Abstract
We describe the characterization of m4, an autosomal recessive, temperature-sensitive paralytic mutant in Drosophila that is associated with shortened lifespan and neurodegeneration. Deletion mapping places the mutation in the gene encoding the glycolytic enzyme, Aldolase. The mutant enzyme contains a single amino acid substitution, which results in decreased steady-state levels of Aldolase with a consequent reduction in ATP levels. Transgenic-rescue experiments with a genomic construct containing the entire Aldolase gene confirm that paralysis, reduced lifespan, and neurodegeneration all result from the same mutation. Tissue- specific rescue and RNAi knockdown experiments indicate that Aldolase function (and presumably glycolysis) is important both in neurons and in glia for normal lifespan and neuronal maintenance over time. Impaired glycolysis in neurons can apparently be rescued in part by glycolytically active glia. However this rescue may depend on the exact physiological state of the neurons and may also vary in different subsets of neurons. Further studies of m4 and related mutants in Drosophila should help elucidate the connections between energy production and utilization in glia and neurons and lead to better understanding of how metabolic defects impair neuronal function and maintenance.
Introduction
In a seminal paper (Siddiqi and Benzer, 1976), Obaid Siddiqi and Seymour Benzer described the electrophysiological analysis of the three temperature-sensitive (ts) paralytic mutants in Drosophila known at that time: paralyzed (para), shibire (shi), and comatose (comt). David Suzuki was the first to isolate ts- paralytic mutants with the discovery of para and shi (Suzuki et al.1971; Grigliatti et al., 1973). comt was subsequently isolated in Benzer’s lab (Siddiqi and Benzer, 1976). Siddiqi and Benzer showed that each mutant had a different electrophysiological defect in the adult flight motor pathway and concluded that “mutants of this kind will indeed be a rich source of material for neurophysiology.” This paper was published in 1976 the year that the senior author of the present paper (B. G.) began postdoctoral studies in Benzer’s lab. By then, the larval neuromuscular junction (NMJ) had become the system of choice for electrophysiological studies in Drosophila because of the finer level of analysis it afforded (Jan and Jan, 1976a, b). With the tools in hand, the hunt was on to find mutants that exhibited electrophysiological defects at the larval NMJ as a means of ultimately dissecting the molecular mechanisms of neural signaling. Given Siddiqi’s precedent, the notion of focusing on ts-paralytic mutants as likely candidates was straightforward. Because the three existing ts-paralytic mutants were X-linked, B. G. embarked on screens for similar mutants on the autosomes, representing 80 percent of the Drosophila genome (Wu et al., 1978). The pursuit of these mutants, deciphering how they affect the nervous system, and identifying their molecular lesions has subsequently kept B. G. and his colleagues occupied for over 30 years. We offer our gratitude and respect to Obaid Siddiqi for his seminal studies and for leaving behind such a rich vein of investigation to be mined.
As Siddiqi and Benzer had inferred (Siddiqi and Benzer, 1976), subsequent studies have shown that ts-paralytic mutants are indeed a rich source of material that have led to the discovery and cloning of genes encoding a variety of ion channels (Loughney et al., 1989; Atkinson et al., 1991; Titus et al., 1997; Wang et al., 1997), components of the synaptic release machinery (Ikeda et al., 1976; Chen et al., 1991; van der Bliek and Meyerowitz, 1991; Pallanck et al., 1995; Littleton et al., 1998; Tolar and Pallanck, 1998), and ion channel regulatory proteins (Ganetzky, 1986; Feng et al., 1995; Reenan et al., 2000; Fergestad et al., 2010). These mutants have provided key insights into molecular mechanisms of neural function and shed important light on a number of human neurological disorders. More recently, ts-paralytic mutants have also proved to be a valuable resource in studies of synaptic development at the larval NMJ (Coyle et al., 2004; O'Connor-Giles et al., 2008; Rodal et al., 2008) and in analyzing mechanisms of age-dependent neuroprotection and neural maintenance (Palladino et al., 2002; Palladino et al., 2003; Fergestad et al., 2006a; Gnerer et al., 2006). Among the previously identified mutants in the latter category, which manifest age-dependent neurodegeneration marked by the pathological vacuolization of the brain, are those encoding the Na+/K+ ATPase and components of the synaptic release machinery (Palladino et al., 2003; Fergestad et al., 2006a). Interestingly however, we also found mutations in this category whose relationship to normal neural function and maintenance was not immediately apparent. One of these mutants, wasted away (wstd), is defective in the glycolytic enzyme triosephosphate isomerase (Tpi), which interconverts dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP) following the production of these compounds via cleavage of fructose 1,6-bisphosphate by Aldolase (Gnerer et al., 2006). Although we had not anticipated finding mutations affecting glycolysis in this screen there was good precedent: triosephosphate isomerase deficiency in humans caused by inheritance of an autosomal recessive mutation (Schneider, 2000) results in shortened lifespan and neurodegeneration similar to what is observed in wstd mutants in Drosophila. Subsequent analysis of wstd has suggested a novel mechanism of neuropathogenesis that could also explain the human syndrome, which has remained puzzling (Schneider, 2000; Gnerer et al., 2006).
Part of the puzzle is that mutations in nearly all glycolytic enzymes have been identified in humans and are generally associated with significant decrements in ATP levels accompanied by acute hemolytic anemia (Valentine and Paglia, 1984). However, only two glycolytic enzymopathies result in neurological defects in humans: Tpi deficiency, which causes the most severe neurodegeneration, and phosphoglycerate kinase (PKG) deficiency. In addition to wstd, it is notable that mutations of PKG in Drosophila also result in ts- paralysis and neurological dysfunction (Wang et al., 2004; Gnerer et al., 2006). Although it remains unclear why other glycolytic enzymopathies are not associated with neurodegeneration in humans, it is clear that neurons are very sensitive to metabolic disruption as revealed by the frequency of mutations in Drosophila affecting energy production among those identified on the basis of ts-paralytic and/or seizure phenotypes (Palladino et al., 2002; Wang et al., 2004; Fergestad et al., 2006a; Liu et al., 2007; Fergestad et al., 2008). Moreover, defects in energy metabolism and the mitochondrial machinery are emerging as prominent players in human neurodegenerative diseases (Valente et al., 2004; Manfredi and Xu, 2005; Gautier et al., 2008; Narendra et al., 2008; Magrane and Manfredi, 2009; Shi et al., 2009; Du et al., 2010; Narendra et al., 2010; Dupuis et al., 2011; Kapogiannis and Mattson, 2011). Still, the mechanistic links among energy metabolism, neural function, and neurodegeneration remain very incompletely understood.
Adding to the complexity is the suggestion that even though glucose represents the predominant fuel in vertebrate brains, other metabolites may provide additional energy, particularly during energetic stress. One model, the astrocyte-neuron lactate shuttle hypothesis (ANLSH), suggests that lactate generated by glycolysis in glia provides a rapidly utilizable metabolic substrate for the neuronal TCA cycle and oxidative phosphorylation (Pellerin et al., 2007). The extent of neuronal lactate metabolism and the degree to which glycolysis in neurons can be substituted by lactate uptake remains controversial (Chih and Roberts, 2003).
Drosophila glycolytic mutants that manifest neuropathological phenotypes can thus provide important tools for investigating neuronal metabolism and its relationship to neuronal function and maintenance. Here, we describe the characterization of m4, a ts-paralytic mutant associated with shortened lifespan and neurodegeneration that maps to the Drosophila gene encoding Aldolase and causes a deficit in steady-state ATP levels. Tissue-specific rescue and RNAi knockdown experiments indicate that Aldolase function (and presumably glycolysis) is important both in neurons and in glia for normal lifespan and neuronal maintenance over time. It appears that impaired glycolysis in neurons can be rescued in part by glycolytically active glia. However this rescue may depend on the exact physiological state of the neurons and may also vary in different subsets of neurons. Further studies of m4 and related mutants in Drosophila should help elucidate the connections between energy production and utilization in glia and neurons and lead to better understanding of how metabolic defects impair neuronal function and maintenance.
Materials and Methods
Fly Stocks
m4 was isolated in a screen for recessive autosomal ts-paralytic following mutagenesis with ethyl methanesulfonate. Deficiency stocks and Gal4 driver stocks were acquired from the Bloomington stock center, except for MIB-Gal4 (a repo-based glial driver), which was a gift from Kate O’Connor-Giles and Aldolase-RNAi obtained from Vienna Drosophila RNAi Center (v47667)
Behavior
Flies were collected one to two days after eclosion and aged for 10 days at room temperature (approximately 22°C). They were then aspirated into empty glass vials that were preheated to 37.5°C in a water bath and the fraction of flies that remained actively moving was scored at two-minute intervals after the temperature shift.
Lifespan analysis
Flies collected one to two days after eclosion were shifted to 28°C (25°C for RNAi experiments), and transferred to fresh vials every three days. The number of flies of each genotype that were still alive was recorded daily. Multiple collections were taken for each genotype, and groups from each collection were aged independently under identical conditions. The percentages of flies surviving for each genotype were then averaged to generate a cumulative lifespan curve with standard errors of the mean.
Histology
Histological analysis was performed on brains from a subset of flies in the lifespan experiments to look for vacuolization indicative of neurodegeneration. Four to eight heads from flies of various ages were removed and fixed overnight at 4°C in 60% ethanol, 30% chloroform, and 10% acetic acid, washed in 70% ethanol, and paraffin embedded. Serial, frontal 5μm sections were obtained and mounted on slides, stained with hematoxylin/eosin and examined by light microscopy.
Molecular cloning and sequencing
The Aldolase gene region was amplified from wild-type and M4 genomic DNA by PCR, using primers specific to exons 1–3, 3–7, and 8–10. These PCR products were sequenced with the PCR primers as well as additional sequencing primers. Aldolase cDNA clone LD37852 was acquired from the Drosophila Genome Resource Center (DGRC), the insert was cloned into a mini-white marked pUAS plasmid and integrated on the second chromosome using standard ψC31 mediated integration techniques (Groth et al., 2004; Bischof et al., 2007).
ATP Assays
Flies were shifted to 28°C two days post-eclosion. After 5 days, flies were snap-frozen on dry ice. 8 thoraces were ground in 6M Guanidine-HCl, in duplicate, followed by two freeze-thaw cycles in liquid nitrogen and boiling for 3 minutes. Debris was removed by centrifugation at 14k at 4C. Supernatants were diluted 1:20 in 100mM Tris, 4mM EDTA, pH 7.8 for protein assay (in triplicate), and 1:300 for ATP assay (in triplicate) using the bioluminescence Molecular Probes ATP Determination Kit (A22066), following the manufacturers recommendations. Plates were read on a Biotek multimode microplate reader, and luminescence values were normalized by protein content. Means and standard errors of the means were determined from normalized values. Statistical significance was determined using a two tailed student’s t- test.
Western blot analysis
Six-day old flies maintained at 28°C were snap frozen in liquid nitrogen, and heads were separated from bodies by centrifugation. Approximately 10 heads were ground in PBS 0.1% Triton-X with protease inhibitors using a mechanized kontex pestle on ice. Debris was removed by centrifugation at 14,000 rpm at 4° C. 30μg of extracted protein were resolved via 10% SDS/PAGE, and transferred to PVDF membranes. Membranes were cut and appropriate regions were probed using primary antbodies: rabbit polyclonal anti-Aldoase (Cell Signalling #3188, 1:1,000) and rabbit polyclonal anti-Tpi (Proteintech Group #10713, 1:1,000), and secondary antibody: IRDye 800CW donkey anti-rabbit (Li-Cor, 1:20,000). Membranes were scanned using a Li-Cor Odyssey™ infrared laser scanner.
Results
Mapping and molecular identification of a new mutant exhibiting age-dependent neurodegeneration
To identify new mutants manifesting neurodegeneration, we re-screened our collection of ts- paralytic mutants by direct histological analysis. This screen revealed that m4, a recessive mutation on the third chromosome, exhibited progressive, age-dependent neurodegeneration (vacuolar pathology in central brain and optic lobes). These phenotypes are associated with a lifespan defect that reduces the survival midpoint at 29°C from 30 days to 10 days (see below).
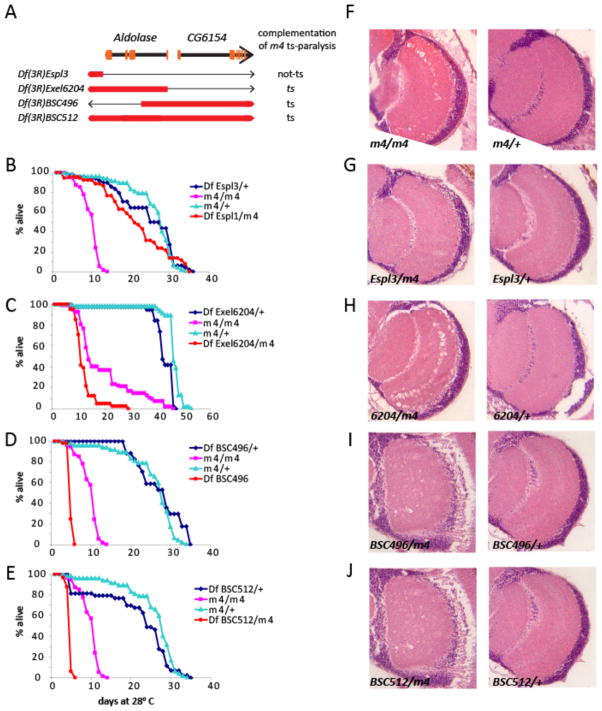
m4 flies become uncoordinated after a one-minute exposure to 37.5°C, immobilized within four minutes, and fully motionless within 8 minutes. Recovery to full mobility begins within minutes after the paralyzed flies are returned to room temperature. We mapped the ts-paralytic phenotype of m4 to the right of Hairless (3-69.5) on the right arm of chromosome 3. We refined the location of m4 by deletion mapping, using large deletions that spanned the relevant region. Df(3R)TI-P, failed to complement the ts-paralytic phenotype of m4, placing the gene in the cytological interval 97A-98A2. We further subdivided this interval using three smaller, overlapping deletions with molecularly defined breakpoints that also failed to complement ts-paralysis of m4 (Fig 1A). All three of these deletions also uncovered the lifespan and neurodegeneration phenotypes of m4 indicating that all three phenotypes are caused by the same mutation (Fig 1 B-J). Moreover, the lifespan defect is more severe in m4/Df flies than in m4/m4 homozygotes suggesting that m4 is a hypomorphic mutation. Two of the small deletions, Df(3R)Exel6204 and Df(3R)BSC496, share only 2.8kb of deleted sequence that corresponds to the 5’UTR and first intron of the gene encoding the glycolytic enzyme, Aldolase (Fig 1 A). Df(3R)Espl1, which deletes a region immediately 3’ to the Aldolase gene fully complements all mutant phenotypes of m4 (Fig 1B, G). These results strongly suggest that a hypomorphic mutation in the Aldolase gene is responsible for the ts-paralytic, lifespan, and neurodegeneration phenotypes of m4. We confirmed this conclusion via full rescue of ts-paralysis (data not shown) and significant rescue of lifespan and neurodegeneration (see below) using a UAS-Aldolase trangene driven by a Tubulin-Gal4 driver. Hereafter, we refer to m4 as Aldoslasem4 (Aldm4).
Figure 1.
Deletion mapping of m4. (A) Schematic diagram of the genomic region containing the Aldolase locus and the neighboring gene, CG6154. Orange bars indicate the approximate locations of the exons for each gene. Shown below are the extents of the chromosomal intervals that are uncovered by several deficiencies that were used in complementation tests to map the ts-paralytic phenotype of m4. Red bars represent the chromosome region that is missing. The thin black line represents the portion of the chromosome still present in that interval for each deficiency. (B–E) Deficiencies that remove or disrupt the Aldolase gene fail to complement the lifespan phenotype of m4, while a deficiency that does not perturb the Aldolase gene (B) fully complements the lifespan phenotype. (F–J) Deficiencies that uncover Aldolase also fail to complement the neurodegeneration phenotype of m4. Brain sections of age matched flies raised at 28° C showing optic lobe region. m4 homozygotes (F) and m4/Df heterozygotes for deficiencies that remove or disrupt Aldolase (H–J) show extensive vacuolization that is indicative of neurodegeneration. This neuropathology is not observed in m4/+ heterozygotes (F), Df/+ heterozygotes (F–J), or in m4/Df heterozygotes for a deficiency that does not uncover Aldolase (G).
Molecular and biochemical analysis of Aldm4
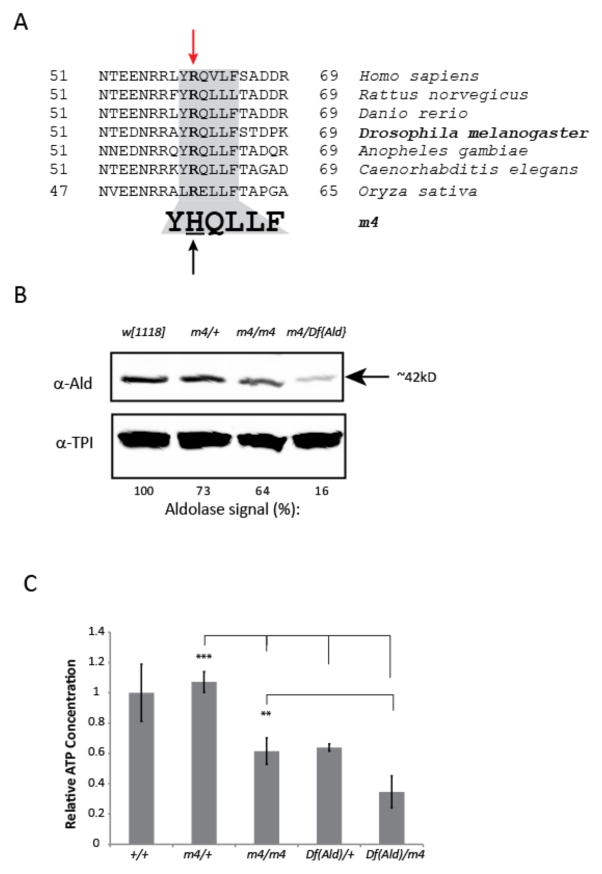
To identify the molecular lesion in Aldm4, we sequenced genomic DNA amplified from single Df(3R)BSC512/Aldm4 flies using primers to DNA sequences within the deleted region. Results from multiple independent analyses reveal a single nucleotide (G > A) change at nucleotide 179 of exon 1, common to all transcript isoforms of Aldolase. This change results in an Arginine (R) to Histidine (H) change at amino acid 60 (R60H), within a highly conserved region of the Aldolase protein (Fig 2A).
Figure 2.
Molecular and biochemical characterization of m4 mutants. (A) Amino acid alignment of a region of the Aldolase protein from various species showing complete conservation in plants, nematodes, insects, zebrafish, and mammals. A single nucleotide change in m4 results in a substitution of Histidine for Arginine in this highly conserved region. (B) Western blot of extracts from heads of flies of the indicated genotypes probed with antibodies against Aldolase or triosephosphate isomerase (Tpi). Steady-state levels of Aldolase are decreased in head extracts of m4 homozygotes and m4/Df hemizygotes. The relative intensity of the Aldolase band in each genotype is shown below the blot. For quantification, Aldolase levels are normalized against the Tpi band for each genotype and these values are shown relative to the levels observed in the control strain (w1118). (C) Steady-state levels of ATP were measured in extracts of flies of the indicated genotypes as described in the Materials and Methods. ATP levels were normalized to total protein and these values are shown relative to the levels measured in the control strain. Data are presented as the mean of the normalized values ± standard error of the mean. **P ≤ 0.01, ***P ≤ 0.001.
Western blot analysis of head extracts reveals a 35% decrease in Aldolase protein levels in Aldm4 homozygotes normalized to the levels of triosephosphate isomerase (Tpi), another glycolytic enzyme that immediately follows Aldolase in the pathway (Fig 2B). In Df/Aldm4 flies, Aldolase levels are decreased by approximately 80%. These results confirm that Aldm4 is a hypomorphic allele and that the R60H mutation affects either the production or stability of the Aldolase mRNA or protein.
Aldolase plays an essential role in glycolysis, catalyzing the conversion of fructose 1,6-bisphosphate to dihydoxyacetone phosphate (DHAP) and glyceradehyde-3-phosphate (GAP), which are further catabolized to pyruvate (the TCA cycle fuel), ATP and NADH. It is expected that any disruption of Aldolase activity should lead to a decreased ability to maintain normal levels of cellular energy. To test whether the Aldm4 R60H mutation causes a reduction of Aldolase activity, we assayed total ATP levels in mutant flies. Aldm4 homozygotes exhibit a roughly 50% reduction in steady-state ATP levels. This reduction is similar to that observed in flies carrying only a single normal allele of Aldolase, Df(Ald)/+ (Fig 2C) consistent with the conclusion that Aldm4 is hypomorphic mutation. Moreover, ATP levels are further reduced in Df(Ald)/Aldm4 flies compared with Df(Ald)/+. Together these data show that the R60H mutation in Aldolase reduces ATP levels, by causing a decrease in Aldolase protein levels and perhaps by reducing enzymatic activity as well.
Tissue-specific rescue of lifespan and neurodegeneration phenotypes in Aldm4
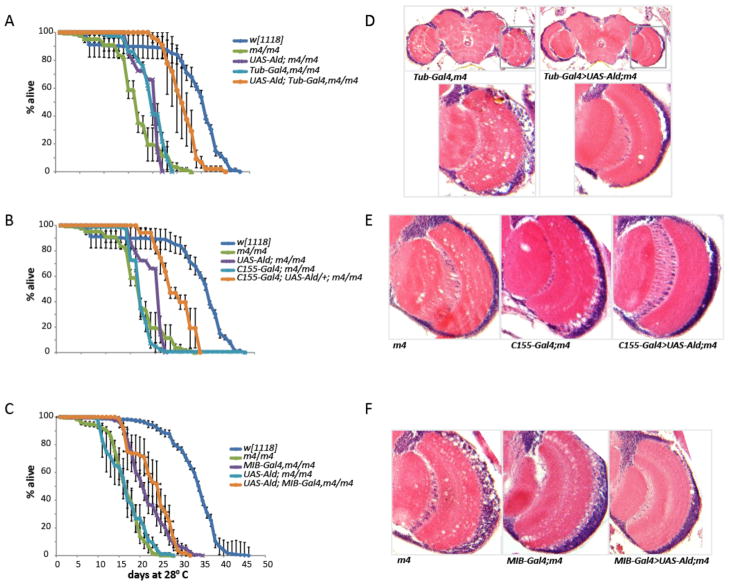
Although the lifespan defect of Aldm4 is significantly rescued by ubiquitous expression of UAS Aldolase via a Tubulin-Gal4 driver, detailed analysis revealed that it was not fully rescued back to normal (Fig 3A, D). One possible explanation is that is the UAS-Aldolase rescue construct is generated from a cDNA corresponding to a single splice variant (Ald-RB) of the Aldolase transcript, although several distinct splice variants are known. Consistent with this possibility, a large genomic P[acman] construct (CH322-98F06 (Venken et al., 2009)) that spans the entire Aldolase gene fully rescues the lifespan defect (data not shown).
Figure 3.
Lifespan and neurodegeneration phenotypes of m4 mutants can be rescued by tissue-specific expression of a UAS-Aldolase transgene. (A–C) Survival curves for flies of the indicated genotypes maintained at 28°C. Each curve shows mean percent survival from at least three independent replicates. Error bars are standard errors of the mean. Ubiquitous (Tubulin-Gal4) and pan-neuronal (C155-Gal4) expression of UAS- Aldolase substantially rescue the m4 lifespan phenotype, whereas glial expression (Mib-Gal4) does not improve life-span as compared with the Mib-Gal4, m4/m4 control. (D–F) Sections of optic lobes from age matched flies of the indicated genotypes, raised at 28°C for 17 days. Ubiquitous (D), pan-neuronal (E), or glial expression (F) markedly rescue the neuropathological appearance of the optic lobes (right panels in each row) in comparison with m4 homozygotes (left panels) or driver controls (middle panels).
Similarly, although ubiquitous expression of UAS-Aldolase substantially rescues age-dependent neurodegeneration in Aldm4 mutants, rescue is incomplete (Fig 3D). Tubulin-Gal4,Aldm4/Aldm4 flies aged to the midpoint of their survival curve exhibit extensive neuropathological vacuolization in the central brain and optic lobes. Age-matched UAS-Aldolase; Tubulin-Gal4 Aldm4/Aldm4 flies exhibit no tissue loss in the central brain and many fewer lesions in the optic lobes but the optic lobes still do not appear fully normal (Fig 3D). As with lifespan, the large genomic P[acman] construct fully rescues neurodegeneration (data not shown), again suggesting that rescue of Aldm4 mutant phenotypes requires proper expression of Aldolase isoforms at the right levels in the right tissues and that this endogenous expression pattern is not accurately replicated by Tubulin-Gal4-driven expression of UAS-Aldolase.
To determine where Aldolase expression is required to rescue the lifespan and neurodegeneration phenotypes of Aldm4 and, in particular, whether restoration of glycolysis in either glia or neurons can rescue these phenotypes, we used several different tissue-specific drivers. Expression of UAS-Aldolase using either of the strong glial drivers MIB-Gal4 (Fig 3C) or Repo-Gal4 (data not shown), had no significant effect on lifespan in Aldm4 flies. However, pan-neuronal expression using C155-Gal4, significantly rescues the lifespan defect in Aldm4 to a similar extent as that observed when UAS-Aldolase is driven ubiquitously by Tubulin-Gal4 (Fig 3B). These results indicate that providing wild-type levels of Aldolase activity in glia but not neurons is insufficient to rescue the lifespan defect of Aldm4 mutants, whereas restoring Aldolase activity to neurons but not glia can rescue this phenotype. Thus, although glia may provide an important source of TCA cycle metabolites and ATP for neurons, these results suggest that neurons also carry out glycolysis, and that its impairment causes deleterious consequences even when glycolysis is relatively normal in glia.
We also examined the effect of glial-specific versus neuronal-specific expression of UAS-Aldolase on neurodegeneration. In this case, expression of UAS-Aldolase either in glia or in neurons substantially rescues age-dependent neurodegeneration compared with Aldm4 mutants and controls (Fig 3E,F). Thus, it seems that normal glycolytic activity in either glia or neurons is sufficient to enable proper neuronal maintenance and integrity as a function of age. However, the fact that Aldolase expression in glia does not appear to rescue the lifespan of Aldm4 mutants suggests either that a deficit in Aldolase affects lifespan and neurodegeneration via separate mechanisms, or that shortened-lifespan Aldm4 flies expressing Aldolase in glia have neuropathological defects in the brain that are not detectable in our histological analysis.
RNAi induced loss of Aldolase in neurons and glia
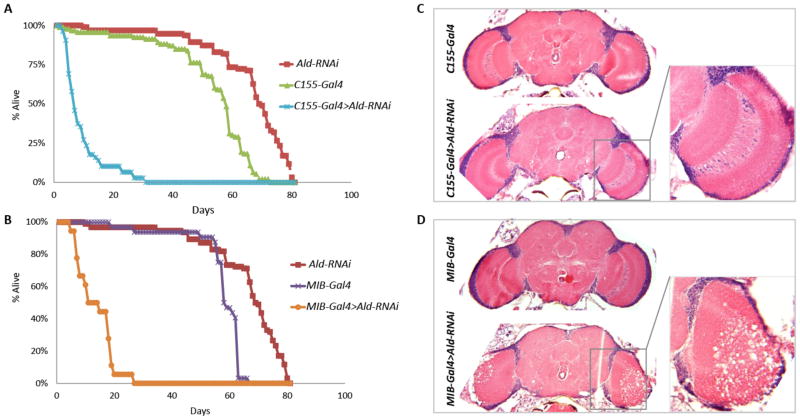
To further investigate the requirement for Aldolase in glia versus neurons for normal lifespan and age-dependent neuroprotection, we used the opposite approach: specifically knocking down Aldolase expression using a UAS-Aldolase-RNAi construct. As expected, ubiquitous expression of UAS-Aldolase-RNAi using the Tubulin-Gal4 driver is lethal. Western blot analysis of flies expressing UAS-Aldolase-RNAi specifically in neurons (using C155-Gal4) revealed that Aldolase protein is reduced by approximately 50% in total-head extract (data not shown).
Aldolase knockdown either in neurons (C155-Gal4) or in glia (MIB-Gal4) results in severely reduced lifespan at 25°C, with 50% survival at 8 days and 12 days respectively compared with around 60 days in control flies (Fig 4A-B). Surprisingly, in contrast with our previous rescue experiments, neuronal-specific knockdown of Aldolase does not result in any overt neuropathological manifestation (Fig 4C). Conversely, flies with glial-specific knockdown exhibit vacuolization in the central brain (data not shown) and optic lobes, which in some cases is very severe (Fig 4D). Thus, contrary to the previous rescue experiments, the knockdown experiments suggest that loss of Aldolase either in glia or neurons is sufficient to cause a reduction in lifespan and that Aldolase is specifically required in glia for neuronal maintenance as a function of age. At this point it remains unclear how to reconcile these conflicting observations but we consider several possible explanations in the Discussion.
Figure 4.
Tissue-specific knockdown of Aldolase expression via RNAi causes reduced lifespan and neurodegeneration. (A) Expression of Aldolase-RNAi in neurons (C155-Gal4) or (B) glia (MIB-Gal4) severely reduces lifespan. (C) Expression of Aldolase-RNAi in neurons does not cause overt neuropathology, whereas expression in glia but not neurons (D), elicits neuropathology in the optic lobes, which in some cases can be very severe.
Discussion
Molecular and genetic analysis of a behavioral mutant, originally discovered on the basis of its ts- paralytic phenotype, has led us to the isolation of the first functional mutation in the Drosophila Aldolase gene. As in the case of a number of other ts-paralytic mutants, we have found that even though the flies appear to behave reasonably normally at permissive temperatures, the causative mutation results in unconditional defects. In this instance, an Arginine-to-Histidine substitution at amino acid 60 results in decreased steady state levels of Aldolase protein and a concurrent reduction in ATP levels. We still do not know why this deficit leads to a ts-paralytic phenotype but the rapid onset of paralysis following a shift to elevated temperature indicates that this shift is exacerbating an underlying (unconditional) defect rather than, for example, causing further loss in Aldolase and an additional decrement in ATP levels, which would likely require more than a minute or two to have phenotypic consequences. We suggest that the steady state reduction in ATP is tolerable at permissive temperatures but as soon as flies are exposed to higher temperatures where there is a greater energy demand, the reduced level of ATP becomes limiting.
Even at nominally permissive temperatures where the flies are not paralyzed, the reduction in Aldolase and ATP levels in Aldm4 homozygotes results in strong phenotypic manifestations as a function of time, including a severe shortening of lifespan and the onset of neurodegeneration in the brain and optic lobes. Complete rescue of paralysis, lifespan, and neurodegeneration with a genomic construct that contains the entire Aldolase gene confirms that all three phenotypes are indeed caused by a single mutation in Aldolase. Ubiquitous expression of a cDNA corresponding to a specific Aldolase splice isoform fully rescues ts- paralysis, and substantially rescues lifespan and neurodegeneration. Incomplete rescue of the lifespan and neurodegeneration phenotypes with the cDNA construct suggests that additional Aldolase splice isoforms are required or that the precise expression pattern of Aldolase—time, place, and level—is critical for a fully normal phenotype and not precisely re-created by the Tub-Gal4-driven transgene. Thus, even an “ordinary” housekeeping gene such as Aldolase may be associated with previously unexpected degrees of regulation that we do not yet fully understand.
In vertebrates, it has been proposed that in addition to glucose derived from blood, neurons may obtain additional fuel for the citric acid cycle and oxidative phosphorylation in the form of lactate (a product of glycolysis) supplied by glia (Figley, 2011; Stridh et al., 2012), especially during periods of high activity (Pierre and Pellerin, 2005; Pellerin et al., 2007). However, this possibility remains the subject of some controversy (Chih and Roberts, 2003; Hertz, 2004). The availability of a hypomorphic allele of Aldolase in Drosophila provides a potentially very useful tool for investigating the requirement for glycolysis in glia versus neurons with respect to normal neuronal function, maintenance and viability. We found that supplying a functional Aldolase gene either to glia or to neurons substantially rescues the Aldm4 neurodegeneration phenotype. This result suggests that both neurons and glia can carry out glycolysis and that when glycolysis is impaired in neurons, the neuropathological consequences can be alleviated by the presence of glia with normal glycolysis. Surprisingly, however, whereas neuronal expression of Aldolase also rescues the lifespan defect of Aldm4, glial expression does not appear to rescue this phenotype. Minimally, this result indicates that the overt neuropathology observed in the central brain in Aldm4 mutants is not the direct caused of the shortened lifespan. The fact that the reduction in lifespan in Aldm4 mutants is rather severe, whereas the neuropathology is relatively modest, is consistent with the idea that these phenotypes represent distinct manifestations of reduced Aldolase activity. If neurodegeneration and shortened lifespan are associated with reduced Aldolase activity in different tissues, one possible explanation for rescue of both phenotypes with the C155-Gal4 driver is that this “pan-neuronal” driver might also be expressed in non- neuronal cells, including glia. Indeed, we and others have observed non-neuronal expression of elav-Gal4 and its derivative C155-Gal4 (Berger et al., 2007). Another more interesting possibility is that impaired glycolysis in different subsets of neurons is responsible for the lifespan and neurodegeneration phenotype of Aldm4 mutants and that glia do not supplement glycolysis equally in both sets of neurons, perhaps simply reflecting a differential density of glia in different parts of the nervous system. In this case, neuronal expression of Aldolase would rescue both lifespan and neurodegeneration but glial expression might be insufficient to restore necessary function to neurons whose activity is required for normal lifespan.
The results of tissue-specific knock down of Aldolase via Aldolase-RNAi are somewhat harder to explain by the above model. In particular, knockdown of Aldolase in glia causes both a severe decrease in lifespan and striking neurodegeneration, suggesting that a decrement in glycolysis in glia alone can affect lifespan and neuronal viability. This contrasts with the results of the neuron-specific rescue experiments (leaving glia impaired) where the rescue of lifespan and neurodegeneration suggested that glycolysis in glia was not necessary for normal lifespan or neuronal maintenance if glycolysis in neurons was restored. One possible explanation to resolve this puzzle that we cannot rule out is that expression of Aldolase-RNAi is causing a more severe phenotype owing to off-target affects. However, we suspect it is more likely related to the degree of impairment of glycolysis in glia in Aldm4 mutants versus knockdown with RNAi. The reduction in Aldolase activity in hypomorphic Aldm4 mutants appears to be relatively mild, whereas the RNAi knockdown is likely causing a much more severe reduction in Aldolase activity. This notion is supported by the observation that neuronal-specific expression of Aldolase-RNAi results in a 50% reduction of Aldolase protein in the total head extract (data not shown), whereas homozygosity for Aldm4 reduces Aldolase by only 40% in head extract (Fig 2B), even though all cells are affected by the mutation. Thus, with the neuronal rescue experiments, glycolysis is approximately normal in neurons and partial in glia. In the knockdown experiments, glycolysis is normal in neurons and severely impaired in glia, which is likely responsible for the severity of the phenotypes. Taken together, our results support a general model where glycolysis plays an important role in both glia and in neurons and the consequences of impairment in one or the other alone will vary depending on the severity of the impairment, the density of glia, and the specific properties of the particular neurons that are affected. Additional work is needed to examine these issues in more detail and to resolve the puzzles that remain.
The discovery that m4 turned out to be a mutation in Aldolase is of particular interest because we previously discovered that another of our mutants associated with ts-paralysis, shortened lifespan, and neurodegeneration was due to a mutation in the gene encoding triosephosphate isomerase (Tpi), the enzyme that catalyzes the next step in glycolysis after Aldolase (Gnerer et al., 2006; Celotto et al., 2006). Remarkably, however, mutations in Aldolase and Tpi appear to be exerting their effect in completely distinct ways. Mutations of Tpi do not significantly impair glycolysis and there is no significant reduction in ATP levels in the Tpi mutants (Gnerer et al., 2006; Celotto et al., 2006). Instead, our results have led us to propose that in Tpi mutants, the toxic compound methylglyoxal accumulates upstream of the enzymatic block leading to production of advanced glycation endproducts (AGEs), which underlie the resultant phenotypes. Interestingly, in humans Tpi Deficiency Syndrome associated with impaired Tpi activity also causes neurodegeneration and early onset lethality independent of any obvious deficit in glycolysis or ATP levels. In contrast, there are no known mutations of Aldolase that are associated with neurodegeneration in humans, possibly because there are three different genes encoding Aldolase in humans only one of which (Aldolase C) is specific to the nervous system (Penhoet et al., 1967).
The importance of energy metabolism and mitochondrial function for normal neuronal function and maintenance is highlighted by the enrichment for mutations affecting enzymes such as Aldolase, Tpi, cytochrome C oxidase, citrate synthase, and others (Celotto et al., 2006; Fergestad et al., 2006b; Gnerer et al., 2006; Liu et al., 2007) among ts-paralytic and bang-sensitive paralytic mutants (Palladino et al., 2002; Wang et al., 2004; Fergestad et al., 2006a; Liu et al., 2007; Fergestad et al., 2008) that also exhibit reduced lifespan and neurodegeneration. Similar links are found for numerous neurological and neurodegenerative disorders in humans. Thus, further studies of mutants such as Aldm4 should continue to provide important new information about the basic cellular and molecular mechanisms that govern neuronal function and maintenance and how perturbations of these mechanisms lead to the resultant neuropathological consequences.
Acknowledgments
We thank Robert Kreber for technical assistance, Aki Ikeda for use of his microtome, and members of the Ganetzky lab for helpful suggestions. This research was supported by NIH grants T32 AG000213 (DLM), R01 NS015390 and R01 AG033620 (BG) and a Hilldale Undergraduate Research Scholarship (CH).
References
- Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- Berger C, Renner S, Luer K, Technau GM. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn. 2007;236:3562–3568. doi: 10.1002/dvdy.21372. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Frank AC, Seigle JL, Palladino MJ. Drosophila model of human inherited triosephosphate isomerase deficiency glycolytic enzymopathy. Genetics. 2006;174:1237–1246. doi: 10.1534/genetics.106.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Obar RA, Schroeder CC, Austin TW, Poodry CA, Wadsworth SC, Vallee RB. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- Chih CP, Roberts EL., Jr Energy substrates for neurons during neural activity: a critical review of the astrocyte-neuron lactate shuttle hypothesis. J Cereb Blood Flow Metab. 2003;23:1263–1281. doi: 10.1097/01.WCB.0000081369.51727.6F. [DOI] [PubMed] [Google Scholar]
- Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer's disease mouse model. Proc Natl Acad Sci U S A. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis L, Pradat P-Fo, Ludolph AC, Loeffler J-P. Energy metabolism in amyotrophic lateral sclerosis. The Lancet Neurology. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- Feng G, Deak P, Chopra M, Hall LM. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell. 1995;82:1001–1011. doi: 10.1016/0092-8674(95)90279-1. [DOI] [PubMed] [Google Scholar]
- Fergestad T, Ganetzky B, Palladino MJ. Neuropathology in Drosophila membrane excitability mutants. Genetics. 2006a;172:1031–1042. doi: 10.1534/genetics.105.050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Bostwick B, Ganetzky B. Metabolic disruption in Drosophila bang-sensitive seizure mutants. Genetics. 2006b;173:1357–1364. doi: 10.1534/genetics.106.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Olson L, Patel KP, Miller R, Palladino MJ, Ganetzky B. Neuropathology in Drosophila mutants with increased seizure susceptibility. Genetics. 2008;178:947–956. doi: 10.1534/genetics.107.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad T, Sale H, Bostwick B, Schaffer A, Ho L, Robertson GA, Ganetzky B. A Drosophila behavioral mutant, down and out (dao), is defective in an essential regulator of Erg potassium channels. Proc Natl Acad Sci U S A. 2010;107:5617–5621. doi: 10.1073/pnas.1001494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley CR. Lactate transport and metabolism in the human brain: implications for the astrocyte-neuron lactate shuttle hypothesis. J Neurosci. 2011;31:4768–4770. doi: 10.1523/JNEUROSCI.6612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B. Neurogenetic analysis of Drosophila mutations affecting sodium channels: synergistic effects on viability and nerve conduction in double mutants involving tip-E. J Neurogenet. 1986;3:19–31. doi: 10.3109/01677068609106892. [DOI] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerer JP, Kreber RA, Ganetzky B. wasted away, a Drosophila mutation in triosephosphate isomerase, causes paralysis, neurodegeneration, and early death. Proc Natl Acad Sci U S A. 2006;103:14987–14993. doi: 10.1073/pnas.0606887103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz L. The astrocyte-neuron lactate shuttle: a challenge of a challenge. J Cereb Blood Flow Metab. 2004;24:1241–1248. doi: 10.1097/00004647-200411000-00008. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ozawa S, Hagiwara S. Synaptic transmission reversibly conditioned by single-gene mutation in Drosophila melanogaster. Nature. 1976;259:489–491. doi: 10.1038/259489a0. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Properties of the larval neuromuscular junction in Drosophila melanogaster. J Physiol. 1976a;262:189–214. doi: 10.1113/jphysiol.1976.sp011592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. L-glutamate as an excitatory transmitter at the Drosophila larval neuromuscular junction. J Physiol. 1976b;262:215–236. doi: 10.1113/jphysiol.1976.sp011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapogiannis D, Mattson MP. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer's disease. The Lancet Neurology. 2011;10:187–198. doi: 10.1016/S1474-4422(10)70277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Chapman ER, Kreber R, Garment MB, Carlson SD, Ganetzky B. Temperature-sensitive paralytic mutations demonstrate that synaptic exocytosis requires SNARE complex assembly and disassembly. Neuron. 1998;21:401–413. doi: 10.1016/s0896-6273(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Liu W, Gnanasambandam R, Benjamin J, Kaur G, Getman PB, Siegel AJ, Shortridge RD, Singh S. Mutations in cytochrome c oxidase subunit VIa cause neurodegeneration and motor dysfunction in Drosophila. Genetics. 2007;176:937–946. doi: 10.1534/genetics.107.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- Magrane J, Manfredi G. Mitochondrial function, morphology, and axonal transport in amyotrophic lateral sclerosis. Antioxid Redox Signal. 2009;11:1615–1626. doi: 10.1089/ars.2009.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi G, Xu Z. Mitochondrial dysfunction and its role in motor neuron degeneration in ALS. Mitochondrion. 2005;5:77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Hadley TJ, Ganetzky B. Temperature-sensitive paralytic mutants are enriched for those causing neurodegeneration in Drosophila. Genetics. 2002;161:1197–1208. doi: 10.1093/genetics/161.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Bower JE, Kreber R, Ganetzky B. Neural dysfunction and neurodegeneration in Drosophila Na+/K+ ATPase alpha subunit mutants. J Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallanck L, Ordway RW, Ganetzky B. A Drosophila NSF mutant. Nature. 1995;376:25. doi: 10.1038/376025a0. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Bouzier-Sore AK, Aubert A, Serres S, Merle M, Costalat R, Magistretti PJ. Activity-dependent regulation of energy metabolism by astrocytes: an update. Glia. 2007;55:1251–1262. doi: 10.1002/glia.20528. [DOI] [PubMed] [Google Scholar]
- Penhoet E, Kochman M, Valentine R, Rutter WJ. The subunit structure of mammalian fructose diphosphate aldolase. Biochemistry. 1967;6:2940–2949. doi: 10.1021/bi00861a039. [DOI] [PubMed] [Google Scholar]
- Pierre K, Pellerin L. Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J Neurochem. 2005;94:1–14. doi: 10.1111/j.1471-4159.2005.03168.x. [DOI] [PubMed] [Google Scholar]
- Reenan RA, Hanrahan CJ, Ganetzky B. The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Motola-Barnes RN, Littleton JT. Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider AS. Triosephosphate isomerase deficiency: historical perspectives and molecular aspects. Baillieres Best Pract Res Clin Haematol. 2000;13:119–140. doi: 10.1053/beha.2000.0061. [DOI] [PubMed] [Google Scholar]
- Shi P, Gal J, Kwinter DM, Liu X, Zhu H. Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2009;1802:45–51. doi: 10.1016/j.bbadis.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi O, Benzer S. Neurophysiological defects in temperature-sensitive paralytic mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1976;73:3253–3257. doi: 10.1073/pnas.73.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stridh MH, Alt MD, Wittmann S, Heidtmann H, Aggarwal M, Riederer B, Seidler U, Wennemuth G, McKenna R, Deitmer JW, Becker HM. Lactate flux in astrocytes is enhanced by a non-catalytic action of carbonic anhydrase II. J Physiol. 2012 doi: 10.1113/jphysiol.2011.220152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus SA, Warmke JW, Ganetzky B. The Drosophila erg K+ channel polypeptide is encoded by the seizure locus. J Neurosci. 1997;17:875–881. doi: 10.1523/JNEUROSCI.17-03-00875.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar LA, Pallanck L. NSF function in neurotransmitter release involves rearrangement of the SNARE complex downstream of synaptic vesicle docking. J Neurosci. 1998;18:10250–10256. doi: 10.1523/JNEUROSCI.18-24-10250.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Valentine WN, Paglia DE. Erythrocyte enzymopathies, hemolytic anemia, and multisystem disease: an annotated review. Blood. 1984;64:583–591. [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Saraswati S, Guan Z, Watkins CJ, Wurtman RJ, Littleton JT. A Drosophila temperature-sensitive seizure mutant in phosphoglycerate kinase disrupts ATP generation and alters synaptic function. J Neurosci. 2004;24:4518–4529. doi: 10.1523/JNEUROSCI.0542-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Reynolds ER, Deak P, Hall LM. The seizure locus encodes the Drosophila homolog of the HERG potassium channel. J Neurosci. 1997;17:882–890. doi: 10.1523/JNEUROSCI.17-03-00882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Ganetzky B, Jan LY, Jan YN, Benzer S. A Drosophila mutant with a temperature-sensitive block in nerve conduction. Proc Natl Acad Sci U S A. 1978;75:4047–4051. doi: 10.1073/pnas.75.8.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]