assessed by checking whether Δ2 ≡ R1(2) – R0(2) ≠ 0.
For all simulations, data are generated under [CB], [Al]–[A3′] and [A5]–[A6], with the restrictions on π, π* described in Web Appendix D. We consider five scenarios representing different amounts of surrogate value: In the first scenario (referred to in Tables 2 and 3 as No1), Δ1 = Δ2 = 0 (ACS fails); in the second (No2), Δ1 = Δ2 < 0 (ACN is violated), so that the biomarker has no surrogate value. In the third scenario (Low), we take Δ1 ≈ 0 and Δ2 < 0; though ACN fails, Δ1 is close to zero and ACS is satisfied, so the biomarker may be said to have some surrogate value. Scenarios 4 and 5 (High1 and High2) both have Δ1 = 0 and Δ2 < 0, with Δ2 larger in magnitude for the latter scenario; since ACN and ACS are satisfied, in both cases the biomarker is a good surrogate.
Table 2.
Bias, power and coverage for Δ̂1 and Δ̂2, estimated via maximum estimated likelihood. n is the total number of subjects in the simulated trial, divided equally between vaccine and placebo recipients. SV = surrogate value scenario.
| n | SV | Δ1 | Bias | Power | Coverage | Δ2 | Bias | Power | Coverage |
|---|---|---|---|---|---|---|---|---|---|
| 2000 | No1 | 0 | −0.0027 | 0.064 | 0.95 | 0 | 0.00041 | 0.072 | 0.96 |
| 10000 | 0 | 0.00035 | 0.048 | 0.94 | 0 | −0.00077 | 0.048 | 0.95 | |
| 30000 | 0 | 0.00059 | 0.032 | 0.98 | 0 | −0.00016 | 0.056 | 0.95 | |
| 2000 | No2 | −0.028 | −0.0018 | 0.14 | 0.96 | −0.028 | −0.0022 | 0.21 | 0.94 |
| 10000 | −0.028 | 0.00087 | 0.39 | 0.94 | −0.028 | −0.00069 | 0.38 | 0.94 | |
| 30000 | −0.028 | 0.00093 | 0.76 | 0.95 | −0.028 | −0.00099 | 0.83 | 0.94 | |
| 2000 | Low | −0.011 | 0.00029 | 0.076 | 0.94 | −0.028 | −0.0013 | 0.18 | 0.94 |
| 10000 | −0.011 | 0.0018 | 0.12 | 0.96 | −0.028 | −0.00071 | 0.4 | 0.95 | |
| 30000 | −0.011 | −2.9e-05 | 0.28 | 0.96 | −0.028 | −0.00022 | 0.73 | 0.94 | |
| 2000 | High1 | 0 | 0.00085 | 0.04 | 0.96 | −0.028 | −0.0039 | 0.18 | 0.95 |
| 10000 | 0 | 9.2e-05 | 0.04 | 0.92 | −0.028 | 2e-05 | 0.36 | 0.93 | |
| 30000 | 0 | −0.00044 | 0.056 | 0.93 | −0.028 | 0.00039 | 0.76 | 0.95 | |
| 2000 | High2 | 0 | 0.00058 | 0.044 | 0.93 | −0.05 | −0.0022 | 0.33 | 0.92 |
| 10000 | 0 | 0.0011 | 0.048 | 0.95 | −0.05 | −0.00062 | 0.84 | 0.94 | |
| 30000 | 0 | −0.00052 | 0.052 | 0.96 | −0.05 | 0.00062 | 1 | 0.94 |
Table 3.
Operating characteristics of PTE, RE, and sensitivity analyses in simulated trials of size n = 10; 000 when . SV = surrogate value scenario, SB = amount of post-randomization selection bias. For PTE and RE, we report and , the median point estimates over 500 simulations, along with the Monte Carlo standard error (σ̂). For the sensitivity analysis, each simulation yields 64 estimates of Δ1 and Δ2 from which the minimum, median, and maximum values are computed. (Min, Med, Max) [Rng] gives the medians of these three order statistics and the median range over 500 simulations. MSIW = Median Sensitivity Interval Width, where the sensitivity interval is the union of the Wald confidence intervals for all the sensitivity parameter settings, using Monte Carlo standard errors computed over the 500 simulations.
| Simulation settings | Statistical surrogate approach | Sensitivity analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SV | Δ1 | Δ2 | SB | (σ̂) | (σ̂) | Δ̂1(Min,Med,Max) [Rng] | MSIW(Δ̂1) | Δ̂2 (Min,Med,Max) [Rng] | MSIW(Δ̂2) |
| No1 | 0 | 0 | None | 0.12 (2e+09) | 0 (0.005) | (0.001, 0.001, 0.003) [0] | 0.08 | (−0.002, −0.001, 0) [0] | 0.08 |
| 0 | 0 | Low | −0.54 (34.81) | 0 (0.004) | (−0.002, 0, 0.008) [0.01] | 0.09 | (−0.008, −0.006, 0.002) [0.01] | 0.09 | |
| 0 | 0 | Mod | −0.9 (68.92) | 0.001 (0.004) | (−0.003, −0.001, 0.008) [0.01] | 0.09 | (−0.005, −0.003, 0.006) [0.01] | 0.09 | |
| 0 | 0 | High | −1.23 (43.35) | 0.003 (0.004) | (0.001, 0.002, 0.009) [0.01] | 0.09 | (−0.002, 0, 0.008) [0.01] | 0.09 | |
| No2 | −0.028 | −0.028 | None | 0.01 (0.13) | −0.037 (0.005) | (−0.028, −0.027, −0.026) [0] | 0.08 | (−0.028, −0.027, −0.026) [0] | 0.08 |
| −0.028 | −0.028 | Low | 0.4 (0.13) | −0.03 (0.005) | (−0.032, −0.03, −0.022) [0.01] | 0.09 | (−0.034, −0.032, −0.025) [0.01] | 0.09 | |
| −0.028 | −0.028 | Mod | 0.41 (0.13) | −0.028 (0.005) | (−0.034, −0.032, −0.024) [0.01] | 0.09 | (−0.03, −0.029, −0.02) [0.01] | 0.09 | |
| −0.028 | −0.028 | High | 0.44 (0.15) | −0.025 (0.005) | (−0.028, −0.026, −0.019) [0.009] | 0.09 | (−0.03, −0.028, −0.02) [0.009] | 0.09 | |
| Low | −0.011 | −0.028 | None | 0.48 (0.2) | −0.023 (0.005) | (−0.011, −0.011, −0.009) [0] | 0.08 | (−0.027, −0.027, −0.026) [0] | 0.08 |
| −0.011 | −0.028 | Low | 0.89 (0.37) | −0.019 (0.005) | (−0.016, −0.014, −0.005) [0.01] | 0.09 | (−0.033, −0.031, −0.024) [0.01] | 0.1 | |
| −0.011 | −0.028 | Mod | 0.98 (0.4) | −0.018 (0.004) | (−0.014, −0.012, −0.005) [0.01] | 0.09 | (−0.032, −0.03, −0.023) [0.01] | 0.09 | |
| −0.011 | −0.028 | High | 1.06 (0.55) | −0.015 (0.005) | (−0.011, −0.009, −0.001) [0.009] | 0.09 | (−0.03, −0.029, −0.022) [0.009] | 0.09 | |
| High1 | 0 | −0.028 | None | 0.97 (2.34) | −0.015 (0.005) | (0, 0.001, 0.002) [0] | 0.08 | (−0.029, −0.029, −0.027) [0] | 0.08 |
| 0 | −0.028 | Low | 1.54 (1.39) | −0.013 (0.004) | (−0.005, −0.004, 0.004) [0.009] | 0.09 | (−0.033, −0.03, −0.023) [0.009] | 0.09 | |
| 0 | −0.028 | Mod | 1.66 (5.2) | −0.012 (0.004) | (−0.003, −0.001, 0.006) [0.009] | 0.09 | (−0.034, −0.032, −0.024) [0.009] | 0.09 | |
| 0 | −0.028 | High | 2.01 (7.09) | −0.009 (0.004) | (0, 0.001, 0.008) [0.009] | 0.09 | (−0.03, −0.029, −0.02) [0.009] | 0.09 | |
| High2 | 0 | −0.05 | None | 1 (0.2) | −0.032 (0.005) | (0.001, 0.001, 0.003) [0] | 0.08 | (−0.053, −0.053, −0.051) [0] | 0.08 |
| 0 | −0.05 | Low | 1.25 (0.24) | −0.027 (0.005) | (−0.002, −0.001, 0.006) [0.009] | 0.09 | (−0.057, −0.054, −0.048) [0.009] | 0.09 | |
| 0 | −0.05 | Mod | 1.3 (0.29) | −0.025 (0.005) | (−0.005, −0.003, 0.005) [0.01] | 0.09 | (−0.055, −0.052, −0.045) [0.01] | 0.09 | |
| 0 | −0.05 | High | 1.42 (0.4) | −0.022 (0.005) | (−0.002, 0, 0.008) [0.01] | 0.09 | (−0.051, −0.049, −0.041) [0.01] | 0.09 | |
6.1 Operating characteristics of estimated likelihood method
Table 2 displays the performance of the maximum estimated likelihood estimators (MELEs) of Δ1 and Δ2 when the sensitivity parameters are correctly specified to match the values used to generate the data (π(j) = π*(j) = 0.995 for j = 1,2) and [CB], [Al]–[A3′], and [A5]–[A6] are assumed. The pre-Τ infection rates are set to 1.4% and 1.9% for vaccine and placebo recipients, respectively. The post-Τ infection rate is set at 7% for placebo recipients; the rate for vaccine recipients depends on Δ1 and Δ2, and ranges from 2.8% to 6.5% across the five surrogate value scenarios. These rates are consistent with what might reasonably be observed in an HIV vaccine trial enrolling a high-risk population.
We focus on the case where higher values of the biomarker are associated with better clinical outcomes, and calculate power based on Wald tests for the hypotheses H0 : Δj ≥ 0 vs. H1 : Δj < 0, j = 1, 2. In each of 250 simulations, percentile-based 95% confidence intervals are computed from 100 bootstrap replicates. Standard errors for Wald tests are based on empirical estimates from the set of 250 simulations. The proposed method yields unbiased estimates and correct coverage probabilities. Tests have approximately correct nominal size, and power increases to 1 with the sample size. We note, however, that large trial sizes are required to achieve adequate power to detect even large effects.
In Web Table A, we present an analog of Table 2 suggested by a reviewer, where data are generated without enforcing the monotonicity assumption [A3′]. When the data generating process does not obey monotonocity but we assume that it does, Δ̂1 and Δ̂2 are somewhat biased, the size of tests is not correct, and confidence interval coverage is degraded. Though the effect of incorrectly assuming [A3′] is relatively small for these simulation settings, additional simulations (results not shown) confirm that the impact of the monotonicity assumption is much more pronounced when the pre-Τ infection rate is higher.
6.2 Assessing surrogate value
We now return to the problem of surrogate endpoint assessment in the presence of pre-Τ treatment effects (i.e. when [A3] does not hold). We consider two approaches to quantifying the surrogate value of a biomarker:
Proportion of Treatment Effect Explained (PTE) and Relative Effect (RE) ["statistical surrogate" approach]: These summary measures are computed by fitting regression models from observed data. The motivation for and properties of PTE and RE are discussed at length in Burzykowski et al. (2005). We fit the logistic models for binary outcomes suggested in Buyse and Molenberghs (1998) - details appear in Web Appendix E.
Sensitivity analysis [“principal surrogate” approach]: This method consists of calculating the marginal risks via estimated likelihood, then computing Δ1 and Δ2 across the entire range of sensitivity parameters consistent with the restrictions described in Web Appendix D. For each simulated dataset, we consider 64 possible sensitivity parameter settings, with each of π(2),π*(2),π*(1) taking on 4 equally spaced values between their minimum and maximum allowed values (note that π(1) is determined by π(2)).
Table 3 summarizes the performance (over 500 simulated trials of size n =10,000) of these surrogate value assessment methods under a variety of settings. We fix the pre-Τ infection rate under assignment to vaccine at . In addition to the five different surrogate value scenarios, we consider four different degrees of post-randomization selection bias: None , Low , Moderate (, small departure of π* from π), and High ( far from π). The parameter values for all the scenarios appear in Web Table B. Web Table C presents a version of Table 3 showing the performance of these methods when .
Since they are based on different paradigms, it is difficult to compare the performance of PTE/RE and the sensitivity analysis. PTE is undefined in scenario No1 (ACN holds, ACS fails), leading to unstable point estimates and large standard errors, but performs well when there is no pre-Τ treatment effect (i.e. ), with values near zero in scenarios with no surrogate value (No2), values near 1 in high surrogate value scenarios High1 and High2, and intermediate values in the Low surrogate value scenario. As expected, the performance of PTE degrades when there is post-randomization selection bias (i.e. ), taking on values near 1 in the Low surrogate value scenario and greater than 1 in the two High surrogate value scenarios. The RE performs poorly, as it is unable to distinguish between scenarios No2 and High2. We note that Buyse and Molenberghs recommend considering the RE together with the adjusted association, but the odds ratio which they propose is undefined in case [CB].
Given estimates of marginal risks, the constraints imposed by Web Appendix D limit the sensitivity parameters to a narrow range of values under most of the simulation settings in Table 3, and hence a “typical” sensitivity analysis yields a narrow range of plausible values for Δ1 and Δ2. However, the uncertainty in estimating Δ1 and Δ2 due to possible post-randomization selection bias is dwarfed by the uncertainty in estimating the marginal risks: the sensitivity intervals (unions of the 64 Wald confidence intervals constructed around the point estimates from the sensitivity analysis) are very wide in comparison to the effect size, indicating that even trials of size 10,000 may have low power to detect surrogate value. Power increases with the infection rate: the sensitivity intervals are much narrower in relation to the effect size for the simulations presented in Web Table C, where the pre-Τ and overall infection rates are larger (approximately 6% and 16%, respectively). However, as shown by the wider ranges for Δ̂1 and Δ̂2 in Web Table C, a higher pre-Τ infection rate carries the potential for greater post-randomization selection bias. Hence, in assessing surrogate value there is a trade-off between the ability to estimate identifiable quantities and the ability to restrict nonidentifiable quantities to a sufficiently narrow range.
7. Discussion
In this article we described how several sets of assumptions contribute (or not) to the identifiability of the joint and marginal risk causal estimands of interest. In addition, to relax the untestable assumptions [A3] and [A4] made in previous work for evaluating a candidate surrogate endpoint, we developed methods that combine assumptions that may be more plausible, a creative study design, and a sensitivity analysis to assess the surrogate value of a biomarker.
The use of counterfactuals has been criticized by some (eg. Dawid (2000)) because of the underlying assumption of determinism (or “fatalism”) required to make certain counterfactual expressions well-defined. Dawid argues that this is a fundamental flaw in the potential outcomes framework, and the use of counterfactuals should therefore be restricted to a small class of problems where no such definitional problems arise. However, we subscribe to the point of view expressed by the majority of the respondents to Dawid's article: Potential outcomes provide a natural way of formulating and answering important scientific questions which are difficult to express without counterfactuals.
Jin and Rubin (2008), among others, employed the principal stratification framework to estimate treatment effects in the presence of partial noncompliance by partitioning individuals into compliance strata (eg. “always-taker”, “never-taker”, “complier”, “defier”). The counterfactual values defining these strata are generally assumed to be well-defined for all individuals, but this assumption may be violated if the outcome can occur before compliance is (fully) measured: for example, consider a clinical trial to assess the survival of late-stage cancer patients where the compliance measure is average weekly pill count over three months. In such setups, the methods we have described for quantifying surrogate value when [A3] is violated can be applied to evaluate compliance-adjusted treatment effects. Of course, certain simplifying assumptions such as Constant Biomarkers and Monotonicity may not apply in some studies where noncompliance is of interest.
To our knowledge, no previous work on surrogate endpoint assessment has addressed the consequences of treatment effects prior to measurement of the biomarker, a situation which is common in practice. In such cases, available data from a closeout vaccine trial are insufficient to identify joint risks, even in the simple case where there is no variability in the biomarker values of placebo recipients. Non-identifiability of joint risks leads naturally to a sensitivity analysis framework, but estimating surrogate value requires the ability to estimate marginal risks precisely and restrict the sensitivity parameter values to a suitably narrow range without relying on implausible assumptions. When non-identifiability makes the assessment of surrogate value intractable, it may be more useful to focus on the information which can be obtained from alternate estimands such as contrasts of marginal risks, which provide clinically relevant information and are statistically identifiable under weaker assumptions.
Supplementary Material
Acknowledgements
The authors wish to thank the reviewers for their comments, which helped clarify our thinking about these difficult issues and resulted in a much improved manuscript. This work was partially supported by NIH grant ROI AI54165-06, as well as Le Fonds quebecois de la recherche sur la nature et les technologies (FQRNT).
Footnotes
Supplementary Materials
Web Appendices and Tables referenced in Sections 3–6 are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
Contributor Information
Julian Wolfson, Department of Biostatistics, University of Washington, F-600 Health Sciences Building, Box 357232, Seattle, WA 98195-7232, julianw@u.washington.edu.
Peter Gilbert, Fred Hutchinson Cancer Research Center, Statistical Center for HIV/AIDS Research and Prevention, 1100 Fairview Avenue North, M2-C801, PO Box 19024, Seattle, WA 98109, pgilbert@scharp.org.
References
- Buchbinder S, Mehrotra D, Duerr A, Fitzgerald D, Mogg R, Li D, Gilbert P, Lama J, Marmor M, Delrio C. Efficacy assessment of a cell-mediated immunity hiv-1 vaccine (the step study): a double-blind, randomised, placebo-controlled, test-of- concept trial. The Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzykowski T, Molenberghs G, Buyse M. The Evaluation of Surrogate Endpoints. Springer; 2005. [Google Scholar]
- Buyse M, Molenberghs G. Criteria for the validation of surrogate endpoints in randomized experiments. Biometrics. 1998;54:1014–1029. [PubMed] [Google Scholar]
- Buyse M, Molenberghs G, Burzykowski T, Renard D, Geys H. The validation of surrogate endpoints in meta-analyses of randomized experiments. Biostatistics. 2000;1:49–67. doi: 10.1093/biostatistics/1.1.49. [DOI] [PubMed] [Google Scholar]
- Dawid AP. Causal inference without counterfactuals. Journal of the American Statistical Association. 2000;95:407–424. [Google Scholar]
- Follmann D. Augmented designs to assess immune response in vaccine trials. Biometrics. 2006;62:1161–1169. doi: 10.1111/j.1541-0420.2006.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis CE, Rubin DB. Principal stratification in causal inference. Biometrics. 2002;58:21–29. doi: 10.1111/j.0006-341x.2002.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gail MH, Pfeifier R, Van Houwelingen HC, Carroll RJ. On meta-analytic assessment of surrogate outcomes. Biostatistics. 2000;1:231–246. doi: 10.1093/biostatistics/1.3.231. [DOI] [PubMed] [Google Scholar]
- Gilbert PB, Hudgens MG. Evaluating candidate principal surrogate endpoints. Biometrics. 2008;64:1146–1154. doi: 10.1111/j.1541-0420.2008.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, Heyward WL, Jobes DV, Popovic V, Self SG, Sinangil F, Burke D, Berman PW. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of hiv-1 infection in a phase 3 hiv-1 preventive vaccine trial. The Journal of infectious diseases. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- Gilbert PB, Qin L, Self SG. Evaluating a surrogate endpoint at three levels, with application to vaccine development. Statistics in Medicine. 2008;27:4758–4778. doi: 10.1002/sim.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy JJ, Weyand CM. T cell development and receptor diversity during aging. Current Opinion in Immunology. 2005;17:468–475. doi: 10.1016/j.coi.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Hudgens MG, Halloran ME. Toward causal inference with interference. Journal of the American Statistical Association. 2008;103:832–842. doi: 10.1198/016214508000000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Rubin DB. Principal stratification for causal inference with extended partial compliance. Journal of the American Statistical Association. 2008;103:101–111. [Google Scholar]
- Joffe MM, Greene T. Related causal frameworks for surrogate outcomes. Biometrics. 2009;65:530–538. doi: 10.1111/j.1541-0420.2008.01106.x. [DOI] [PubMed] [Google Scholar]
- Lacroix-Desmazes S, Mouthon L, Kaveri SV, Kazatchkine MD, Weksler ME. Stability of natural self-reactive antibody repertoires during aging. Journal of Clinical Immunology. 1999;19:26–34. doi: 10.1023/a:1020510401233. [DOI] [PubMed] [Google Scholar]
- Lauritzen SL. Discussion on causality. Scandinavian Journal of Statistics. 2004;31:189–193. [Google Scholar]
- Pearl J. Causality : Models, Reasoning, and Inference. Cambridge University Press; 2000. [Google Scholar]
- Pepe MS, Fleming TR. A nonparametric method for dealing with mismeasured covariate data. Journal of the American Statistical Association. 1991;86:108. [Google Scholar]
- Prentice RL. Surrogate endpoints in clinical trials: Definition and operational criteria. Statistics in Medicine. 1989;8:431–440. doi: 10.1002/sim.4780080407. [DOI] [PubMed] [Google Scholar]
- Vanderweele T. Simple relations between principal stratification and direct and indirect effects. Statistics & Probability Letters. 2008;78:2957–2962. [Google Scholar]
- Weir CJ, Walley RJ. Statistical evaluation of biomarkers as surrogate endpoints: a literature review. Stat Med. 2006;25:183–203. doi: 10.1002/sim.2319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.